Metagenomics Response of Anaerobic Ammonium Oxidation (anammox) Bacteria to Bio-Refractory Humic Substances in Wastewater
Abstract
1. Introduction
2. Materials and Methods
2.1. Reactor Operation
2.2. Sample Collection
2.3. Metagenomic Sequencing and Bioinformatics Analysis
2.4. Chemical Analyses
3. Results and Discussion
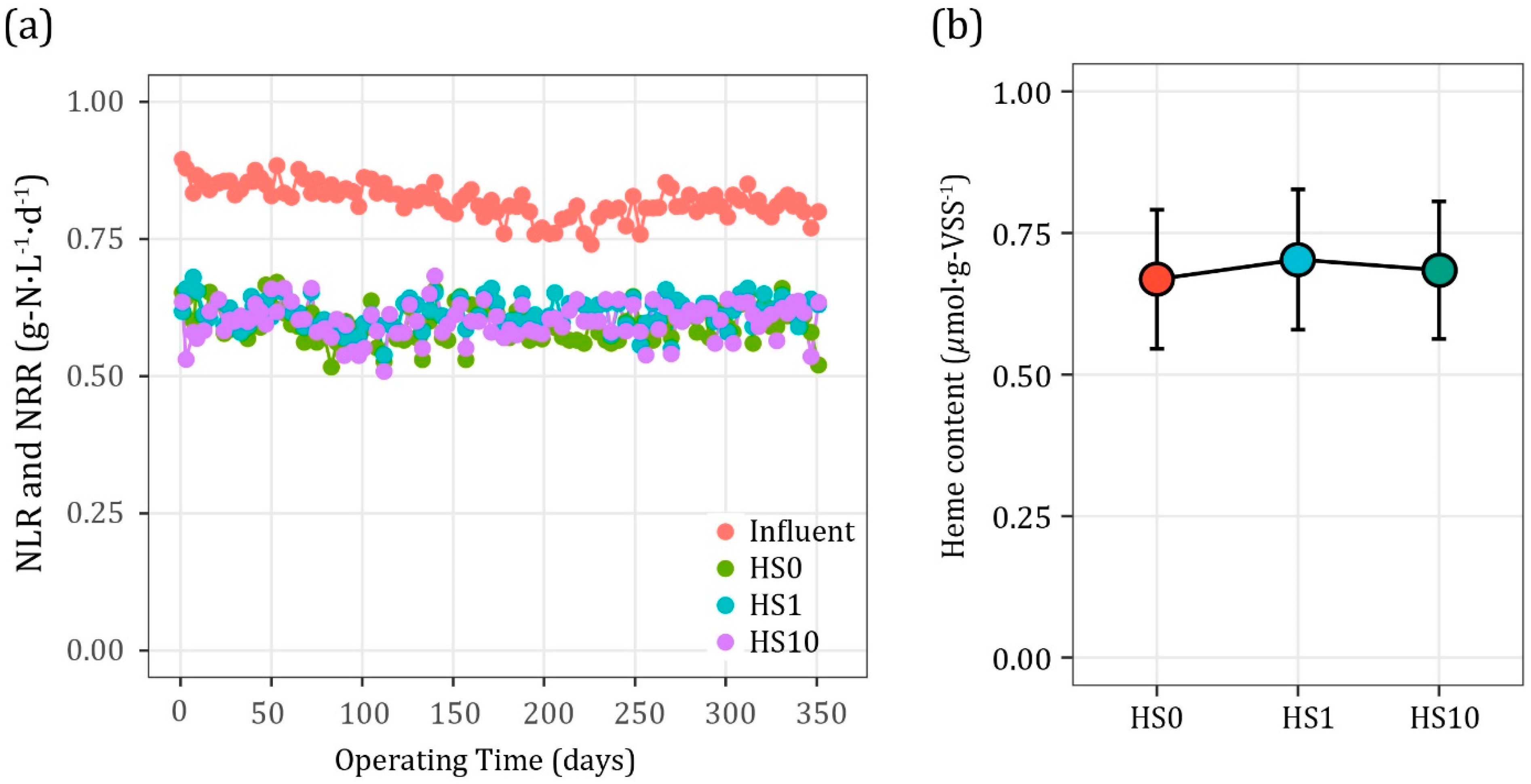
3.1. Reactor Operation
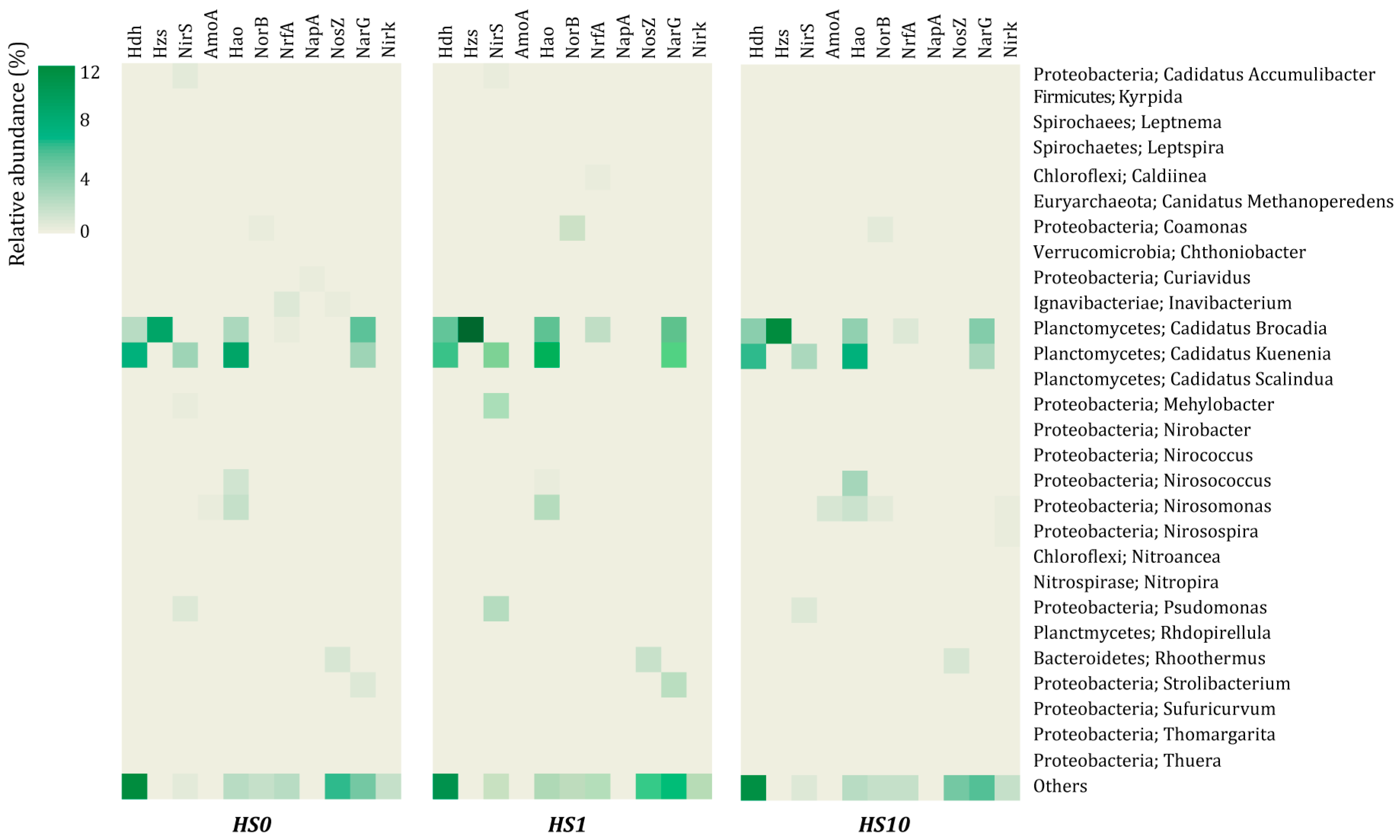
3.2. Microbial Community Structure
3.3. Enrichment of Biological Functions
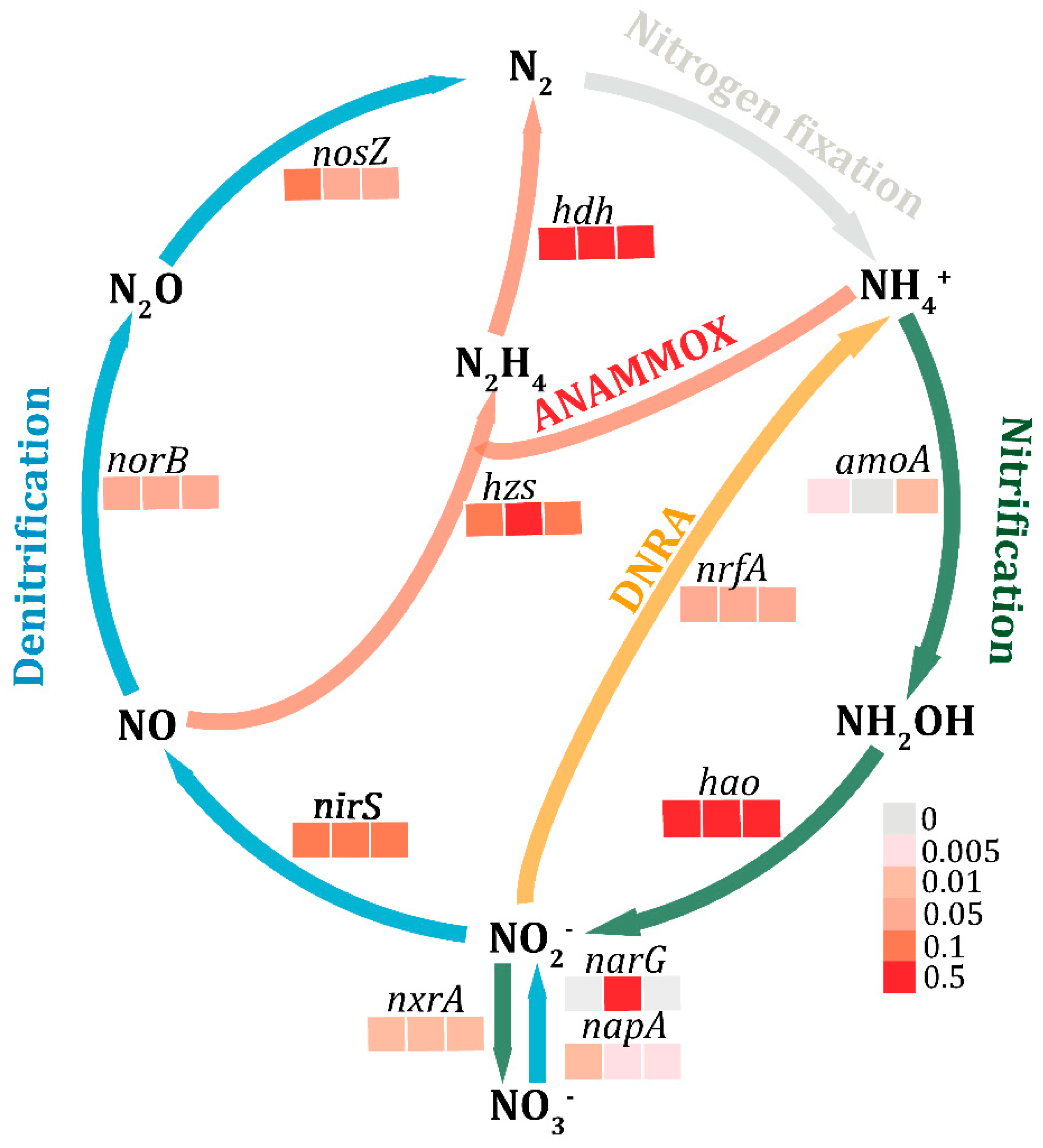
3.4. Functions in Nitrogen Metabolism
3.5. Quantification of Nitrogen-Related Genes
3.6. Implications of This Work
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hu, B.-L.; Shen, L.-D.; Xu, X.-Y.; Zheng, P. Anaerobic ammonium oxidation (anammox) in different natural ecosystems. Biochem. Soc. Trans. 2011, 39, 1811–1816. [Google Scholar] [CrossRef] [PubMed]
- Dalsgaard, T.; Thamdrup, B. Factors Controlling Anaerobic Ammonium Oxidation with Nitrite in Marine Sediments. Appl. Environ. Microbiol. 2002, 68, 3802–3808. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Lotti, T.; de Kreuk, M.; Kleerebezem, R.; van Loosdrecht, M.; Kruit, J.; Jetten, M.S.M.; Kartal, B. Nitrogen Removal by a Nitritation-Anammox Bioreactor at Low Temperature. Appl. Environ. Microbiol. 2013, 79, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Kampschreur, M.J.; van der Star, W.R.L.; Wielders, H.A.; Mulder, J.W.; Jetten, M.S.M.; van Loosdrecht, M.C.M. Dynamics of nitric oxide and nitrous oxide emission during full-scale reject water treatment. Water Res. 2008, 42, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Strous, M.; Heijnen, J.; Kuenen, J.; Jetten, M. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 1998, 50, 589–596. [Google Scholar] [CrossRef]
- Jin, R.C.; Yang, G.F.; Yu, J.-J.; Zheng, P. The inhibition of the Anammox process: A review. Chem. Eng. J. 2012, 197, 67–79. [Google Scholar] [CrossRef]
- Obarska-Pempkowiak, H.; Gajewska, M.; Wojciechowska, E.; Pempkowiak, J. Treatment Wetlands for Environmental Pollution Control; Rowiński, P., Ed.; GeoPlanet: Earth and Planetary Sciences; Springer International Publishing: New York, NY, USA, 2015; p. 169. [Google Scholar]
- Dapena-Mora, A.; Fernandez, I.; Campos, J.L.; Mosquera-Corral, A.; Mendez, R.; Jetten, M.S.M. Evaluation of activity and inhibition effects on Anammox process by batch tests based on the nitrogen gas production. Enzyme Microb. Technol. 2007, 40, 859–865. [Google Scholar] [CrossRef]
- Guven, D.; Dapena, A.; Kartal, B.; Schmid, M.C.; Maas, B.; van de Pas-Schoonen, K.; Sozen, S.; Mendez, R.; Op den Camp, H.J.; Jetten, M.S.; et al. Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Appl. Environ. Microbiol. 2005, 71, 1066–1071. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Vahala, R.K.; Wang, Y.; Smets, B.F. Microbes in biological processes for municipal landfill leachate treatment: Community, function and interaction. Int. Biodeterior. Biodegrad. 2016, 113, 88–96. [Google Scholar] [CrossRef]
- Kang, K.-H.; Shin, H.S.; Park, H. Characterization of humic substances present in landfill leachates with different landfill ages and its implications. Water Res. 2002, 36, 4023–4032. [Google Scholar] [CrossRef]
- Artiola-Fortuny, J.; Fuller, W.H. Humic Substances in Landfill Leachates: I. Humic Acid Extraction and Identification1. J. Environ. Qual. 1982, 11, 663–669. [Google Scholar] [CrossRef]
- Dang, H.; Chen, R.; Wang, L.; Guo, L.; Chen, P.; Tang, Z.; Tian, F.; Li, S.; Klotz, M.G. Environmental Factors Shape Sediment Anammox Bacterial Communities in Hypernutrified Jiaozhou Bay, China. Appl. Environ. Microbiol. 2010, 76, 7036–7047. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.M.; Thamdrup, B.; Dalsgaard, T. Effects of specific inhibitors on anammox and denitrification in marine sediments. Appl. Environ. Microbiol. 2007, 73, 3151–3158. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.T.; McKnight, D.M.; Blunt-Harris, E.L.; Kolesar, S.E.; Lovley, D.R. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ. Sci. Technol. 1998, 32, 2984–2989. [Google Scholar] [CrossRef]
- Visser, S.A. Effect of humic acids on numbers and activities of micro-organisms within physiological groups. Org. Geochem. 1985, 8, 81–85. [Google Scholar] [CrossRef]
- Pellissier, F. Allelopathic effect of phenolic acids from humic solutions on two spruce mycorrhizal fungi: Cenococcum graniforme andLaccaria laccata. J. Chem. Ecol. 1993, 19, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Awata, T.; Zhang, D.; Zhang, C.; Li, Z.; Katayama, A. Enhanced denitrification of Pseudomonas stutzeri by a bioelectrochemical system assisted with solid-phase humin. J. Biosci. Bioeng. 2016, 122, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Aulenta, F.; Di Maio, V.; Ferri, T.; Majone, M. The humic acid analogue antraquinone-2, 6-disulfonate (AQDS) serves as an electron shuttle in the electricity-driven microbial dechlorination of trichloroethene to cis-dichloroethene. Bioresource Technol. 2010, 101, 9728–9733. [Google Scholar] [CrossRef]
- Ji, V.T.; Sun, Y.; Coates, J.D. Microbial Interactions with Humic Substances1. Adv. Appl. Microbiol. 2006, 60, 55–96. [Google Scholar]
- Lovley, D.R.; Coates, J.D.; Bluntharris, E.L.; Phillips, E.J.P.; Woodward, J.C. Humic substances as electron acceptors for microbial respiration. Nature 1996, 382, 445–448. [Google Scholar] [CrossRef]
- Trellu, C.; Pechaud, Y.; Oturan, N.; Mousset, E.; Huguenot, D.; van Hullebusch, E.D.; Esposito, G.; Oturan, M.A. Comparative study on the removal of humic acids from drinking water by anodic oxidation and electro-Fenton processes: Mineralization efficiency and modelling. Appl. Catal. B Environ. 2016, 194, 32–41. [Google Scholar] [CrossRef]
- Strous, M.; Kuenen, J.G.; Jetten, M.S.M. Key Physiology of Anaerobic Ammonium Oxidation. Appl. Environ. Microbiol. 1999, 65, 3248–3250. [Google Scholar] [PubMed]
- Speth, D.R.; Hu, B.; Bosch, N.; Keltjens, J.T.; Stunnenberg, H.G.; Jetten, M. Comparative genomics of two independently enriched “Candidatus Kuenenia stuttgartiensis” anammox bacteria. Front. Microbiol. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. R. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Y.; Speth, D.R.; Francoijs, K.J.; Quan, Z.X.; Jetten, M.S.M. Metagenome analysis of a complex community reveals the metabolic blueprint of anannmox bacterium “Candidatus Jettenia asiatica”. Front. Microbiol. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oshiki, M.; Shinyako-Hata, K.; Satoh, H.; Okabe, S. Draft Genome Sequence of an Anaerobic Ammonium-Oxidizing Bacterium, “Candidatus Brocadia sinica”. Genome Announc. 2015, 3, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Speth, D.R.; Russ, L.; Kartal, B.; Hj, O.D.C.; Dutilh, B.E.; Jetten, M.S. Draft Genome Sequence of Anammox Bacterium “Candidatus Scalindua brodae,” Obtained Using Differential Coverage Binning of Sequencing Data from Two Reactor Enrichments. Genome Announc. 2015, 3, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Maalcke, W.J.; de Almeida, N.M.; Cirpus, I.; Gloerich, J.; Geerts, W.; den Camp, H.J.O.; Harhangi, H.R.; Janssen-Megens, E.M.; Francoijs, K.-J.; et al. Molecular mechanism of anaerobic ammonium oxidation. Nature 2011, 479, 127–130. [Google Scholar] [CrossRef]
- Speth, D.R.; Guerrero-Cruz, S.; Dutilh, B.E.; Jetten, M.S.M. Genome-based microbial ecology of anammox granules in a full-scale wastewater treatment system. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Ni, S.Q.; Lee, P.H.; Fessehaie, A.; Gao, B.Y.; Sung, S. Enrichment and biofilm formation of Anammox bacteria in a non-woven membrane reactor. Bioresource Technol. 2010, 101, 1792–1799. [Google Scholar] [CrossRef]
- Van de Graaf, A.A.; de Bruijn, P.; Robertson, L.A.; Jetten, M.S.M.; Kuenen, J.G. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 1996, 142, 2187–2196. [Google Scholar] [CrossRef]
- Bis, M.; Montusiewics, A.; Ozonek, J.; Pasieczna-Patkowska, S. Application of hydrodynamic cavitation to improve the biodegradability of mature landfill leachate. Ultrason. Sonochem. 2015, 26, 378–387. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Lukashin, A.V.; Borodovsky, M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998, 26, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Lüke, C.; Speth, D.R.; Kox, M.A.R.; Villanueva, L.; Jetten, M.S.M. Metageomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. Peer J. 2016, 4, 1924. [Google Scholar] [CrossRef]
- Chen, L.X.; Hu, M.; Huang, L.N.; Hua, Z.S.; Kuang, J.L.; Li, S.J.; Shu, W.S. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2015, 9, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater, 19th ed.; American Public Health Association: Washington, DC, USA, 1995. [Google Scholar]
- Berry, E.A.; Trumpower, B.L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 1987, 161, 1–15. [Google Scholar] [CrossRef]
- Tang, C.J.; Zheng, P.; Wang, C.H.; Mahmood, Q.; Zhang, J.Q.; Chen, X.G.; Zhang, L.; Chen, J.W. Performance of high-loaded ANAMMOX UASB reactors containing granular sludge. Water Res. 2011, 45, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-J.; Zhu, X.-L.; Zhang, Q.-Q.; Cheng, Y.-F.; Xu, L.-Z.-J.; Zhu, Y.-H.; Ji, Z.-Q.; Jin, R.-C. Roles of MnO2 on performance, sludge characteristics and microbial community in anammox system. Sci. Total. Environ. 2018, 633, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Li, M.; Chen, Y. Inherent humic substance promotes microbial denitrification of landfill leachate via shifting bacterial community, improving enzyme activity and up-regulating gene. Sci. Rep. 2017, 7, 12215. [Google Scholar] [CrossRef]
- Chatterjee, P.; Ghangrekar, M.M.; Rao, S. Development of anammox process for removal of nitrogen from wastewater in a novel self-sustainable biofilm reactor. Bioresource Technol. 2016, 218, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.K.; Ashbolt, N.J. Adaptation of anaerobic ammonium-oxidising consortium to synthetic coke-ovens wastewater. Appl. Microbiol. Biotechnol. 2002, 59, 344–352. [Google Scholar]
- Guo, J.H.; Peng, Y.Z.; Fan, L.; Zhang, L.; Ni, B.J.; Kartal, B.; Feng, X.; Jetten, M.S.M.; Yuan, Z.G. Metagenomic analysis of anammox communities in three different microbial aggregates. Environ. Microbiol. 2015, 18, 2979–2993. [Google Scholar] [CrossRef]
- Kuenen, J.G. Anammox bacteria: From discovery to application. Nat. Rev. Microbiol. 2008, 6, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gu, J.-D. The diversity and distribution of anammox bacteria in the marine aquaculture zones. Appl. Microbiol. Biotechnol. 2016, 100, 8943–8953. [Google Scholar] [CrossRef]
- Gori, F.; Tringe, S.G.; Kartal, B.; Marchiori, E.; Jetten, M.S.M. The metagenomic basis of anammox metabolism in Candidatus ‘Brocadia fulgida’. Biochem. Soc. Trans. 2011, 39, 1799–1804. [Google Scholar] [CrossRef] [PubMed]
- Oshiki, M.; Shimokawa, M.; Fujii, N.; Satohl, H.; Okabe, S. Physiological characteristics of the anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia sinica’. Microbiology 2011, 157, 1706–1713. [Google Scholar] [CrossRef]
- Kartal, B.; Van Niftrik, L.; Rattray, J.; Van De Vossenberg, J.L.; Schmid, M.C.; Sinninghe Damsté, J.; Jetten, M.S.; Strous, M. Candidatus ‘Brocadia fulgida’: An autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 2008, 63, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.-L.; Zheng, P.; Tang, C.-J.; Chen, J.-W.; van der Biezen, E.; Zhang, L.; Ni, B.-J.; Jetten, M.S.; Yan, J.; Yu, H.-Q. Identification and quantification of anammox bacteria in eight nitrogen removal reactors. Water Res. 2010, 44, 5014–5020. [Google Scholar] [CrossRef] [PubMed]
- Theng, K.G.; Orchard, V.A.; Huang, P.M.; Berthelin, J.; Bollag, J.M.; Mcgill, W.B.; Page, A.L. Interactions of clays with microorganisms and bacterial survival in soil: A physicochemical perspective. In Environmental Impact of Soil Componenent Interactions, Vol II. Metals, Other Inorganics, and Microbial Activities; Huang, P.M., Berthelin, J., Bollag, J.M., McGill, W.B., Page, A.L., Eds.; CRC Lewis: Boca Raton, FL, USA, 1995; p. 123. [Google Scholar]
- Anielak, A.M.; Polus, M.; Lominska, D.; Zaba, T. Biodegradation of fulvic acids with using activated sludge enriched with archea. Przem. Chem. 2016, 95, 110–113. [Google Scholar]
- Xue, Y.; Yu, Z.; Chen, H.; Yang, J.R.; Liu, M.; Liu, L.; Huang, B.; Yang, J. Cyanobacterial bloom significantly boosts hypolimnelic anammox bacterial abundance in a subtropical stratified reservoir. FEMS Microbiol. Ecol. 2017, 93, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Tsushima, I.; Ogasawara, Y.; Shimokawa, M.; Ozaki, N.; Satoh, H.; Okabe, S. In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl. Environ. Microbiol. 2007, 73, 4931–4939. [Google Scholar] [CrossRef] [PubMed]
- Byrne-Bailey, K.G.; Weber, K.A.; Chair, A.H.; Bose, S.; Knox, T.; Spanbauer, T.L.; Chertkov, O.; Coates, J.D. Completed Genome Sequence of the Anaerobic Iron-Oxidizing Bacterium Acidovorax ebreus Strain TPSY. J. Bacteriol. 2010, 192, 1475–1476. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, J.J.; Liang, Y.L.; Liu, X.D.; Yin, H.Q. Metagenome-scale analysis yields insights into the structure and function of microbial communities in a copper bioleaching heap. BMC Genet. 2016, 17, 1–12. [Google Scholar] [CrossRef]
- Wu, L.; Li, Z.; Zhao, C.; Liang, D.; Peng, Y. A novel partial-denitrification strategy for post-anammox to effectively remove nitrogen from landfill leachate. Sci. Total. Environ. 2018, 633, 745–751. [Google Scholar] [CrossRef]
- Wei, H.; Wang, L.; Hassan, M.; Xie, B. Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresource Technol. 2018, 256, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, S.; Xu, X.; Guo, Y.; Yang, F.; Wang, D. Role of cyclic diguanylate in affecting microbial community shifts at different pH during the operation of simultaneous partial nitrification, anammox and denitrification process. Sci. Total Environ. 2018, 637–638, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.T.; He, Y.L.; Yue, H.; Gao, J.L.; Wang, Q.Y.; Yang, S.C. Enhanced long-term nitrogen removal by organotrophic anammox bacteria under different C/N ratio constraints: Quantitative molecular mechanism and microbial community dynamics. RSC Adv. 2016, 6, 87593–87606. [Google Scholar] [CrossRef]
- Tikhonov, V.V.; Yakushev, A.V.; Zavgorodnyaya, Y.A.; Byzov, B.A.; Demin, V.V. Effects of humic acids on the growth of bacteria. Eurasian Soil. Sci. 2010, 43, 305–313. [Google Scholar] [CrossRef]
- Pan, F.; Chapman, S.J.; Li, Y.; Yao, H. Straw amendment to paddy soil stimulates denitrification but biochar amendment promotes anaerobic ammonia oxidation. J. Soils Sediment. 2017, 17, 2428–2437. [Google Scholar] [CrossRef]
- Kartal, B.; Tan, N.C.G.; Van de Biezen, E.; Kampschreur, M.J.; Van Loosdrecht, M.C.M.; Jetten, M.S.M. Effect of Nitric Oxide on Anammox Bacteria. Appl. Environ. Microbiol. 2010, 76, 6304–6306. [Google Scholar] [CrossRef]
- Harhangi, H.R.; Le Roy, M.; van Alen, T.; Hu, B.-L.; Groen, J.; Kartal, B.; Tringe, S.G.; Quan, Z.-X.; Jetten, M.S.M.; Op den Camp, H.J.M. Hydrazine Synthase, a Unique Phylomarker with Which To Study the Presence and Biodiversity of Anammox Bacteria. Appl. Environ. Microbiol. 2012, 78, 752–758. [Google Scholar] [CrossRef]
- Visser, S.A. Physiological action of humic substances on microbial cells. Soil. Biol. Biochem. 1985, 17, 457–462. [Google Scholar] [CrossRef]
- Smith, C.J.; Nedwell, D.B.; Dong, L.F.; Osborn, A.M. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 2007, 73, 3612–3622. [Google Scholar] [CrossRef]
- Kartal, B.; Kuypers, M.M.M.; Lavik, G.; Schalk, J.; den Camp, H.J.M.O.; Jetten, M.S.M.; Strous, M. Anammox bacteria disguised as denitrifiers: Nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 2007, 9, 635–642. [Google Scholar] [CrossRef]
- Arshad, A.; Martins, P.D.; Frank, J.; Jetten, M.S.M.; den Camp, H.J.M.O.; Welte, C.U. Mimicking microbial interactions under nitrate-reducing conditions in an anoxic bioreactor: Enrichment of novel Nitrospirae bacteria distantly related to Thermodesulfovibrio. Environ. Microbiol. 2017, 19, 4965–4977. [Google Scholar] [CrossRef] [PubMed]
- Lipczynska-Kochany, E.; Kochany, J. Application of aerobic respirometry: Studies on the impact of humate on biological treatment of municipal sewage. Environ. Technol. 2008, 29, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, D.; Ruscalleda, M.; Ficara, E.; Balaguer, M.D.; Colprim, J. Response to high nitrite concentrations of anammox biomass from two SBR fed on synthetic wastewater and landfill leachate. Chem. Eng. J. 2012, 209, 62–68. [Google Scholar] [CrossRef]
- Wang, C.C.; Lee, P.H.; Kumar, M.; Huang, Y.T.; Sung, S.; Lin, J.G. Simultaneous partial nitrification, anaerobic ammonium oxidation and denitrification (SNAD) in a full-scale landfill-leachate treatment plant. J. Hazard. Mater. 2010, 175, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Tian, B.H.; Zhang, X.; Ghula, A.; Fang, C.R.; He, R. Investigation on characteristics of leachate and concentrated leachate in three landfill leachate treatment plants. Waste Manag. 2013, 33, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Z.; Novak, J.T.; Goldsmith, C.D. Evaluation of on-site biological treatment for landfill leachates and its impact: A size distribution study. Water Res. 2012, 46, 3837–3848. [Google Scholar] [CrossRef] [PubMed]


| Sample | High-Quality Reads (M) | Assembled Reads (%) | Contigs > 300 bp | Contigs N50 (bp) | ORFs Total | Annotated ORFs by COG (%) | Annotated ORFs by KEGG (%) |
|---|---|---|---|---|---|---|---|
| HS0 | 1.80 | 83.23 | 68,869 | 7752 | 206,324 | 61.09 | 70.56 |
| HS1 | 1.74 | 83.23 | 72,451 | 5105 | 202,417 | 61.50 | 70.55 |
| HS10 | 1.78 | 81.26 | 84,185 | 4518 | 230,257 | 61.76 | 71.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.; Huang, L.-N.; Meng, F. Metagenomics Response of Anaerobic Ammonium Oxidation (anammox) Bacteria to Bio-Refractory Humic Substances in Wastewater. Water 2019, 11, 365. https://doi.org/10.3390/w11020365
Meng Y, Huang L-N, Meng F. Metagenomics Response of Anaerobic Ammonium Oxidation (anammox) Bacteria to Bio-Refractory Humic Substances in Wastewater. Water. 2019; 11(2):365. https://doi.org/10.3390/w11020365
Chicago/Turabian StyleMeng, Yabing, Li-Nan Huang, and Fangang Meng. 2019. "Metagenomics Response of Anaerobic Ammonium Oxidation (anammox) Bacteria to Bio-Refractory Humic Substances in Wastewater" Water 11, no. 2: 365. https://doi.org/10.3390/w11020365
APA StyleMeng, Y., Huang, L.-N., & Meng, F. (2019). Metagenomics Response of Anaerobic Ammonium Oxidation (anammox) Bacteria to Bio-Refractory Humic Substances in Wastewater. Water, 11(2), 365. https://doi.org/10.3390/w11020365