Trophic Niches, Trophic Positions, and Niche Overlaps between Non-Native and Native Fish Species in a Subalpine Lake
Abstract
:1. Introduction
2. Materials and Methods
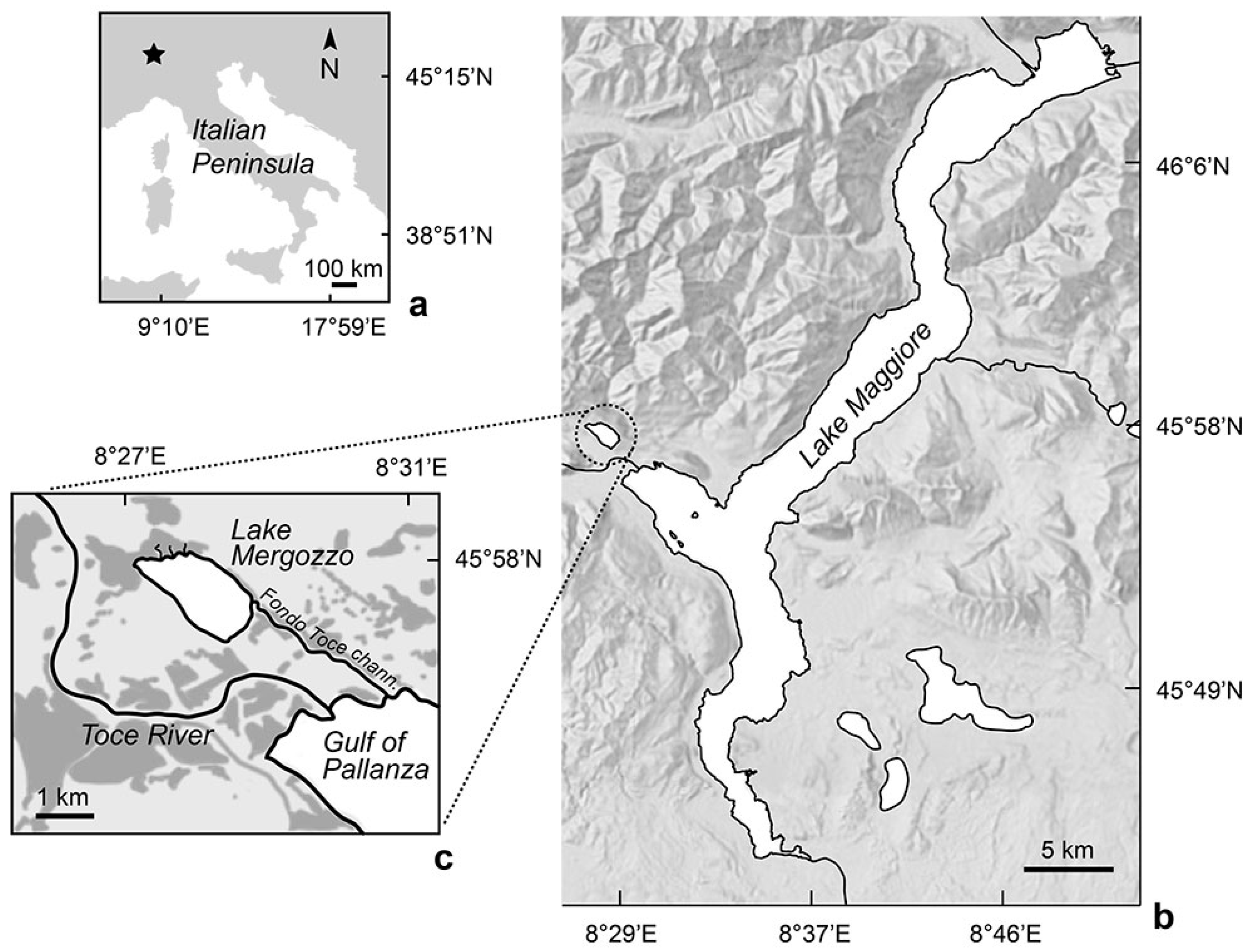
2.1. Study Site
2.2. Zooplankton, Zoobenthos, and Fish Samples
2.3. Stable Isotope Analysis and Trophic Positions
2.4. Isotopic Niche Metrics, Asymmetrical Overlaps and Statistical Analyses
3. Results
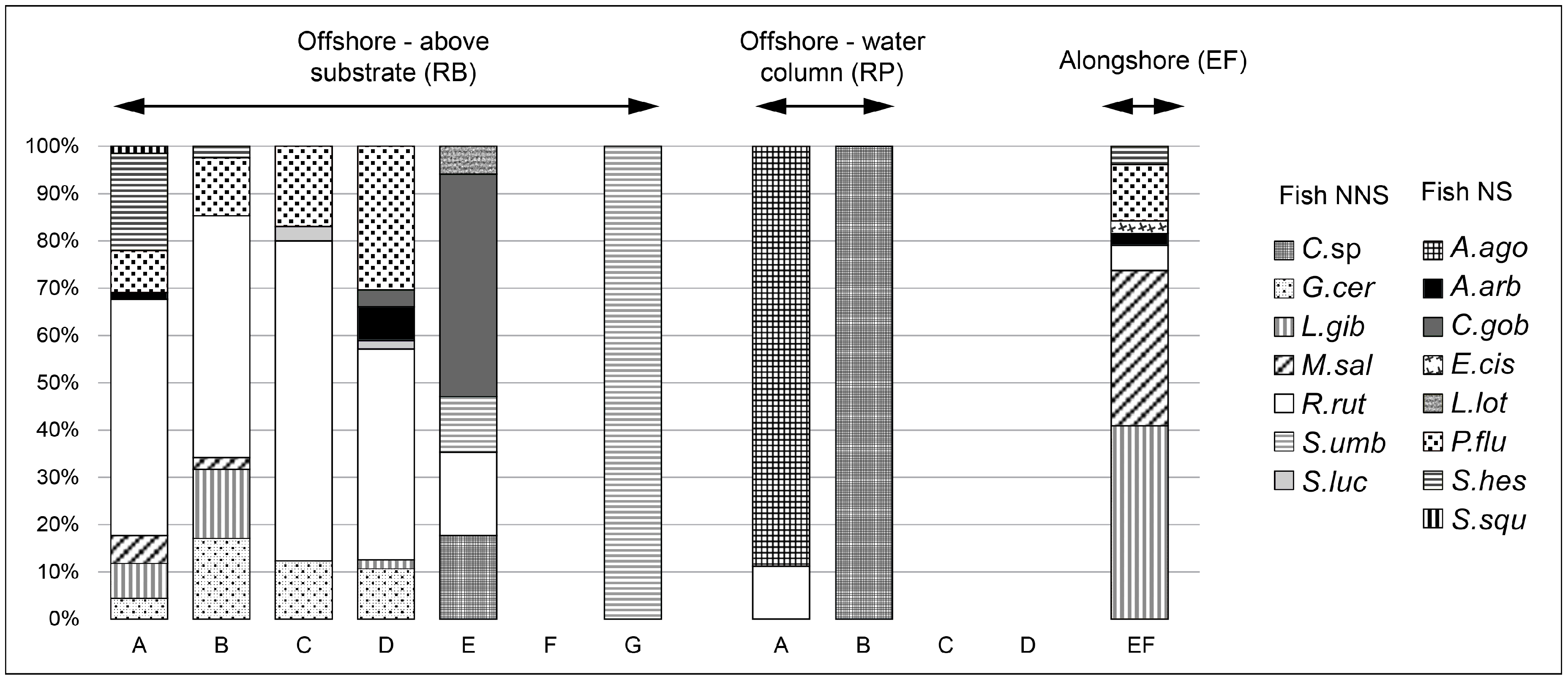
3.1. Zooplankton, Zoobenthos, and Fish Samples
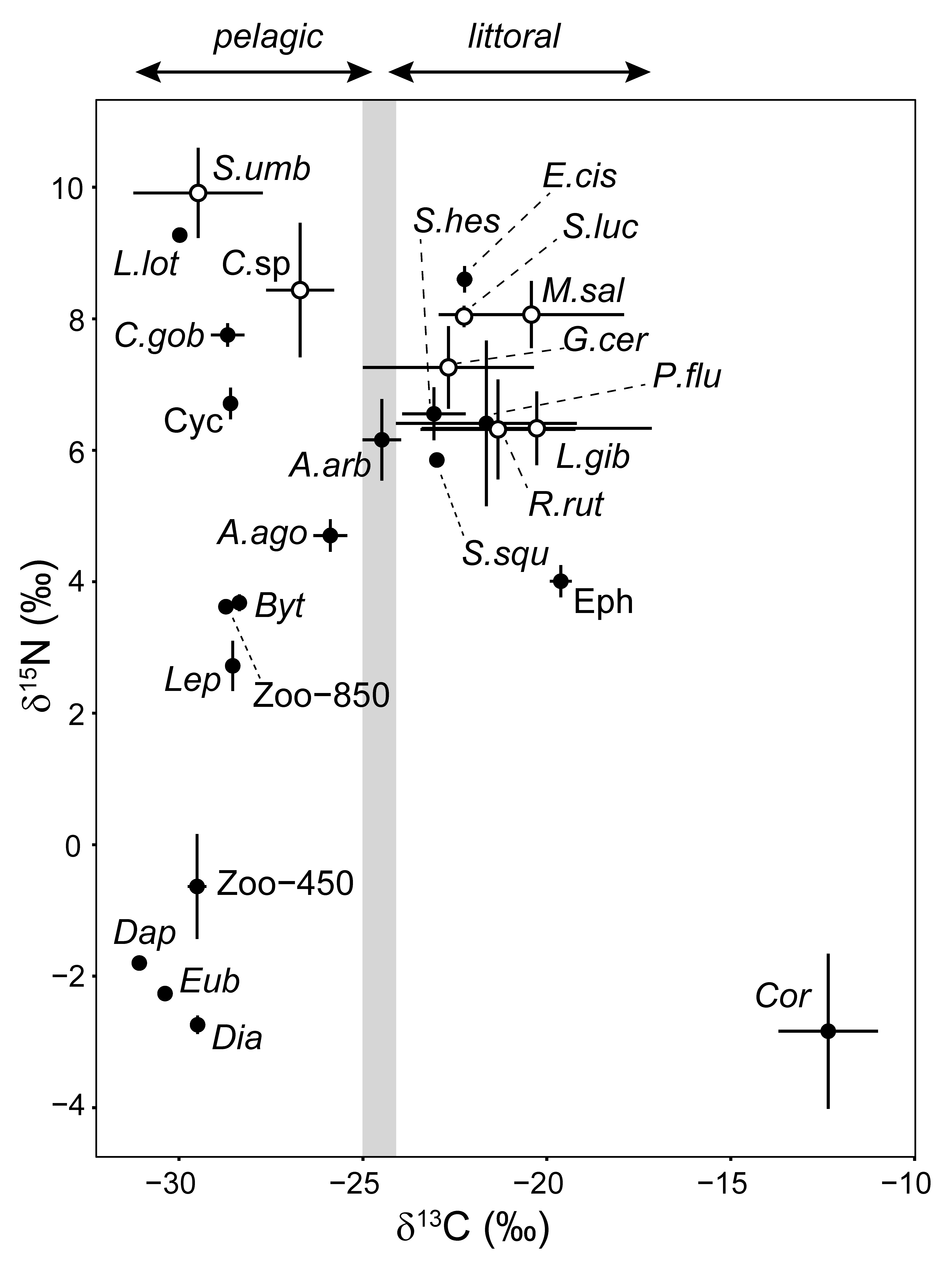
3.2. Stable Isotope Analysis and Trophic Positions
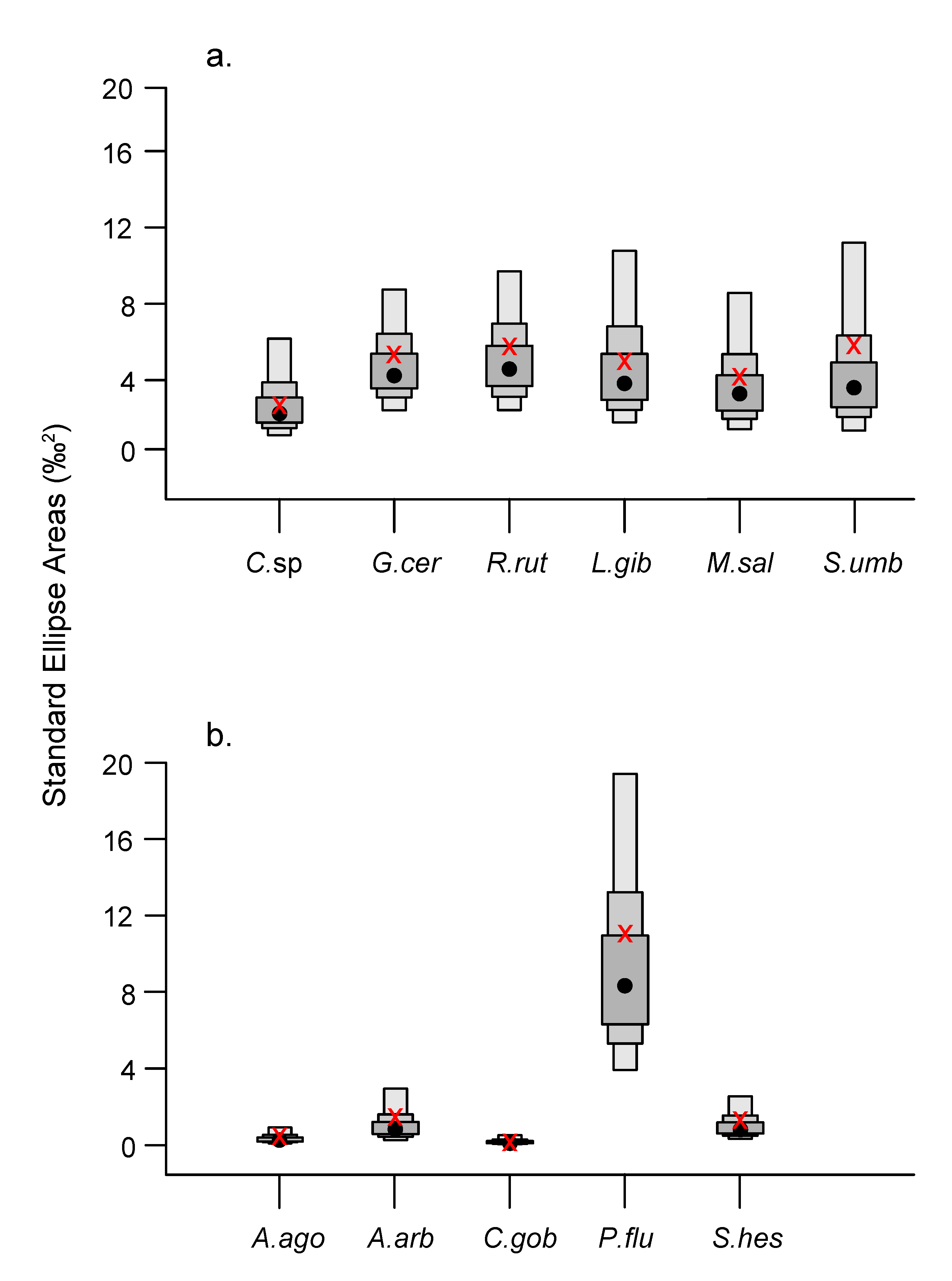
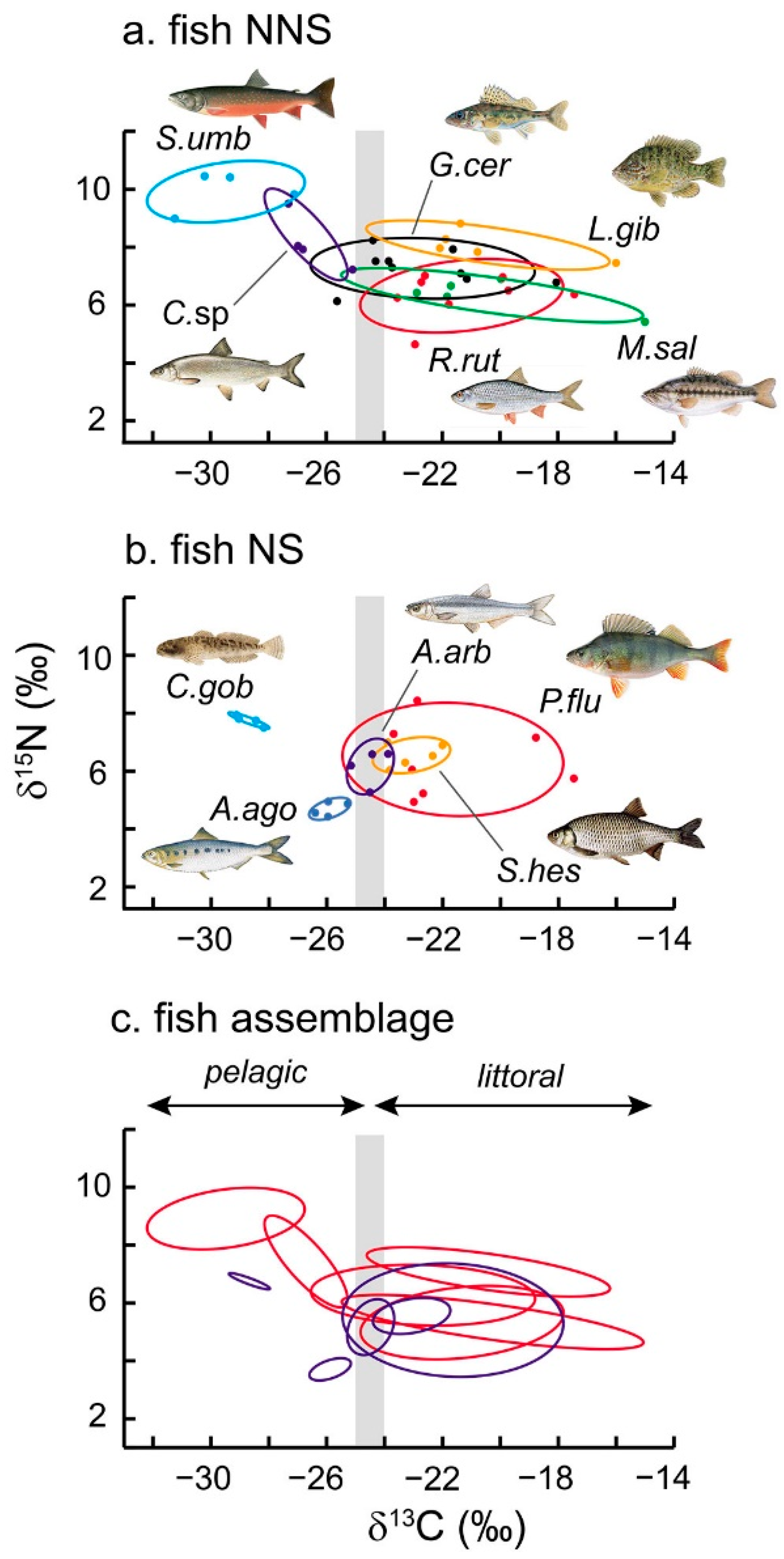
3.3. Isotopic Niche Metrics and Overlaps
4. Discussion
4.1. The Fish Community of Lake Mergozzo
4.2. Foraging Habitats and Food Ecology
4.3. Trophic Generalism and Niche Overlaps
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| TL (cm) | BPUE (g ind. m−2 or g ind. dip−1) | NPUE (n. ind. m−2 or n. ind. dip−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fish Species | RB | RP | EF | RB | RP | EF | RB | RP | EF | ||
| Fish NNS | |||||||||||
| Coregonus sp. | 11.1 ± 2.3 | 39.1 ± 4.3 | - | 0.01 ± 0.05 | 0.312 ± 0.863 | 0.00 ± 0.00 | 0.0011 ± 0.0056 | 0.0006 ± 0.0017 | 0.00 ± 0.00 | ||
| G. cernua | 12.3 ± 3.3 | - | - | 0.26 ± 0.53 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0087 ± 0.0159 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| L. gibbosus | 13.2 ± 2.3 | - | 11.5 ± 3.8 | 0.2 ± 0.52 | 0.000 ± 0.000 | 18.65 ± 77.31 | 0.0045 ± 0.0104 | 0.0000 ± 0.0000 | 0.42 ± 1.10 | ||
| M. salmoides | 19.5 ± 6.2 | - | 13.9 ± 7.9 | 0.20 ± 0.70 | 0.000 ± 0.000 | 30.19 ± 104.70 | 0.0019 ± 0.0064 | 0.0000 ± 0.0000 | 0.34 ± 0.69 | ||
| R. rutilus | 17.4 ± 6.8 | 13.8 | 9.7 ± 3.9 | 4.04 ± 6.74 | 0.003 ± 0.014 | 0.73 ± 4.57 | 0.0481 ± 0.0762 | 0.0002 ± 0.0006 | 0.05 ± 0.23 | ||
| S. umbla | 22.8 ± 4.4 | - | - | 0.20 ± 0.62 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0019 ± 0.0054 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| S. lucioperca | 56.0, 65.0 | - | - | 1.92 ± 11.21 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0011 ± 0.0056 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| Overall means ± SD | 0.98 ± 5.11 | 0.045 ± 0.336 | 7.08 ± 50.25 | 0.0096 ± 0.0337 | 0.0001 ± 0.0007 | 0.12 ± 0.52 | |||||
| Fish NS | |||||||||||
| A. agone | - | 11.9 ± 0.7 | - | 0.00 ± 0.00 | 0.019 ± 0.035 | 0.00 ± 0.00 | 0.0000 ± 0.0000 | 0.0012 ± 0.0024 | 0.00 ± 0.00 | ||
| A. arborella | 8.2 ± 0.7 | - | - | 0.01 ± 0.03 | 0.000 ± 0.000 | 0.05 ± 0.37 | 0.0019 ± 0.0082 | 0.0000 ± 0.0000 | 0.03 ± 0.16 | ||
| C. gobio | 7.1 ± 2.0 | - | - | 0.02 ± 0.06 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0038 ± 0.0134 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| E. cisalpinus | - | - | - | 0.00 ± 0.00 | 0.000 ± 0.000 | 40.2 ± 248.77 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.03 ± 0.16 | ||
| L. lota | 31.3 | - | - | 0.06 ± 0.40 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0004 ± 0.0025 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| P. fluviatilis | 16.1 ± 8.7 | - | 12.4 ± 3.9 | 1.35 ± 3.48 | 0.000 ± 0.000 | 4.01 ± 20.24 | 0.0148 ± 0.0292 | 0.0000 ± 0.0000 | 0.12 ± 0.37 | ||
| S. hesperidicus | 19.1 ± 4.2 | - | 16.9 ± 2.0 | 0.49 ± 1.91 | 0.000 ± 0.000 | 2.77 ± 14.19 | 0.0057 ± 0.0262 | 0.0000 ± 0.0000 | 0.04 ± 0.2.0 | ||
| S. squalus | 24.6 | - | - | 0.05 ± 0.36 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.0004 ± 0.0025 | 0.0000 ± 0.0000 | 0.00 ± 0.00 | ||
| Overall means ± SD | 0.25 ± 1.47 | 0.002 ± 0.014 | 5.88 ± 88.83 | 0.0034 ± 0.0156 | 0.0002 ± 0.0009 | 0.03 ± 0.17 | |||||
| Grand means | 0.59 ± 3.67 | 0.022 ± 0.230 | 6.44 ± 73.36 | 0.0063 ± 0.0258 | 0.0001 ± 0.0008 | 0.07 ± 0.38 | |||||
| Species | TL (cm) | Status | Common Food Source | Foraging Habitat | References |
|---|---|---|---|---|---|
| Coregonus sp. | 8–44 | NNS | zooplankton, zoobenthos | pelagic, demersal | [51,68,99] |
| G. cernua | 5–18 | NNS | zooplankton, zoobenthos, (fishes) | littoral, pelagic, demersal | [55,97,111,112] |
| L. gibbosus | 5–25 | NNS | (plant material), zooplankton, zoobenthos, (fishes) | littoral, demersal | [99,113,114] |
| M. salmoides | 5–30 | NNS | zooplankton, zoobenthos, macroinvertebrates, fishes | littoral, demersal | [99,115,116] |
| R. rutilus | 6–33 | NNS | detritus, plant material, zooplankton, zoobenthos | littoral, pelagic | [54,117] |
| S. umbla | 16–28 | NNS | zoobenthos, macroinvertebrates, fishes | pelagic, demersal deep waters | [64] |
| S. lucioperca | 37–73 | NNS | (zoobenthos), fishes | littoral, demersal | [118,119] |
| A. agone | 11–13 | NS | zooplankton | pelagic | [68,120] |
| A. arborella | 7–9 | NS | algae, zooplankton | pelagic | [121,122] |
| C. gobio | 4–10 | NS | zoobenthos | benthic | [123] |
| E. cisalpinus | 56, 65 | NS | fishes | littoral | [115,124] |
| L. lota | 31 | NS | macroinvertebrates, fishes | benthic, deep waters | [106,125] |
| P. fluviatilis | 6–41 | NS | zooplankton, zoobenthos, fishes | littoral, pelagic | [97,103] |
| S. hesperidicus | 15–33 | NS | detritus, algae, aquatic macrophytes, terrestrial plant matter, zooplankton, (zoobenthos) | littoral | [126] |
| S. squalus | 25 | NS | plant material, zoobenthos, terrestrial epineuston, fishes | littoral | [127,128] |

References
- Simberloff, D. Non‒natives: 141 Scientists object. Nature 2011, 475, 36. [Google Scholar] [CrossRef] [PubMed]
- Simberloff, D.; Martin, J.L.; Genovesi, P.; Maris, V.; Wardle, D.A.; Aronson, J.; Courchamp, F.; Galil, B.; Garcìa-Berthou, E.; Pascal, M.; et al. Impacts of biological invasions: What’s what and the way forward. Trends Ecol. Evol. 2013, 28, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallardo, B.; Clavero, M.; Sànchez, M.I.; Vilà, M. Global ecological impacts of invasive species in aquatic ecosystems. Glob. Chang. Biol. 2016, 22, 151–163. [Google Scholar] [CrossRef] [PubMed]
- ISSG (Invasive Species Specialist Group) Species Survival Commission. IUCN Guidelines for the Prevention of Biodiversity Loss Caused by Alien Invasive Species; International Union for the Conservation of Nature (IUCN): Gland, Switzerland, 2000; pp. 1–25. [Google Scholar]
- Leprieur, F.; Brosse, S.; García-Berthou, E.; Oberdorff, T.; Olden, J.D.; Townsend, C.R. Scientific uncertainty and the assessment of risks posed by non-native freshwater fishes. Fish Fish. 2009, 10, 88–97. [Google Scholar] [CrossRef]
- Mooney, H.A.; Hobbs, R.J. Invasive Species in a Changing World; Island Press: Washington, DC, USA, 2000; pp. 1–465. [Google Scholar]
- Jackson, D.A. Ecological effects of Micropterus introductions: The dark side of black bass. In Black Bass: Ecology, Conservation, and Management, Proceedings of Black Bass 2000: Ecology, Conservation, and Management of Black Bass in North America, St. Louis, MO, USA, 21–24 August 2000; Symposium 31; Philipp, D.P., Ridgway, M.S., Eds.; American Fisheries Society: Bethesda, MD, USA, 2003; pp. 221–232. [Google Scholar]
- Townsend, C.R. Individual, population, community, and ecosystem consequences of a fish invader in New Zealand streams. Conserv. Biol. 2003, 17, 38–47. [Google Scholar] [CrossRef]
- Hooper, D.U.; Chapin III, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, P.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Dudgeon, D.; Arthington, A.H.; Gessner, M.O.; Kawabata, Z.I.; Knowler, D.J.; Lévêque, C.; Naiman, R.J.; Prieur-Richard, A.H.; Soto, D.; Stiassny, M.L.J.; et al. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biol. Rev. 2006, 81, 163–182. [Google Scholar] [CrossRef]
- Gherardi, F.; Bertolino, S.; Bodon, M.; Casellato, S.; Cianfanelli, S.; Ferragutti, M.; Lori, E.; Mura, G.; Nocita, A.; Riccardi, N.; et al. Animal xenodiversity in Italian inland waters: Distribution, models of arrivals, and pathways. Biol. Invasions 2008, 10, 435–454. [Google Scholar] [CrossRef]
- Mooney, H.A.; Cleland, E.E. The evolutionary impact of invasive species. Proc. Natl. Acad. Sci. USA 2001, 98, 5446–5451. [Google Scholar] [CrossRef] [Green Version]
- Shea, K.; Chesson, P. Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 2002, 17, 170–176. [Google Scholar] [CrossRef]
- Olden, J.D.; LeRoy Poff, N.; Bestgen, K.R. Life-history strategies predict fish invasions and extirpations in the Colorado river basin. Ecol. Monogr. 2006, 76, 25–40. [Google Scholar] [CrossRef] [Green Version]
- David, P.; Thébeault, E.; Anneville, O.; Duyck, P.F.; Chapuis, E.; Loeuille, N. Impacts of invasive species on food webs: A review of empirical data. Adv. Ecol. Res. 2017, 56, 1–60. [Google Scholar]
- Britton, J.R.; Ruiz-Navarro, A.; Verreycken, H.; Amat-Trigo, F. Trophic consequences of introduced species: Comparative impacts of increased interspecific versus intraspecific competitive interactions. Funct. Ecol. 2018, 32, 486–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simberloff, D. Community effects of introduced species. In Biotic Crises in Ecological and Evolutionary Time, Proceedings of the Third Annual Spring Systematics Symposium, Field Museum of Natural History, Chicago, IL, USA, 10 May 1980; Nitecki, M.H., Ed.; Academic Press Inc.: New York, NY, USA, 1981; pp. 53–81. [Google Scholar]
- Herbold, B.; Moyle, P.B. Introduced species and vacant niches. Am. Nat. 1986, 128, 751–760. [Google Scholar] [CrossRef]
- Elton, C.S. The Ecology of Invasions by Animals and Plants, 1st ed.; (Reprint 1972); Chapman and Hall Ltd.: London, UK, 1958; pp. 1–181. [Google Scholar]
- Golani, D. Trophic adaptation of Red Sea fishes to the eastern Mediterranean environment—review and new data. Isr. J. Zool. 1993, 39, 391–402. [Google Scholar]
- Karlson, A.M.L.; Gorokhova, E.; Elmgren, R. Do deposit-feeders compete? Isotopic niche analysis of an invasion in a species-poor system. Sci. Rep. UK 2015, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Correia, A.M. Niche breadth and trophic diversity: Feeding behaviour of the red swamp crayfish (Procambarus clarkii) towards environmental availability of aquatic macroinvertebrates in a rice field (Portugal). Acta Oecol. 2002, 23, 421–429. [Google Scholar] [CrossRef]
- Petren, K.; Case, T.J. An experimental demonstration of exploitation competition in an ongoing invasion. Ecology 1996, 77, 118–132. [Google Scholar] [CrossRef]
- Holway, D.A. Competitive mechanisms underlying the displacement of native ants by the invasive Argentine ant. Ecology 1999, 80, 238–251. [Google Scholar] [CrossRef]
- Byers, J.E. Competition between two estuarine snails: Implications for invasions of exotic species. Ecology 2000, 81, 1225–1239. [Google Scholar] [CrossRef]
- Newsome, S.D.; Martinez del Rio, C.; Bearhop, S.; Phillips, D.L. A niche for isotopic ecology. Front. Ecol. Environ. 2007, 5, 429–436. [Google Scholar] [CrossRef]
- Rau, G. Carbon‒13 depletion in a subalpine lake: Carbon flow implications. Science 1978, 201, 901–902. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Schelske, C.L.; Hoyer, M.V. Stable isotopes of carbon and nitrogen as indicators of diet and trophic structure of the fish community in a shallow hypereutrophic lake. J. Fish Biol. 1996, 49, 1233–1243. [Google Scholar] [CrossRef]
- Vander Zanden, M.J.; Casselman, J.M.; Rasmussen, J.B. Stable isotope evidence for the food web consequences of species invasions in lakes. Nature 1999, 401, 464–467. [Google Scholar] [CrossRef]
- Croll, D.A.; Maron, J.L.; Estes, J.A.; Danner, E.M.; Byrd, G.V. Introduced predators transform subarctic islands from grassland to tundra. Science 2005, 307, 1959–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagouis, A.; Cucherousset, J.; Villéger, S.; Santoul, F.; Boulêtreau, S. Non-native species modify the isotopic structure of freshwater fish communities across the globe. Ecography 2015, 38, 979–985. [Google Scholar] [CrossRef]
- Layman, C.A.; Arrington, D.A.; Montaña, C.G.; Post, D.M. Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology 2007, 88, 42–48. [Google Scholar] [CrossRef]
- Jackson, A.L.; Inger, R.; Parnell, A.P.; Bearhop, S. Comparing isotopic niche widths among and within communities: SIBER—Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 2011, 80, 595–602. [Google Scholar] [CrossRef]
- Jackson, M.C.; Donohue, I.; Jackson, A.L.; Britton, J.R.; Harper, D.M.; Grey, J. Population-level metrics of trophic structure based on stable isotopes and their application to invasion ecology. PLoS ONE 2012, 7, e31757. [Google Scholar] [CrossRef]
- Lowe, S.; Browne, M.; Boudjelas, S.; de Poorter, M. 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database; Invasive Species Specialist Group (ISSG, Species Survival Commission of the International Union for the Conservation of Nature): Auckland, New Zealand, 2004; pp. 1–12. [Google Scholar]
- García-Berthou, E. The characteristics of invasive fishes: What has been learned so far? J. Fish Biol. 2007, 71, 33–55. [Google Scholar] [CrossRef]
- Gozlan, R.E.; Britton, J.R.; Cowx, I.; Copp, G.H. Current knowledge on non-native freshwater fish introductions. J. Fish Biol. 2010, 76, 751–786. [Google Scholar] [CrossRef]
- Cucherousset, J.; Olden, J.D. Ecological impacts of non-native freshwater fishes. Fisheries 2011, 36, 215–230. [Google Scholar] [CrossRef]
- Maceda-Veiga, A.; Escribano-Alacid, J.; de Sostoa, A.; García-Berthou, E. The aquarium trade as a potential source of fish introductions in southwestern Europe. Biol. Invasions 2013, 15, 2707–2716. [Google Scholar] [CrossRef]
- Marr, S.M.; Olden, J.D.; Leprieur, F.; Arismendi, I.; Ćaleta, M.; Morgan, D.L.; Nocita, A.; Šanda, R.; Tarkan, A.S.; García-Berthou, E. A global assessment of freshwater fish introductions in mediterranean-climate regions. Hydrobiologia 2013, 719, 317–329. [Google Scholar] [CrossRef] [Green Version]
- Copp, G.H.; Britton, J.R.; Guo, Z.; Edmonds‒Brown, V.R.; Pegg, J.; Vilizzi, L.; Davison, P.I. Trophic consequences of non‒native pumpkinseed Lepomis gibbosus for native pond fishes. Biol. Invasions 2017, 19, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Ogutu‒Ohwayo, R.; Hecky, R.E. Fish introductions in Africa and some of their implications. Can. J. Fish. Aquat. Sci. 1991, 48, 8–12. [Google Scholar] [CrossRef]
- Eby, L.A.; Roach, W.J.; Crowder, L.B.; Stanford, J.A. Effects of stocking-up freshwater food webs. Trends Ecol. Evol. 2006, 21, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Crutchfield, J.U.; Schiller, D.H.; Herlong, D.D.; Mallin, M.A. Establishment and impact of redbelly tilapia in a vegetated cooling reservoir. J. Aquat. Plant Manag. 1992, 30, 28–35. [Google Scholar]
- Zambrano, L.; Scheffer, M.; Martínez-Ramos, M. Catastrophic response of lakes to benthivorous fish introduction. Oikos 2003, 94, 344–350. [Google Scholar] [CrossRef] [Green Version]
- Volta, P.; Jeppesen, E.; Leoni, B.; Campi, B.; Sala, P.; Garibaldi, L.; Lauridsen, L.T.; Winfield, I.J. Recent invasion by a non-native cyprinid (common bream Abramis brama) is followed by major changes in the ecological quality of a shallow lake in southern Europe. Biol. Invasions 2013, 15, 2065–2079. [Google Scholar] [CrossRef] [Green Version]
- Vander Zanden, M.J.; Chandra, S.; Allen, B.C.; Reuter, J.E.; Goldman, C. Historical food web structure and restoration of native aquatic communities in the lake Tahoe (California-Nevada) basin. Ecosystems 2003, 6, 274–288. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, M.M.; Haffner, G.D.; Legler, N.D.; Rush, S.A.; Fisk, A.T. Fifty years later: Trophic ecology and niche overlap of a native and non-indigenous fish species in the western basin of Lake Erie. Biol. Invasions 2013, 15, 1695–1711. [Google Scholar] [CrossRef]
- Córdova-Tapia, F.; Contreras, M.; Zambrano, L. Trophic niche overlap between native and non‒native fishes. Hydrobiologia 2015, 746, 291–301. [Google Scholar] [CrossRef]
- Wang, J.; Chapman, D.; Xu, J.; Wang, Y.; Gu, B. Isotope niche dimension and trophic overlap between bigheaded carps and native filter-feeding fish in the lower Missouri River, USA. PLoS ONE 2018, 13, e0199805. [Google Scholar] [CrossRef] [Green Version]
- Giussani, G. Prime indagini sulla fauna ittica del Lago di Mergozzo. Report CNR 1994, 3, 1–12. [Google Scholar]
- Bianco, P.G. Freshwater fish transfers in Italy: History, local modification of fish composition, and a prediction on the future of native populations. In Stocking and Introductions of Fishes; Cowx, I.G., Ed.; Fishing New Books: Oxford, UK; Blackwell Science: Oxford, UK,, 1998; Chapter 14; pp. 167–185. [Google Scholar]
- Bianco, P.G. An update on the status of native and exotic freshwater fishes of Italy. J. Appl. Ichthyol. 2014, 30, 62–77. [Google Scholar] [CrossRef]
- Volta, P.; Jepsen, N. The recent invasion of Rutilus rutilus (L.) (Pisces: Cyprinidae) in a large South-Alpine lake: Lago Maggiore. J. Limnol. 2008, 67, 163–170. [Google Scholar] [CrossRef]
- Volta, P.; Jeppesen, E.; Campi, B.; Sala, P.; Emmrich, M.; Winfield, I.J. The population biology and life history traits of Eurasian ruffe [Gymnocephalus cernuus (L.), Pisces: Percidae] introduced into eutrophic and oligotrophic lakes in Northern Italy. J. Limnol. 2013, 72, 280–290. [Google Scholar] [CrossRef] [Green Version]
- Volta, P.; Jeppesen, E.; Sala, P.; Galafassi, S.; Foglini, C.; Puzzi, C.; Winfield, I.J. Fish assemblages in deep Italian subalpine lakes: History and present status with an emphasis on non-native species. Hydrobiologia 2018, 824, 255–270. [Google Scholar] [CrossRef]
- Barbanti, L. Osservazioni sul Lago Maggiore. Lineamenti del territorio del Verbano; Circolo del Pallanzotto: Comune di Verbania, Italy, 1994; pp. 1–113. [Google Scholar]
- Council Directive 92/43/EEC. On the conservation of natural habitats and of wild fauna and flora. O. J. L 1992, 206, 7–50. [Google Scholar]
- Post, D.M. Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 2002, 83, 703–718. [Google Scholar] [CrossRef]
- Mazzoni, M.; Ferrario, C.; Bettinetti, R.; Piscia, R.; Cicala, D.; Volta, P.; Borgå, K.; Valsecchi, S.; Polesello, S. Trophic magnification of legacy (PCB, DDT and Hg) and emerging pollutants (PFAS) in the fish community of a small protected southern alpine lake (Lake Mergozzo, Northern Italy). Water 2020, 12, 1591. [Google Scholar] [CrossRef]
- Bettinetti, R.; Quadroni, S.; Manca, M.; Piscia, R.; Volta, P.; Guzzella, L.; Roscioli, C.; Galassi, S. Seasonal fluctuations of DDTs and PCBs in zooplankton and fish of Lake Maggiore (Northern Italy). Chemosphere 2012, 88, 344–351. [Google Scholar] [CrossRef]
- Piscia, R.; Mazzoni, M.; Bettinetti, R.; Caroni, R.; Cicala, D.; Manca, M. Stable Isotope Analysis and persistent organic pollutants in crustacean zooplankton: The role of size and seasonality. Water 2019, 11, 1490. [Google Scholar] [CrossRef] [Green Version]
- Copp, G.H.; Garner, P. Evaluating microhabitat use of fish larvae and juveniles with point abundance sampling. Folia Zool. 1995, 44, 145–158. [Google Scholar]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Kottelat: Cornol, Switzerland; Freyhof: Berlin, Germany, 2007; pp. 1–646. [Google Scholar]
- Bianco, P.G.; Delmastro, G.B. Recenti novità tassonomiche riguardanti i pesci d’acqua dolce autoctoni in Italia e descrizione di una nuova specie di luccio. In Researches on Wildlife Conservation; De Filippo, G., Ed.; Istituto di Gestione della Fauna (IGF Publishing): USA, 2011; Volume 2, pp. 1–14. [Google Scholar]
- Lucentini, L.; Puletti, M.E.; Ricciolini, C.; Gigliarelli, L.; Fontaneto, D.; Lanfaloni, L.; Bilò, F.; Natali, M.; Panara, F. Molecular and phenotypic evidence of a new species of genus Esox (Esocidae, Esociformes, Actinopterygii): The southern pike, Esox flaviae. PLoS ONE 2011, 6, e25218. [Google Scholar] [CrossRef]
- Berg, A.; Grimaldi, E. Biologia delle due forme di Coregone (Coregonus sp.) del Lago Maggiore. Mem. Ist. Ital. Idrobiol. 1965, 18, 25–196. [Google Scholar]
- Berg, A.; Grimaldi, E. Ecological relationships between planktophagic fish species in the Lago Maggiore. Verh. Internat. Verein. Limnol. 1966, 16, 1065–1073. [Google Scholar] [CrossRef]
- Borroni, I.; Grimaldi, E. Fattori e tendenze di modificazione dell’ittiofauna italiana d’acqua dolce. Ital. J. Zool. (Boll. Zool.) 1978, 45 Suppl. S2, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Hudson, A.G.; Vonlanthen, P.; Seehausen, O. Rapid parallel adaptive radiations from a single hybridogenic ancestral population. Proc. R. Soc. Ser. B Bio. 2011, 278, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Tiberti, R.; Splendiani, A. Management of a highly unlikely native fish: The case of Arctic charr Salvelinus alpinus from the Southern Alps. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 312–320. [Google Scholar] [CrossRef]
- Lorenzoni, M.; Dörr, A.J.M.; Erra, R.; Giovinazzo, G.; Mearelli, M.; Selvi, S. Growth and reproduction of largemouth bass (Micropterus salmoides Lacépède, 1802) in Lake Trasimeno (Umbria, Italy). Fish. Res. 2002, 56, 89–95. [Google Scholar] [CrossRef]
- Treer, T.; Šprem, N.; Torcu-Koc, H.; Sun, Y.; Piria, M. Length-weight relationships of freshwater fishes of Croatia. J. Appl. Ichthyol. 2008, 24, 626–628. [Google Scholar] [CrossRef]
- Giannetto, D.; Pompei, L.; Carosi, A.; Dolciami, R.; Ghetti, L.; Lorenzoni, M. Length–weight relationships of three fish species from Trasimeno Lake, Italy. J. Appl. Ichthyol. 2016, 32, 1273–1275. [Google Scholar] [CrossRef]
- Darnaude, A.M.; Salen-Picard, C.; Polunin, N.V.C.; Harmelin-Vivien, M.L. Trophodynamic linkage between river runoff and coastal fishery yield elucidated by stable isotope data in the Gulf of Lions (NW Mediterranean). Oecologia 2004, 138, 325–332. [Google Scholar] [CrossRef]
- Visconti, A.; Fadda, A.; Di Guardo, A.; Manca, M. Seasonality, littoral versus pelagic carbon sources, and stepwise 15N‒enrichment of pelagic food web in a deep subalpine lake: The role of planktivorous fish. Can. J. Fish. Aquat. Sci. 2014, 71, 436–446. [Google Scholar] [CrossRef] [Green Version]
- Cicala, D.; Calizza, E.; Careddu, G.; Fiorentino, F.; Sporta Caputi, S.; Rossi, L.; Costantini, M.L. Spatial variation in the feeding strategies of Mediterranean fish: Flatfish and mullet in the Gulf of Gaeta (Italy). Aquat. Ecol. 2019, 53, 529–541. [Google Scholar] [CrossRef]
- France, R.L. Differentiation between littoral and pelagic lakes using stable carbon isotopes. Limnol. Oceanogr. 1995, 40, 1310–1313. [Google Scholar] [CrossRef]
- Hayden, B.; Massa‒Gallucci, A.; Harrod, C.; O’Grady, M.; Caffrey, J.; Kelly‒Quinn, M. Trophic flexibility by roach Rutilus rutilus in novel habitats facilitates rapid growth and invasion success. J. Fish Biol. 2014, 84, 1099–1116. [Google Scholar] [CrossRef]
- Verstijnen, Y.J.M.; Lucassen, E.C.H.E.T.; van der Gaag, M.; Wagenvoort, A.J.; Castelijns, H.; Ketelaars, H.A.M.; van der Velde, G.; Smolders, A.J.P. Trophic relationships in Dutch reservoirs recently invaded by Ponto-Caspian species: Insights from fish trends and stable isotope analysis. Aquat. Invasions 2019, 14, 280–298. [Google Scholar] [CrossRef]
- Majdi, N.; Bardon, L.; Gilbert, F. Quantification of sediment reworking by the Asiatic clam Corbicula fluminea Müller, 1774. Hydrobiologia 2014, 732, 85–92. [Google Scholar] [CrossRef]
- Post, D.M.; Layman, C.A.; Albrey, D.A.; Takimoto, G.; Quattrochi, J.; Montaña, C.G. Getting to the fat of the matter: Models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 2007, 152, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Layman, C.A.; Araujo, M.S.; Boucek, R.; Hammerschlag-Peyer, C.M.; Harrison, E.; Jud, Z.R.; Matich, P.; Rosenblatt, A.D.; Vaudo, J.J.; Yeager, L.A.; et al. Applying stable isotopes to examine food-web structure: An overview of analytical tools. Biol. Rev. 2012, 87, 545–562. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: www.R‒project.org (accessed on 20 January 2019).
- Perkins, M.J.; Inger, R.; Bearhop, S.; Sanders, D. Multichannel feeding by spider functional groups is driven by feeding strategies and resource availability. Oikos 2017, 127, 23–33. [Google Scholar] [CrossRef]
- Bašić, T.; Copp, G.H.; Edmonds‒Brown, V.R.; Keskin, E.; Davison, P.I.; Britton, J.R. Trophic consequences of an invasive, small‒bodied non‒native fish, sunbleak Leucaspius delineatus, for native pond fishes. Biol. Invasions 2019, 21, 261–275. [Google Scholar] [CrossRef] [Green Version]
- Lysy, M.; Stasko, A.D.; Swanson, H.K. nicheROVER: (Niche) (R)egion and Niche (Over)lap Metrics for Multidimensional Ecological Niches (Version 1.0). Available online: https://cran.r‒project.org/web/packages/nicheROVER/nicheROVER.pdf (accessed on 20 February 2019).
- Swanson, H.K.; Lysy, M.; Power, M.; Stasko, A.D.; Johnson, J.D.; Reist, J.D. A new probabilistic method for quantifying n‒dimensional ecological niches and niche overlap. Ecology 2015, 96, 318–324. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 31 October 2020).
- Calderoni, A.; Monti, C.; Polli, B. Andamento Della Pesca Professionale sul Lago Maggiore nel Periodo 1979–2011; Commissione Italo-Svizzera per la Pesca, 2013; Volume 1, pp. 1–84. [Google Scholar]
- Jeppesen, E.; Mehner, T.; Winfield, I.J.; Kangur, K.; Sarvala, J.; Gerdeaux, D.; Rask, M.; Malmquist, H.J.; Holmgren, K.; Volta, P.; et al. Impacts of climate warming on the long-term dynamics of key fish species in 24 European lakes. Hydrobiologia 2012, 694, 1–39. [Google Scholar] [CrossRef] [Green Version]
- Volta, P.; Yan, N.D.; Gunn, J.M. Past, present and future of the fish community of Lake Orta (Italy), one of the world’s largest acidified lakes. J. Limnol. 2016, 75, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Milardi, M.; Lanzoni, M.; Gavioli, A.; Fano, E.A.; Castaldelli, G. Long-term fish monitoring underlines a rising tide of temperature tolerant, rheophilic, benthivore and generalist exotics, irrespective of hydrological conditions. J. Limnol. 2018, 77, 1–20. [Google Scholar] [CrossRef]
- Stefani, F.; Schiavon, A.; Tirozzi, P.; Gomarasca, S.; Marziali, L. Functional response of fish communities in a multistressed freshwater world. Sci. Total. Environ. 2020, 740, 139902. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, F.; Orjuela, R.L.; Magalhaes, M.F.; Collares-Pereira, M.J. Variability in feeding ecology of a South American cichlid: A reason for successful invasion in Mediterranean-type rivers? Ecol. Freshw. Fish 2007, 16, 559–569. [Google Scholar] [CrossRef]
- Gallagher, J.; Hammerschlag, N.; Cooke, S.J.; Costa, D.P.; Irschick, D.J. Evolutionary theory as a tool for predicting extinction risk. Trends Ecol. Evol. 2015, 30, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Lorenzoni, M.; Carosi, A.; Pedicillo, G.; Trusso, A. Comparative study on the feeding competition of the European perch Perca fluviatilis L. and the ruffe Gymnocephalus cernuus (L.) in Lake Piediluco (Umbria, Italy). B. Fr. Pêche Piscic. 2007, 387, 35–57. [Google Scholar] [CrossRef] [Green Version]
- Vejřıková, I.; Eloranta, A.P.; Vejřík, L.; Smejkal, M.; Čech, M.; Sajdlova, Z.; Frouzova, J.; Kiljunen, M.; Peterka, J. Macrophytes shape trophic niche variation among generalist fishes. PLoS ONE 2017, 12, e0177114. [Google Scholar] [CrossRef]
- Costantini, M.L.; Carlino, P.; Calizza, E.; Careddu, G.; Cicala, D.; Sporta Caputi, S.; Fiorentino, F.; Rossi, L. The role of alien fish (the centrarchid Micropterus salmoides) in lake food webs highlighted by stable isotope analysis. Freshw. Biol. 2018, 63, 1130–1142. [Google Scholar] [CrossRef]
- Linzmaier, S.M.; Twardochleb, L.A.; Olden, J.O.; Mehner, T.; Arlinghaus, R. Size-dependent foraging niches of European Perch Perca fluviatilis (Linnaeus, 1758) and North American Yellow Perch Perca flavescens (Mitchill, 1814). Environ. Biol. Fish. 2018, 101, 23–37. [Google Scholar] [CrossRef] [Green Version]
- Emmrich, M.; Pédron, S.; Brucet, S.; Winfield, I.J.; Jeppesen, E.; Volta, P.; Argillier, C.; Lauridsen, T.L.; Holmgren, K.; Hesthagen, T.; et al. Geographical patterns in the size structure of European lake fish communities along abiotic and biotic gradients. J. Biogeogr. 2014, 41, 2221–2233. [Google Scholar] [CrossRef] [Green Version]
- FAO FishFinder. Available online: www.fao.org/fishery/fishfinder/en (accessed on 15 October 2020).
- Svanbäck, R.; Quevedo, M.; Olsson, J.; Eklöv, P. Individuals in food webs: The relationships between trophic position, omnivory and among-individual diet variation. Oecologia 2015, 178, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, A.A. Phenotypic plasticity in the interactions and evolution of species. Science 2001, 294, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Gould, S.J.; Vrba, E.S. Exaptation‒a missing term in the science of form. Paleobiology 1982, 8, 4. [Google Scholar] [CrossRef]
- McPhail, J.D.; Paragamian, V.L. Burbot biology and life history. In Burbot: Biology, Ecology and Management; Paragamian, V.L., Willis, D.W., Eds.; Fisheries Management Section of the American Fisheries Society: New York, NY, USA, 2000; Volume 128, pp. 11–23. [Google Scholar]
- Haubrock, P.J.; Balzani, P.; Azzini, M.; Inghilesi, A.F.; Veselý, L.; Guo, W.; Tricarico, E. Shared histories of co-evolution may affect trophic interactions in a freshwater community dominated by alien species. Front. Ecol. Evol. 2019, 7, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zambrano, L.; Valiente, E.; Vander Zanden, M.J. Food web overlap among native axolotl (Ambystoma mexicanum) and two exotic fishes: Carp (Cyprinus carpio) and tilapia (Oreochromis niloticus) in Xochimilco, Mexico City. Biol. Invasions 2010, 12, 3061–3069. [Google Scholar] [CrossRef]
- Calizza, E.; Costantini, M.L.; Careddu, G.; Rossi, L. Effect of habitat degradation on competition, carrying capacity, and species assemblage stability. Ecol. Evol. 2017, 7, 5784–5796. [Google Scholar] [CrossRef]
- Coulter, A.; Swanson, H.K.; Goforth, R.R. Seasonal variation in resource overlap of invasive and native fishes revealed by stable isotopes. Biol. Invasions 2019, 21, 315–321. [Google Scholar] [CrossRef]
- Gutsch, M.; Hoffman, J. A review of ruffe (Gymnocephalus cernua) life history in its native versus non-native range. Rev. Fish Biol. Fisher. 2016, 26, 213–233. [Google Scholar] [CrossRef]
- Eloranta, A.P.; Vejřıková, I.; Čech, M.; Vejřík, L.; Holubová, M.; Šmejkal, M.; Frouzová, J.; Kiljunen, M.; Jones, R.I.; Peterka, J. Some like it deep: Intraspecific niche segregation in ruffe (Gymnocephalus cernua). Freshw. Biol. 2017, 62, 1401–1409. [Google Scholar] [CrossRef]
- Osenberg, C.W.; Werner, E.E.; Mittelbach, G.G.; Hall, D.J. Growth patterns in bluegill (Lepomis macrochirus) and pumpkinseed (L. gibbosus) sunfish: Environmental variation and the importance of ontogenetic niche shifts. Can. J. Fish. Aquat. Sci. 1988, 45, 17–26. [Google Scholar] [CrossRef]
- Godinho, F.N.; Ferreira, M.T.; Cortes, R.V. The environmental basis of diet variation in pumpkinseed sunfish, Lepomis gibbosus, and largemouth bass, Micropterus salmoides, along an Iberian river basin. Environ. Biol. Fishes 1997, 50, 105–115. [Google Scholar] [CrossRef]
- Lorenzoni, M.; Corboli, M.; Dörr, A.J.M.; Giovinazzo, G.; Selvi, S.; Mearelli, M. Diets of Micropterus salmoides Lac. and Esox lucius L. in Lake Trasimeno (Umbria, Italy) and their diet overlap. Bull. Fr. Pêche Piscic. 2002, 365/366, 537–547. [Google Scholar] [CrossRef]
- Marinelli, A.; Scalici, M.; Gibertini, G. Diet and reproduction of largemouth bass in a recently introduced population, lake Bracciano (central Italy). Bull. Fr. Pêche Piscic. 2007, 385, 53–68. [Google Scholar] [CrossRef] [Green Version]
- García-Berthou, E. Spatial heterogeneity in roach (Rutilus rutilus) diet among contrasting basins within a lake. Archiv Fur Hydrobiol. 1999, 146, 239–256. [Google Scholar] [CrossRef]
- Nolan, E.T.; Britton, J.R. Diet of invasive pikeperch Sander lucioperca: Developing non‒destructive tissue sampling for stable isotope analysis with comparisons to stomach contents analysis. Knowl. Manag. Aquat. Ecosyst. 2018, 419, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Adámek, Z.; Mikl, L.; Šlapanský, L.; Jurajda, P.; Halačka, K. The diet of predatory fish in drinking water reservoirs—how can they contribute to biomanipulation efforts? Folia Zool. 2019, 68, 215–224. [Google Scholar] [CrossRef]
- Volta, P. Regime alimentare dell’agone (Alosa fallax lacustris Fatio, 1890) nel Lago Maggiore. Studi Trent. Sci. Nat. 2010, 87, 261–263. [Google Scholar]
- Giussani, G.; Ruffoni, T. Biology and growth of the bleak Alburnus alburnus alborella in two lakes (lago Maggiore and lago di Varese) characterized by a different trophic level. Mem. Ist. Ital. Idrobiol. 1985, 43, 147–159. [Google Scholar]
- Šorić, V.M. Redescription and some ecological characteristics of Alburnus arborella (Bonapartae, 1844). Kragujev. J. Sci. 2006, 28, 83–92. [Google Scholar]
- Števove, B.; Babeľová, M.; Haruštiaková, D.; Kováč, V. When a river is affected by a reservoir: Trophic interactions and flexibility in feeding strategies of alpine bullhead (Cottus poecilopus) and European bullhead (Cottus gobio). Sci. Total. Environ. 2019, 651, 1903–1912. [Google Scholar] [CrossRef]
- Jacobson, P.; Bergström, U.; Eklöf, J. Size-dependent diet composition and feeding of Eurasian perch (Perca fluviatilis) and northern pike (Esox lucius) in the Baltic Sea. Boreal Environ. Res. 2019, 24, 137–153. [Google Scholar]
- McBaine, K.E.; Klein, Z.B.; Quist, M.C.; Rhea, D.T. Diet of burbot and implications for sampling. Interm. J. Sci. 2018, 24, 1–13. [Google Scholar]
- Ravera, O.; Jamet, J.L. The diet of the rudd (Scardinius erythrophthalmus, L.) in relation to the possible consequences for the removal of this species from an eutrophic lake. Archiv. Fur Hydrobiol. 1991, 123, 99–109. [Google Scholar]
- Pignalberi, C. Importanza dei diversi gruppi di organismi nell’alimentazione estiva delle specie ittiche litorali del Lago Maggiore. Mem. Ist. Ital. Idrobiol. 1967, 21, 89–103. [Google Scholar]
- Balestrieri, A.; Prigioni, C.; Remonti, L.; Sgrosso, S.; Priore, G. Feeding ecology of Leuciscus cephalus and Rutilus rubilio in southern Italy. Ital. J. Zool. 2006, 73, 129–135. [Google Scholar] [CrossRef]

| TL (cm) | Biomass | Number of Fish Individuals | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Common Name | Range | RBb | RB%b | RPb | RP%b | EFb | EF%b | TOTb | TOT%b | RBn | RB%n | RPn | RP%n | EFn | EF%n | TOTn | TOT%n |
| Fish NNS§ | ||||||||||||||||||
| Coregonus sp. | European whitefish | 8.4–43.6 | 29 | 0.1 | 2024 | 93.4 | 0 | 0.0 | 2053 | 6.3 | 3 | 1.2 | 4 | 30.8 | 0 | 0.0 | 7 | 2.1 |
| G. cernua | Ruffe | 5.2–17.8 | 751 | 3.2 | 0 | 0.0 | 0 | 0.0 | 751 | 2.3 | 24 | 9.6 | 0 | 0.0 | 0 | 0.0 | 24 | 7.1 |
| L. gibbosus | Pumpkinseed | 4.6–25.0 | 537 | 2.3 | 0 | 0.0 | 1380 | 19.3 | 1917 | 5.9 | 12 | 4.8 | 0 | 0.0 | 31 | 40.8 | 43 | 12.7 |
| M. salmoides | Largemouth bass | 5.4–30.5 | 537 | 2.3 | 0 | 0.0 | 2234 | 31.2 | 2771 | 8.5 | 5 | 2.0 | 0 | 0.0 | 25 | 32.9 | 30 | 8.8 |
| R. rutilus | Roach | 5.8–33.0 | 10,657 | 45.7 | 22 | 1.0 | 54 | 0.8 | 10,733 | 32.9 | 127 | 50.8 | 1 | 7.7 | 4 | 5.3 | 132 | 38.9 |
| S. umbla | Char | 16.3–28.3 | 520 | 2.2 | 0 | 0.0 | 0 | 0.0 | 520 | 1.6 | 5 | 2.0 | 0 | 0.0 | 0 | 0.0 | 5 | 1.5 |
| S. lucioperca | Pikeperch | 37.4–73.0 | 5080 | 21.8 | 0 | 0.0 | 0 | 0.0 | 5080 | 15.6 | 3 | 1.2 | 0 | 0.0 | 0 | 0.0 | 3 | 0.9 |
| Fish NS | ||||||||||||||||||
| A. agone | Shad | 10.7–13.0 | 0 | 0.0 | 121 | 5.6 | 0 | 0.0 | 121 | 0.4 | 0 | 0.0 | 8 | 61.5 | 0 | 0.0 | 8 | 2.4 |
| A. arborella | Bleak | 5.0–8.8 | 19 | 0.1 | 0 | 0.0 | 4 | 0.1 | 23 | 0.1 | 5 | 2.0 | 0 | 0.0 | 2 | 2.6 | 7 | 2.1 |
| C. gobio | Bullhead | 4.0–9.7 | 46 | 0.2 | 0 | 0.0 | 0 | 0.0 | 46 | 0.1 | 10 | 4.0 | 0 | 0.0 | 0 | 0.0 | 10 | 2.9 |
| E. cisalpinus | Southern pike | 56.0, 65.0 | 0 | 0.0 | 0 | 0.0 | 2975 | 41.6 | 2975 | 9.1 | 0 | 0.0 | 0 | 0.0 | 2 | 2.6 | 2 | 0.6 |
| L. lota | Burbot | 31.3 | 161 | 0.7 | 0 | 0.0 | 0 | 0.0 | 161 | 0.5 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.3 |
| P. fluviatilis | Perch | 6.5–41.1 | 3554 | 15.2 | 0 | 0.0 | 297 | 4.2 | 3851 | 11.8 | 39 | 15.6 | 0 | 0.0 | 9 | 11.8 | 48 | 14.2 |
| S. hesperidicus | Italian rudd | 15.1–33.0 | 1304 | 5.6 | 0 | 0.0 | 205 | 2.9 | 1509 | 4.6 | 15 | 6.0 | 0 | 0.0 | 3 | 3.9 | 18 | 5.3 |
| S. squalus | Italian chub | 24.6 | 144 | 0.6 | 0 | 0.0 | 0 | 0.0 | 144 | 0.4 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.3 |
| StotMET | 23,339 | 71.5 | 2167 | 6.6 | 7149 | 21.9 | 32,655 | 100.0 | 250 | 73.7 | 13 | 3.8 | 76 | 22.4 | 339 | 100.0 | ||
| Species | Sample | δ13C | δ15N |
|---|---|---|---|
| Corbicula fluminea | AS | −12.35 ± 1.35 | −2.84 ± 1.18 |
| Ephemeroptera | AS | −19.62 ± 0.30 | 4.01 ± 0.25 |
| Bythotrephes longimanus | OS | −28.43, −28.29 | 3.77, 3.59 |
| Cyclopoida | OS | −28.73, −28.47 | 6.88, 6.54 |
| Daphnia sp. | OS | −31.08 | −1.80 |
| Diaphanosoma brachyurum | OS | −29.47, −29.52 | −2.64, −2.84 |
| Eubosmina longispina | OS | −30.39, −30.37 | −2.27, −2.26 |
| Leptodora kindtii | OS | −28.61, −28.47 | 2.45, 2.99 |
| Zoo-450 | OS | −29.33, −29.69 | −0.07, −1.20 |
| Zoo-850 | OS | −28.82, −28.63 | 3.57, 3.67 |
| Species | ss | δ15N | δ13C | TP |
|---|---|---|---|---|
| Fish NNS | ||||
| Coregonus sp. | 4 | 8.17 ± 1.26 | −26.56 ± 0.99 | 4.94 |
| G. cernua | 9 | 7.26 ± 0.63 | −22.68 ± 2.32 | 5.01 |
| L. gibbosus | 5 | 6.33 ± 0.56 | −20.27 ± 3.13 | 4.78 |
| M. salmoides | 5 | 8.06 ± 0.51 | −20.42 ± 2.52 | 5.22 |
| R. rutilus | 8 | 6.32 ± 0.76 | −21.33 ± 2.11 | 4.59 |
| S. umbla | 4 | 9.90 ± 0.69 | −29.48 ± 1.76 | 5.75 |
| S. lucioperca | 2 | 7.92, 8.15 | −22.24, −22.26 | 4.92 |
| Fish NS | ||||
| A. agone | 4 | 4.49 ± 0.39 | −26.06 ± 0.22 | 4.12 |
| A. arborella | 4 | 6.15 ± 0.62 | −24.48 ± 0.52 | 2.83 |
| C. gobio | 4 | 7.75 ± 0.18 | −28.68 ± 0.46 | 3.29 |
| E. cisalpinus | 2 | 8.46, 8.75 | −22.00, −22.30 | 5.06 |
| L. lota | 1 | 9.67 | −29.98 | 5.15 |
| P. fluviatilis | 7 | 6.40 ± 1.26 | −21.64 ± 2.46 | 4.95 |
| S. hesperidicus | 5 | 6.55 ± 0.40 | −23.07 ± 0.86 | 4.61 |
| S. squalus | 1 | 5.85 | −22.99 | 4.11 |
| Species | ss | CR * | NR | CD | SEAc | SEAB | MNND * | SDNND ** |
|---|---|---|---|---|---|---|---|---|
| Fish NNS | ||||||||
| Coregonus sp. | 4 | 2.22 | 2.28 | 1.02 | 2.70 | 2.52 | 0.94 | 0.86 |
| G. cernua | 9 | 7.57 | 2.10 | 1.98 | 5.22 | 5.11 | 0.90 | 0.98 |
| L. gibbosus | 5 | 7.88 | 1.47 | 2.32 | 4.86 | 5.00 | 1.75 | 2.00 |
| M. salmoides | 5 | 6.08 | 1.36 | 1.85 | 3.99 | 4.32 | 1.48 | 1.88 |
| R. rutilus | 8 | 6.11 | 2.37 | 1.88 | 5.68 | 5.56 | 0.96 | 0.74 |
| S. umbla | 4 | 4.13 | 1.47 | 1.45 | 5.42 | 5.79 | 1.46 | 0.70 |
| Mean values ± SD | 5.67 ± 2.15 | 1.84 ± 0.46 | 1.75 ± 0.45 | 4.64 ± 1.12 | 4.72 ± 1.19 | 1.25 ± 0.36 | 1.19 ± 0.59 | |
| Fish NS | ||||||||
| A. agone | 4 | 1.12 | 0.52 | 0.42 | 0.46 | 1.13 | 0.53 | 0.10 |
| A. arborella | 4 | 1.28 | 1.32 | 0.68 | 1.47 | 2.67 | 0.76 | 0.28 |
| C. gobio | 4 | 0.94 | 0.43 | 0.41 | 0.16 | 0.07 | 0.26 | 0.12 |
| P. fluviatilis | 7 | 6.23 | 3.50 | 2.40 | 11.69 | 12.34 | 1.20 | 0.63 |
| S. hesperidicus | 5 | 1.90 | 0.96 | 0.81 | 1.39 | 1.19 | 0.62 | 0.18 |
| Mean values | 2.29 ± 2.23 | 1.35 ± 1.26 | 0.94 ± 0.83 | 3.03 ± 4.87 | 3.48 ± 5.04 | 0.67 ± 0.35 | 0.26 ± 0.22 | |
| A.arb | A.ago | C.gob | C.sp | G.cer | L.gib | M.sal | P.flu | R.rut | S.umb | S.hes | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A.arb | 0.025 CI | NA | 0.00 | 0.00 | 0.00 | 8.12 | 0.00 | 0.00 | 29.22 | 5.55 | 0.00 | 2.34 |
| 0.975 CI | NA | 22.56 | 0.00 | 82.45 | 99.88 | 86.53 | 12.12 | 100.00 | 100.00 | 0.00 | 90.78 | |
| Mean | NA | 2.95 | 0.04 | 16.94 | 56.69 | 32.31 | 1.82 | 89.59 | 75.69 | 0.26 | 41.16 | |
| A.ago | 0.025 CI | 0.00 | NA | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 0.975 CI | 84.87 | NA | 0.00 | 15.89 | 6.0 | 0.00 | 0.00 | 100.00 | 100.00 | 0.00 | 5.38 | |
| Mean | 11.08 | NA | 0.00 | 1.11 | 1.04 | 0.22 | 0.01 | 35.02 | 22.51 | 0.00 | 1.05 | |
| C.gob | 0.025 CI | 0.00 | 0.00 | NA | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 0.975 CI | 0.00 | 0.00 | NA | 19.11 | 100.00 | 100.00 | 0.00 | 100.00 | 7.77 | 94.33 | 0.00 | |
| Mean | 0.05 | 0.00 | NA | 2.52 | 24.48 | 11.23 | 0.80 | 11.07 | 1.26 | 5.87 | 0.03 | |
| C.sp | 0.025 CI | 0.00 | 0.00 | 0.00 | NA | 3.64 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 0.975 CI | 22.98 | 22.34 | 1.13 | NA | 86.28 | 53.65 | 64.87 | 96.34 | 46.47 | 73.81 | 13.55 | |
| Mean | 4.03 | 0.10 | 0.08 | NA | 40.02 | 11.08 | 9.82 | 28.84 | 6.38 | 21.62 | 1.72 | |
| G.cer | 0.025 CI | 1.94 | 1.00 | 0.00 | 1.34 | NA | 11.89 | 4.54 | 56.44 | 21.32 | 0.00 | 7.52 |
| 0.975 CI | 39.76 | 39.56 | 1.16 | 27.43 | NA | 84.34 | 66.99 | 100.00 | 96.77 | 6.66 | 58.99 | |
| Mean | 11.15 | 0.05 | 0.25 | 10.39 | NA | 41.60 | 25.45 | 85.02 | 58.91 | 0.74 | 24.61 | |
| L.gib | 0.025 CI | 0.00 | 0.00 | 0.00 | 0.00 | 19.46 | NA | 0.00 | 48.55 | 37.44 | 0.00 | 5.03 |
| 0.975 CI | 25.8 | 25.56 | 1.34 | 17.34 | 96.02 | NA | 33.23 | 100.00 | 99.99 | 2.55 | 50.05 | |
| Mean | 6.39 | 0.01 | 0.16 | 4.07 | 58.87 | NA | 2.93 | 83.26 | 74.25 | 0.17 | 22.12 | |
| M.sal | 0.025 CI | 0.00 | 0.00 | 0.00 | 0.00 | 7.57 | 0.00 | NA | 12.39 | 0.00 | 0.00 | 0.00 |
| 0.975 CI | 11.28 | 0.00 | 0.00 | 12.33 | 97.33 | 25.71 | NA | 100.00 | 90.06 | 12.25 | 15.47 | |
| Mean | 0.83 | 0.00 | 0.01 | 2.00 | 57.29 | 2.66 | NA | 67.35 | 28.61 | 1.29 | 1.42 | |
| P.flu | 0.025 CI | 1.00 | 0.00 | 0.00 | 0.00 | 21.56 | 17.45 | 2.45 | NA | 31.27 | 0.00 | 6.65 |
| 0.975 CI | 27.43 | 4.65 | 0.00 | 16.56 | 85.33 | 68.44 | 40.89 | NA | 94.84 | 4.56 | 46.43 | |
| Mean | 8.05 | 0.58 | 0.06 | 3.69 | 52.23 | 38.95 | 14.70 | NA | 64.60 | 0.49 | 18.69 | |
| R.rut | 0.025 CI | 1.00 | 0.00 | 0.00 | 0.00 | 22.43 | 26.78 | 0.00 | 68.33 | NA | 0.00 | 6.77 |
| 0.975 CI | 31.85 | 3.76 | 0.00 | 14.88 | 95.44 | 86.54 | 36.55 | 100.00 | NA | 0.00 | 54.72 | |
| Mean | 8.72 | 0.66 | 0.02 | 2.25 | 59.54 | 54.76 | 9.58 | 91.85 | NA | 0.15 | 24.67 | |
| S.umb | 0.025 CI | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | NA | 0.00 |
| 0.975 CI | 0.00 | 0.00 | 1.44 | 51.22 | 10.55 | 3.79 | 36.72 | 24.44 | 0.00 | NA | 0.00 | |
| Mean | 0.02 | 0.00 | 0.09 | 15.19 | 1.15 | 0.25 | 2.24 | 2.29 | 0.06 | NA | 0.01 | |
| S.hes | 0.025 CI | 2.99 | 0.00 | 0.00 | 0.00 | 47.43 | 25.11 | 0.00 | 88.91 | 55.38 | 0.00 | NA |
| 0.975 CI | 89.64 | 1.85 | 0.00 | 34.33 | 100.00 | 100.00 | 29.45 | 100.00 | 100.00 | 0.00 | NA | |
| Mean | 26.47 | 0.18 | 0.01 | 3.97 | 88.08 | 73.53 | 3.34 | 98.61 | 93.84 | 0.18 | NA | |
| Grand-mean overlaps | 7.68 | 0.18 | 0.08 | 5.02 | 42.52 | 26.03 | 7.65 | 55.92 | 38.94 | 3.39 | 10.48 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cicala, D.; Polgar, G.; Mor, J.R.; Piscia, R.; Brignone, S.; Zaupa, S.; Volta, P. Trophic Niches, Trophic Positions, and Niche Overlaps between Non-Native and Native Fish Species in a Subalpine Lake. Water 2020, 12, 3475. https://doi.org/10.3390/w12123475
Cicala D, Polgar G, Mor JR, Piscia R, Brignone S, Zaupa S, Volta P. Trophic Niches, Trophic Positions, and Niche Overlaps between Non-Native and Native Fish Species in a Subalpine Lake. Water. 2020; 12(12):3475. https://doi.org/10.3390/w12123475
Chicago/Turabian StyleCicala, Davide, Gianluca Polgar, Jordi René Mor, Roberta Piscia, Stefano Brignone, Silvia Zaupa, and Pietro Volta. 2020. "Trophic Niches, Trophic Positions, and Niche Overlaps between Non-Native and Native Fish Species in a Subalpine Lake" Water 12, no. 12: 3475. https://doi.org/10.3390/w12123475








