Screening and Immobilizing the Denitrifying Microbes in Sediment for Bioremediation
Abstract
1. Introduction
2. Material and Methods
2.1. Detection and Analysis of Sediment
2.2. Immobilization of Active Breads
2.3. Simulation Experiment of Sediment Remediation by Activated Beads
3. Results and Discussion
3.1. Diversity Analysis of Microflora in Sediment
3.2. Screening and Culture of Dominant Denitrifying Bacteria
3.3. Preparation of Microbial Activated Beads
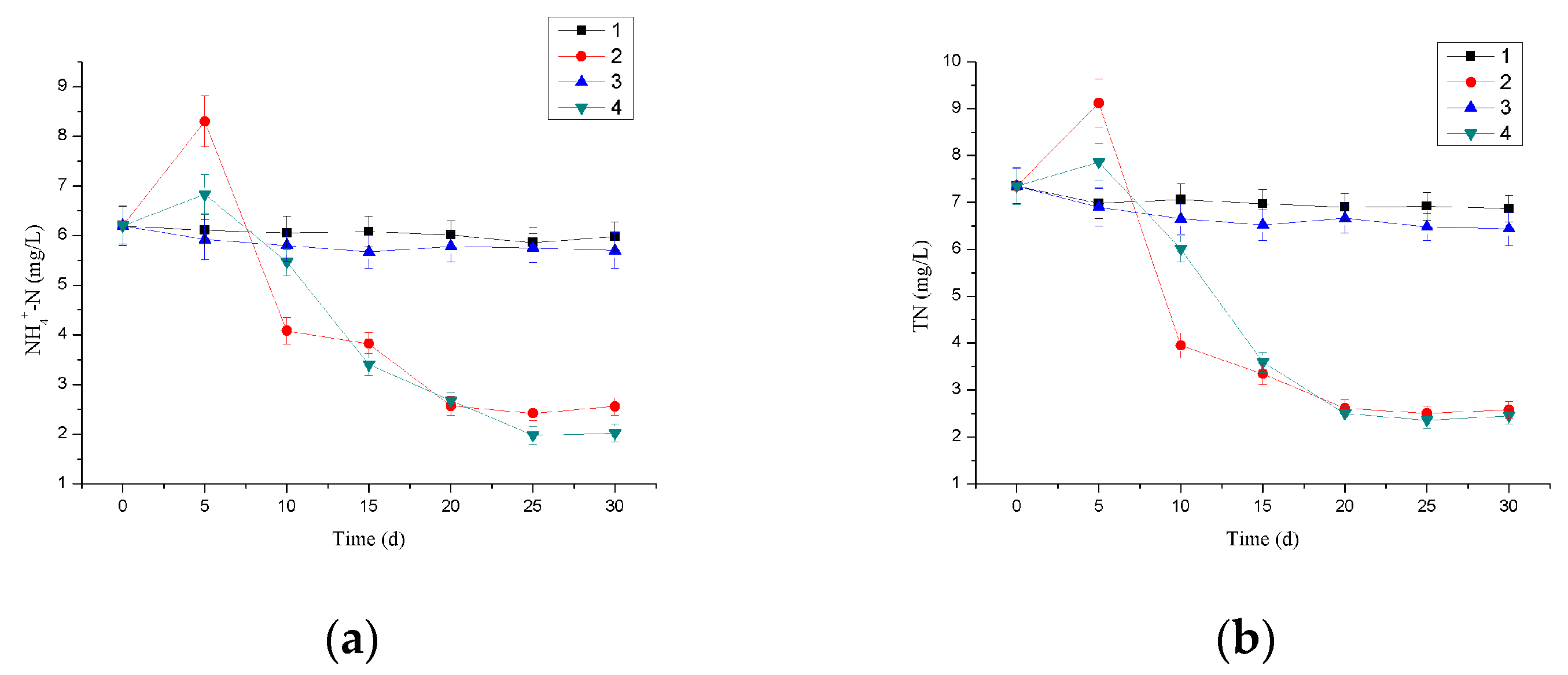
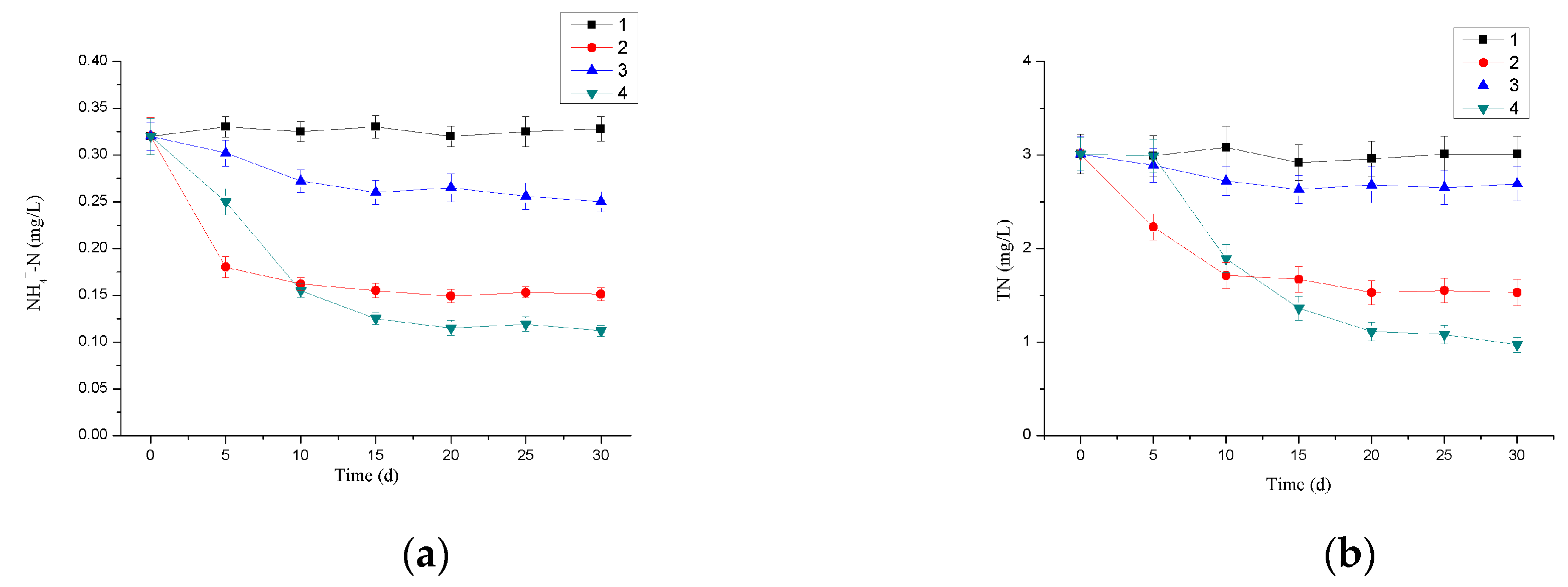
3.4. Nitrogen Removal Efficiency of Immobilized Microbial Beads
3.5. Practical Applications and Future Research Aspects of Current Study
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kanzari, F.; Syakti, A.D.; Asia, L.; Malleret, L.; Piram, A.; Mille, G.; Doumenq, P. Distributions and sources of persistent organic pollutants (aliphatic hydrocarbons, PAHs, PCBs and pesticides) in surface sediments of an industrialized urban river (Huveaune), France. Sci. Total Environ. 2014, 478, 141–151. [Google Scholar] [CrossRef]
- Ahn, Y.H. Sustainable nitrogen elimination biotechnologies: A review. Process. Biochem. 2006, 41, 1709–1721. [Google Scholar] [CrossRef]
- Yang, C.; Wang, S.; Jin, X.; Wu, F. Nitrogen and phosphorus mineralization in sediments of Taihu Lake after the removal of light fraction organic matter. Environ. Earth Sci. 2010, 59, 1437–1446. [Google Scholar] [CrossRef]
- Huo, S.; Zhang, J.; Xi, B.; Zan, F.; Jing, S.; Hong, Y. Distribution of nitrogen forms in surface sediments of lakes from different regions, China. Environ. Earth Sci. 2014, 71, 2167–2175. [Google Scholar] [CrossRef]
- Hou, L.; Rong, W.; Yin, G.; Min, L.; Zheng, Y. Nitrogen fixation in the intertidal sediments of the Yangtze Estuary: Occurrence and environmental implications. J. Geophys. Res. Biogeosci. 2018, 123, 936–944. [Google Scholar] [CrossRef]
- Porubsky, W.P.; Weston, N.B.; Joye, S.B. Benthic metabolism and the fate of dissolved inorganic nitrogen in intertidal sediments. Estuar. Coast. Shelf Sci. 2009, 83, 392–402. [Google Scholar] [CrossRef]
- Xia, X.; Liu, T.; Yang, Z.; Michalski, G.; Liu, S.; Jia, Z.; Zhang, S. Enhanced nitrogen loss from rivers through coupled nitrification-denitrification caused by suspended sediment. Sci. Total Environ. 2016, 579, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bae, H.S.; Reddy, K.R.; Ogram, A. Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res. 2016, 106, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Hou, L.; Zong, H.; Ding, P.; Min, L.; Zhang, S.; Cheng, X.; Zhou, J. Denitrification and Anaerobic Ammonium Oxidization Across the Sediment–Water Interface in the Hypereutrophic Ecosystem, Jinpu Bay, in the Northeastern Coast of China. Estuar. Coast. 2015, 38, 211–219. [Google Scholar] [CrossRef]
- Panepinto, D.; Genon, G. Modeling of Po River Water Quality in Torino. Water Resour. Manag. 2010, 24, 2937–2958. [Google Scholar] [CrossRef]
- Mallavarapu, M.; Balasubramanian, R.; Kadiyala, V.; Nambrattil, S.; Ravi, N. Bioremediation approaches for organic pollutants: A critical perspective. Environ. Int. 2011, 37, 1362–1375. [Google Scholar]
- Perelo, L.W. Review: In situ and bioremediation of organic pollutants in aquatic sediments. J. Hazard. Mater. 2010, 177, 81–89. [Google Scholar] [CrossRef]
- Chisti, Y. Bioremediation—Keeping the Earth clean. Biotechnol. Adv. 2005, 23, 371–372. [Google Scholar] [CrossRef]
- Jeong, D.; Cho, K.; Lee, C.H.; Lee, S.; Bae, H. Integration of forward osmosis process and a continuous airlift nitrifying bioreactor containing PVA/alginate-immobilized cells. Chem. Eng. J. 2016, 306, 1212–1222. [Google Scholar] [CrossRef]
- Hu, J.; Qi, Y. Microbial degradation of di-n-butyl phthalate by Micrococcus sp. immobilized with polyvinyl alcohol. Desalin. Water Treat. 2015, 56, 2457–2463. [Google Scholar] [CrossRef]
- Wen, Q.; Chen, Z.; Zhao, Y.; Zhang, H.; Feng, Y. Biodegradation of polyacrylamide by bacteria isolated from activated sludge and oil-contaminated soil. J. Hazard. Mater. 2010, 175, 955–959. [Google Scholar] [CrossRef]
- Bao, M.; Chen, Q.; Li, Y.; Jiang, G. Biodegradation of partially hydrolyzed polyacrylamide by bacteria isolated from production water after polymer flooding in an oil field. J. Hazard. Mater. 2010, 184, 105–110. [Google Scholar] [CrossRef]
- Magrí, A.; Vanotti, M.B.; Szögi, A.A. Anammox sludge immobilized in polyvinyl alcohol (PVA) cryogel carriers. Bioresour. Technol. 2012, 114, 231–240. [Google Scholar] [CrossRef]
- Ali, M.; Oshiki, M.; Rathnayake, L.; Ishii, S.; Satoh, H.; Okabe, S. Rapid and successful start-up of anammox process by immobilizing the minimal quantity of biomass in PVA-SA gel beads. Water Res. 2015, 79, 147–157. [Google Scholar] [CrossRef]
- Xu, X.; Lv, C.; You, X.; Wang, B.; Ji, F.; Hu, B. Nitrogen removal and microbial diversity of activated sludge entrapped in modified poly(vinyl alcohol)–sodium alginate gel. Int. Biodeter. Biodegr. 2017, 125, 243–250. [Google Scholar] [CrossRef]
- Kai, L.; Jian, L.; Qiao, H.; Lin, A.; Wang, G. Immobilization of Chlorella sorokiniana GXNN 01 in alginate for removal of N and P from synthetic wastewater. Bioresour. Technol. 2012, 114, 26–32. [Google Scholar]
- Liu, P.W.G.; Yang, D.S.; Tang, J.Y.; Hsu, H.W.; Chen, C.H.; Lin, I.K. Development of a cell immobilization technique with polyvinyl alcohol for diesel remediation in seawater. Int. Biodeter. Biodegr. 2016, 113, 397–407. [Google Scholar] [CrossRef]
- Bae, H.; Choi, M.; Lee, C.; Chung, Y.C.; Yoo, Y.J.; Lee, S. Enrichment of ANAMMOX bacteria from conventional activated sludge entrapped in poly(vinyl alcohol)/sodium alginate gel. Chem. Eng. J. 2015, 281, 531–540. [Google Scholar] [CrossRef]
- Kazuichi, I.; Yuya, K.; Tomoko, Y.; Toshifumi, O.; Satoshi, T. Complete autotrophic denitrification in a single reactor using nitritation and anammox gel carriers. Bioresour. Technol. 2013, 147, 96–101. [Google Scholar]
- Jia, Y.; Mike, J.; Jinlong, R.; Yongyou, H. Comparison of the effects of different salts on aerobic ammonia oxidizers for treating ammonium-rich organic wastewater by free and sodium alginate immobilized biomass system. Chemosphere 2010, 81, 669–673. [Google Scholar]
- Gangli, Z.; Yongyou, H.; Qirui, W. Nitrogen removal performance of anaerobic ammonia oxidation co-culture immobilized in different gel carriers. Water Sci. Technol. 2009, 59, 2379. [Google Scholar]
- Chen, G.H.; Li, J.; Tabassum, S.; Zhang, Z.J. Anaerobic ammonium oxidation (ANAMMOX) sludge immobilized by waterborne polyurethane and its nitrogen removal performance-a lab scale study. RSC Adv. 2015, 5, 25372–25381. [Google Scholar] [CrossRef]
- Ansari, S.A.; Husain, Q. Potential applications of enzymes immobilized on/in nano materials: A review. Biotechnol. Adv. 2012, 30, 512–523. [Google Scholar] [CrossRef]
- Chen, M.; Wang, W.; Ye, F.; Zhu, X.; Zhou, H.; Tan, Z.; Li, X. Impact resistance of different factors on ammonia removal by heterotrophic nitrification–aerobic denitrification bacterium Aeromonas sp. HN-02. Bioresour. Technol. 2014, 167, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, X.; Li, H.; Tu, W. Immobilization of Acidithiobacillus ferrooxidans with complex of PVA and sodium alginate. Polym. Degrad. Stabil. 2006, 91, 2408–2414. [Google Scholar]
- Shi, S.; Qu, Y.; Fang, M.; Zhou, J. Bioremediation of coking wastewater containing carbazole, dibenzofuran, dibenzothiophene and naphthalene by a naphthalene-cultivated Arthrobacter sp. W1. Bioresour. Technol. 2014, 164, 28–33. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Xiao, J.; Zhu, C.; Sun, D.; Guo, P. Removal of ammonium-N from ammonium-rich sewage using an immobilized Bacillus subtilis AYC bioreactor system. J. Environ. Sci. China 2011, 23, 1279–1285. [Google Scholar] [CrossRef]
- Qiao, X.; Zhe, L.; Liu, Z.; Zeng, Y.; Zhang, Z. Immobilization of activated sludge in poly(ethylene glycol) by UV technology and its application in micro-polluted wastewater. Biochem. Eng. J. 2010, 50, 71–76. [Google Scholar]
- Bai, Y.; Shi, Q.; Wen, D.; Li, Z.; Jefferson, W.A.; Feng, C.; Tang, X. Bacterial communities in the sediments of Dianchi Lake, a partitioned eutrophic waterbody in China. PLoS ONE 2012, 7, e37796. [Google Scholar] [CrossRef] [PubMed]
- Spring, S.; Schulze, R. Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: Molecular and cultivation studies. FEMS Microbiol. Rev. 2000, 24, 573–590. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zeng, J.; Yan, W.; Wang, J.; Ma, T.; Wang, M.; Wu, Q.L. Diversity analysis of bacterial community compositions in sediments of urban lakes by terminal restriction fragment length polymorphism (T-RFLP). World J. Microbiol. Biotechnol. 2012, 28, 3159–3170. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Paul, V.J. Climate change: Links to global expansion of harmful cyanobacteria. Water Res. 2012, 46, 1349–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, S. Screening and Cultivation of Oligotrophic Aerobic Denitrifying Bacteria. In Water Pollution and Water Quality Control of Selected Chinese Reservoir Basins; Springer: Cham, Switzerland, 2016; pp. 451–473. [Google Scholar]
- Bai, X.; Ye, Z.; Li, Y.; Zhou, L.; Yang, L. Preparation of crosslinked macroporous PVA foam carrier for immobilization of microorganisms. Process. Biochem. 2010, 45, 60–66. [Google Scholar] [CrossRef]
- Xian, L.; Ren, Y.X.; Lei, Y.; Zhao, S.Q.; Xia, Z.H. Characteristics of Nitrogen Removal by a Heterotrophic Nitrification-Aerobic Denitrification Bacterium YL. Environ. Sci. 2015, 36, 1749–1756. [Google Scholar]
- Joo, H.S.; Hirai, M.; Shoda, M. Characteristics of Ammonium Removal by Heterotrophic Nitrification-Aerobic Denitrification by Alcaligenes faecalis No.4. J. Biosci. Bioeng. 2005, 100, 184–191. [Google Scholar] [CrossRef]
- Huang, C.; Wang, L.A.; Cui, Z.Q.; Liu, Y.Y. Application of Sodium Alginate-immobilized Microbes to Treatment of Methanol Wastewater. China Water Wastewater 2008, 24, 78–81. [Google Scholar]
- Bae, H.; Choi, M.; Chung, Y.C.; Lee, S.; Yoo, Y.J. Core-shell structured poly(vinyl alcohol)/sodium alginate bead for single-stage autotrophic nitrogen removal. Chem. Eng. J. 2017, 322, 408–416. [Google Scholar] [CrossRef]
- Xu, P.; Xiao, E.R.; Xu, D.; Zhou, Y.; He, F.; Liu, B.Y.; Zeng, L.; Wu, Z.B. Internal nitrogen removal from sediments by the hybrid system of microbial fuel cells and submerged aquatic plants. PLoS ONE 2017, 12, e172757. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Huang, T.; Yuan, B.; Liao, X. Remediation of Nitrogen-Contaminated Sediment Using Bioreactive, Thin-layer Capping with Biozeolite. J. Soil. Contam. 2016, 25, 89–100. [Google Scholar] [CrossRef]
- Hui, B.; Zhang, Y.; Ye, L. Preparation of PVA hydrogel beads and adsorption mechanism for advanced phosphate removal. Chem. Eng. J. 2014, 235, 207–214. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Xu, D.; Huang, J.; Sun, J. Application potential of aerobic denitrifiers coupled with a biostimulant for nitrogen removal from urban river sediment. Environ. Sci. Pollut. Res. Int. 2018, 25, 1–14. [Google Scholar] [CrossRef] [PubMed]

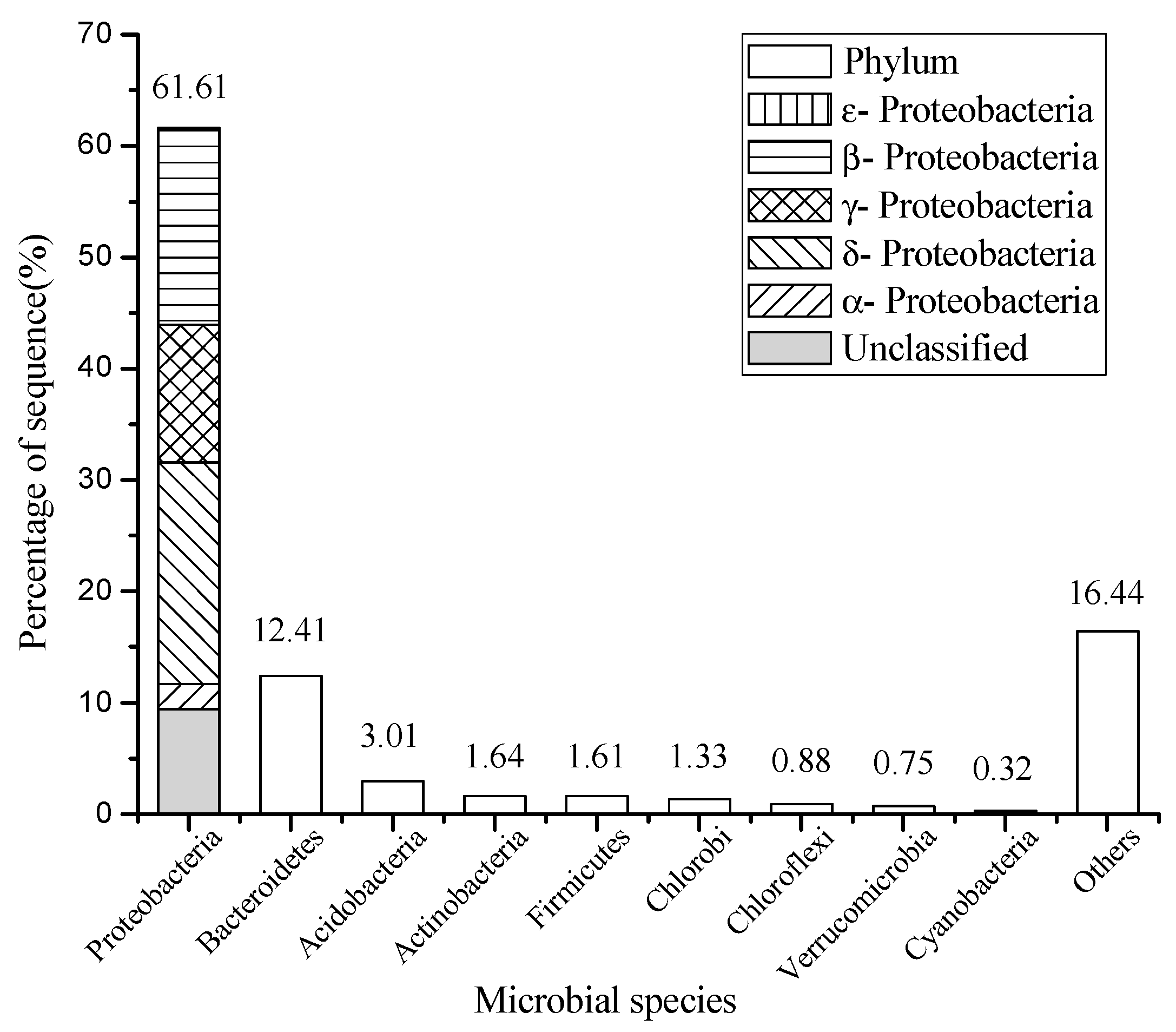

| Genera | Proportion % | Function |
|---|---|---|
| Steroidobacter | 2.64 | Anaerobic denitrification |
| Dechloromonas | 0.65 | Anaerobic denitrification |
| Methylotenera | 0.61 | Anaerobic denitrification |
| Thiobacillus | 0.60 | Denitrification |
| Pseudomonas | 0.38 | Aerobic denitrification, heterotrophic nitrification |
| Acinetobacter | 0.21 | Denitrification, heterotrophic nitrification |
| Paracoccus | 0.18 | Aerobic denitrification |
| Thauera | 0.08 | Heterotrophic nitrification, aerobic denitrification |
| Hyphomicrobium | 0.06 | Aerobic denitrification |
| Nitrospira | 0.05 | Nitrifying bacteria |
| Halomonas | 0.02 | Low temperature high salt denitrification |
| Bacillus | 0.01 | Aerobic denitrification, heterotrophic nitrification |
| Propionibacterium | 0.01 | Aerobic denitrification |
| Rhizobium | 0.01 | Aerobic denitrification |
| Corynebacterium | 0.01 | Heterotrophic nitrification |
| Rhodococcus | 0.01 | Aerobic denitrification, heterotrophic nitrification |
| Category | Proteobacteria% | Firmicutes% | Chlorobi% | Actinobacteria% | Bacteroidetes% | |
|---|---|---|---|---|---|---|
| Sample | ||||||
| Shedu sediment | 15.31 | 0.65 | 0.53 | 2.72 | 1.04 | |
| 5% inoculation | 89.12 | 8.98 | 4.84 | 3.55 | 2.55 | |
| 10% inoculation | 94.33 | 3.98 | 3.98 | 3.62 | 2.66 | |
| Category | Pseudomonas% | Alcaligenes% | Proteiniclasticum% | Achromobacter% | Methylobacillus% | |
|---|---|---|---|---|---|---|
| Sample | ||||||
| Shedu sediment | 0.38 | Nil | 0.01 | Nil | 0.08 | |
| 5 % inoculation | 49.87 | 19.98 | 8.77 | 3.89 | 0.22 | |
| 10% inoculation | 49.22 | 40.87 | 3.82 | 0.26 | 0.26 | |
| No | PVA Concentration (%) | Adhesion | Stability | Mass Transfer Rate A406 |
|---|---|---|---|---|
| a | 8 | General | Good | 0.182 |
| b | 9 | Easy | Good | 0.191 |
| c | 10 | Easy | Good | 0.205 |
| d | 11 | Easy | Good | 0.198 |
| No | Sodium Alginate Concentration (%) | Adhesion | Stability | Mass Transfer Rate A406 |
|---|---|---|---|---|
| a | 0.25 | General | General | 0.193 |
| b | 0.5 | General | Good | 0.182 |
| c | 1 | Easy | Good | 0.179 |
| d | 2 | Easy | Bad | 0.181 |
| e | 3 | Easy | Bad | 0.154 |
| No | Microbial Biomass (%) | Adhesion | Stability | Mass Transfer Rate A406 |
|---|---|---|---|---|
| a | 5 | Easy | General | 0.190 |
| b | 7.5 | General | Good | 0.182 |
| c | 10 | General | Good | 0.179 |
| d | 12.5 | Difficult | Good | 0.171 |
| e | 15 | Easy | General | 0.169 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Fu, D.; Shi, J. Screening and Immobilizing the Denitrifying Microbes in Sediment for Bioremediation. Water 2019, 11, 614. https://doi.org/10.3390/w11030614
Yan Y, Fu D, Shi J. Screening and Immobilizing the Denitrifying Microbes in Sediment for Bioremediation. Water. 2019; 11(3):614. https://doi.org/10.3390/w11030614
Chicago/Turabian StyleYan, Yixin, Dafang Fu, and Jiayuan Shi. 2019. "Screening and Immobilizing the Denitrifying Microbes in Sediment for Bioremediation" Water 11, no. 3: 614. https://doi.org/10.3390/w11030614
APA StyleYan, Y., Fu, D., & Shi, J. (2019). Screening and Immobilizing the Denitrifying Microbes in Sediment for Bioremediation. Water, 11(3), 614. https://doi.org/10.3390/w11030614





