Exploring Bacterial Communities in Aquaponic Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collected in This Study and Samples Preparation
2.1.1. Sump Samples
2.1.2. Biofilter Samples
2.2. DNA Extraction
2.3. Sequencing
- Forward V1-V3
- 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCCTGGCTCAG-3’
- Reverse V1-V3
- 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATTACCGCGGCTGCTGG-3’
2.4. Bioinformatics
2.5. Statistics
3. Results
3.1. Metagenome Sequencing
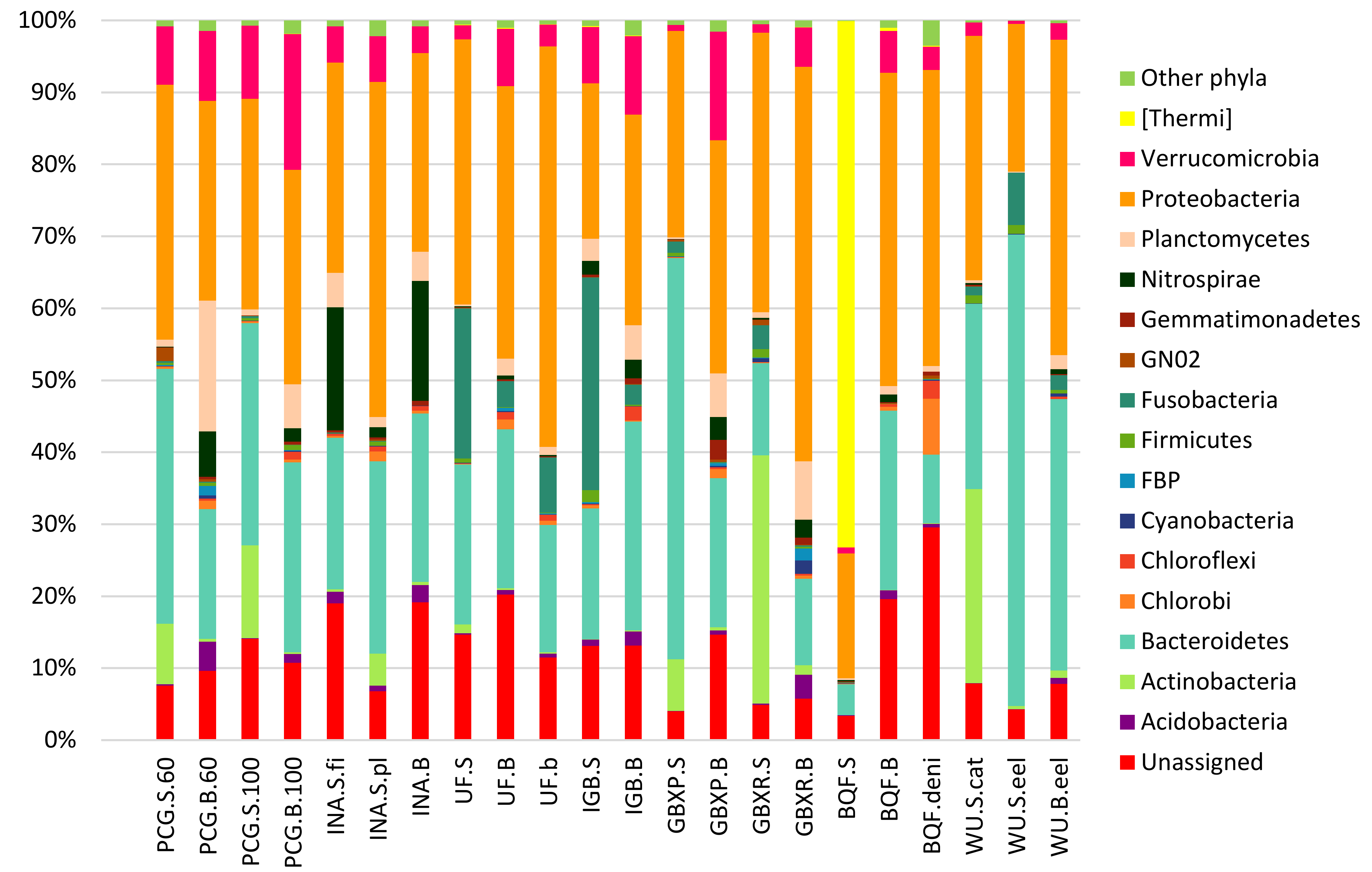
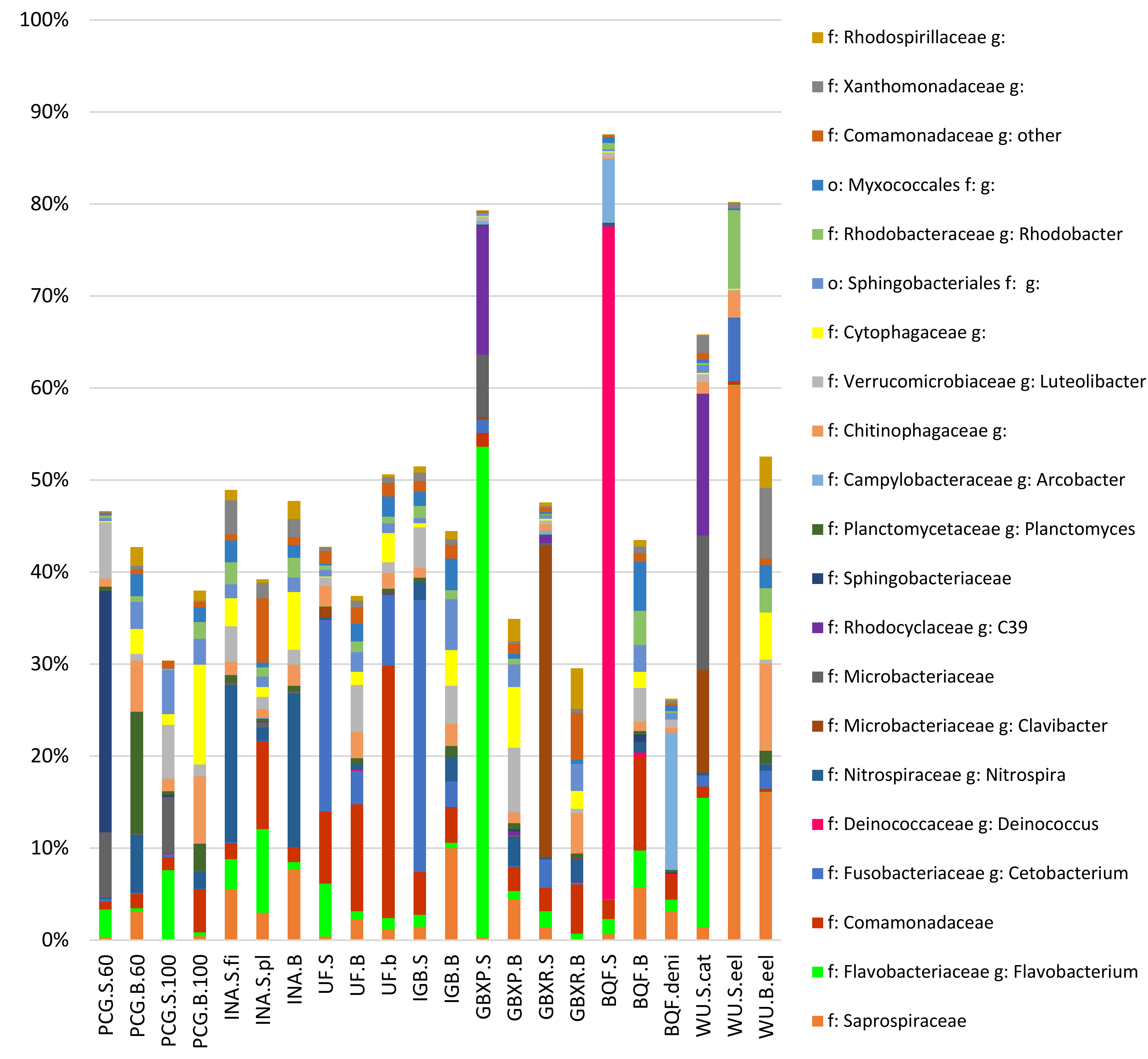
3.2. Taxonomic Assignment of Reads
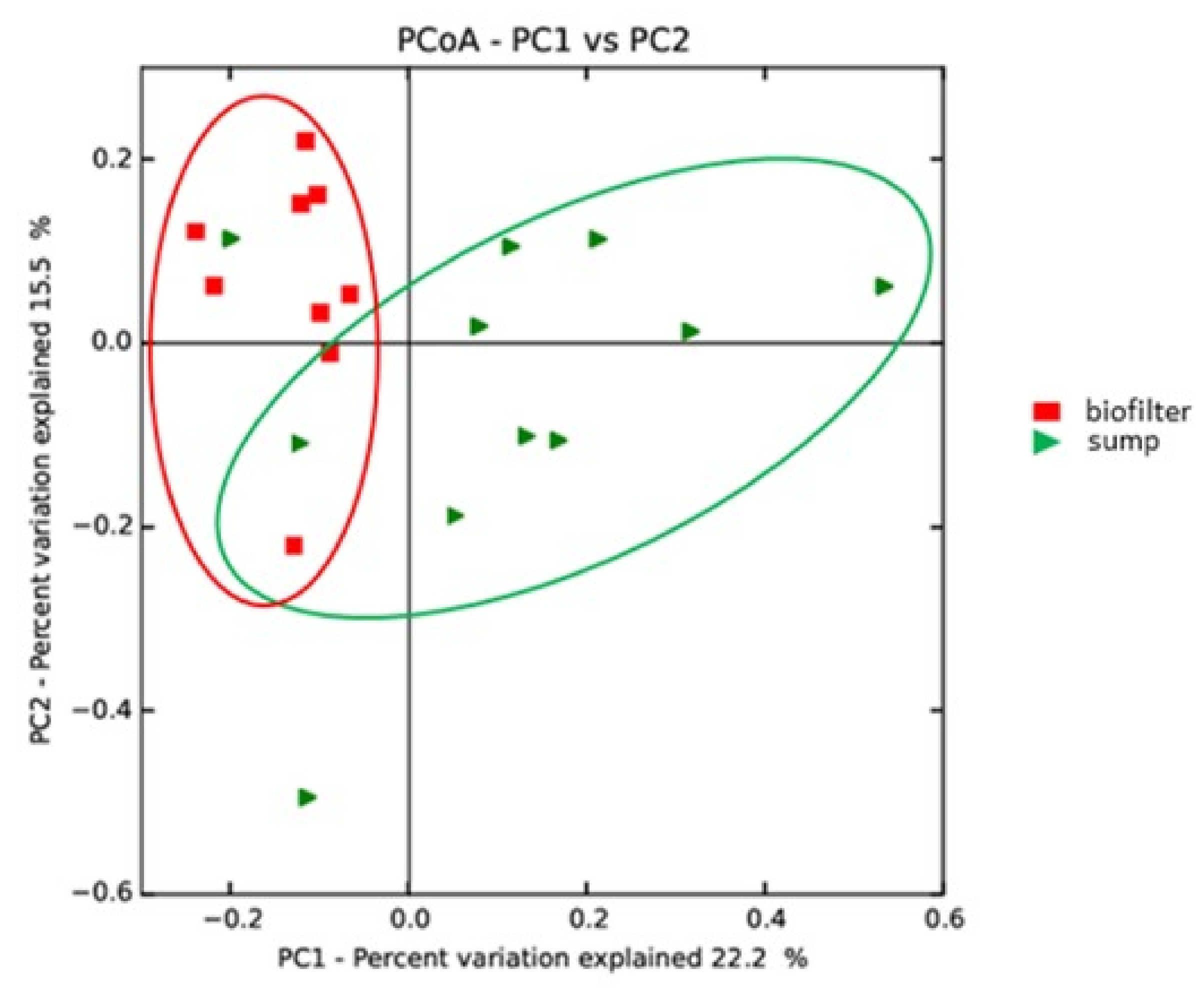
3.3. Potential Influence of the System Design on Microbial Communities
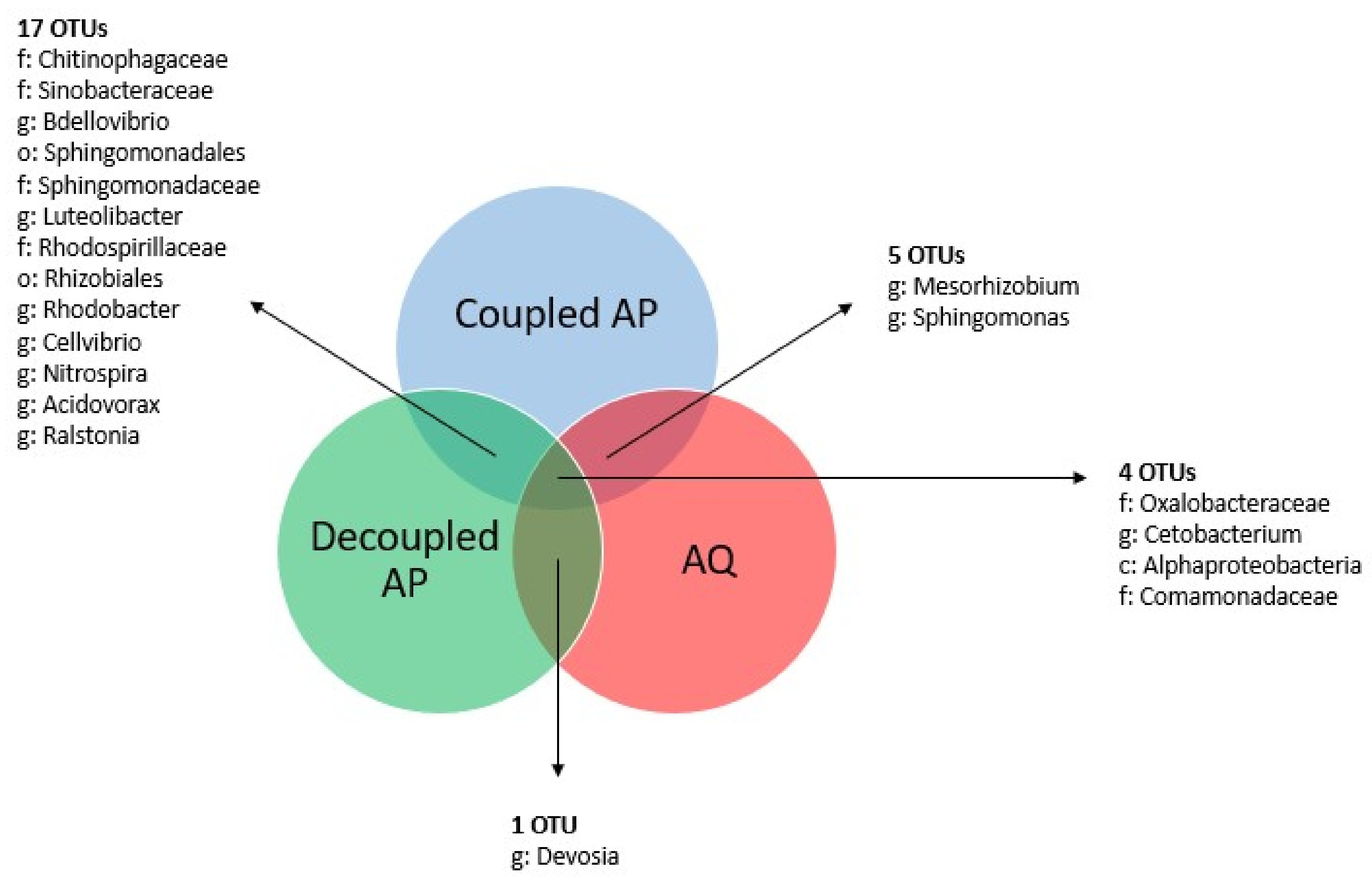
3.4. Core Microbiomes
3.4.1. General Core Microbiome
3.4.2. System-Specific Core Microbiomes
3.4.3. Sampling Site-Specific Core Microbiomes
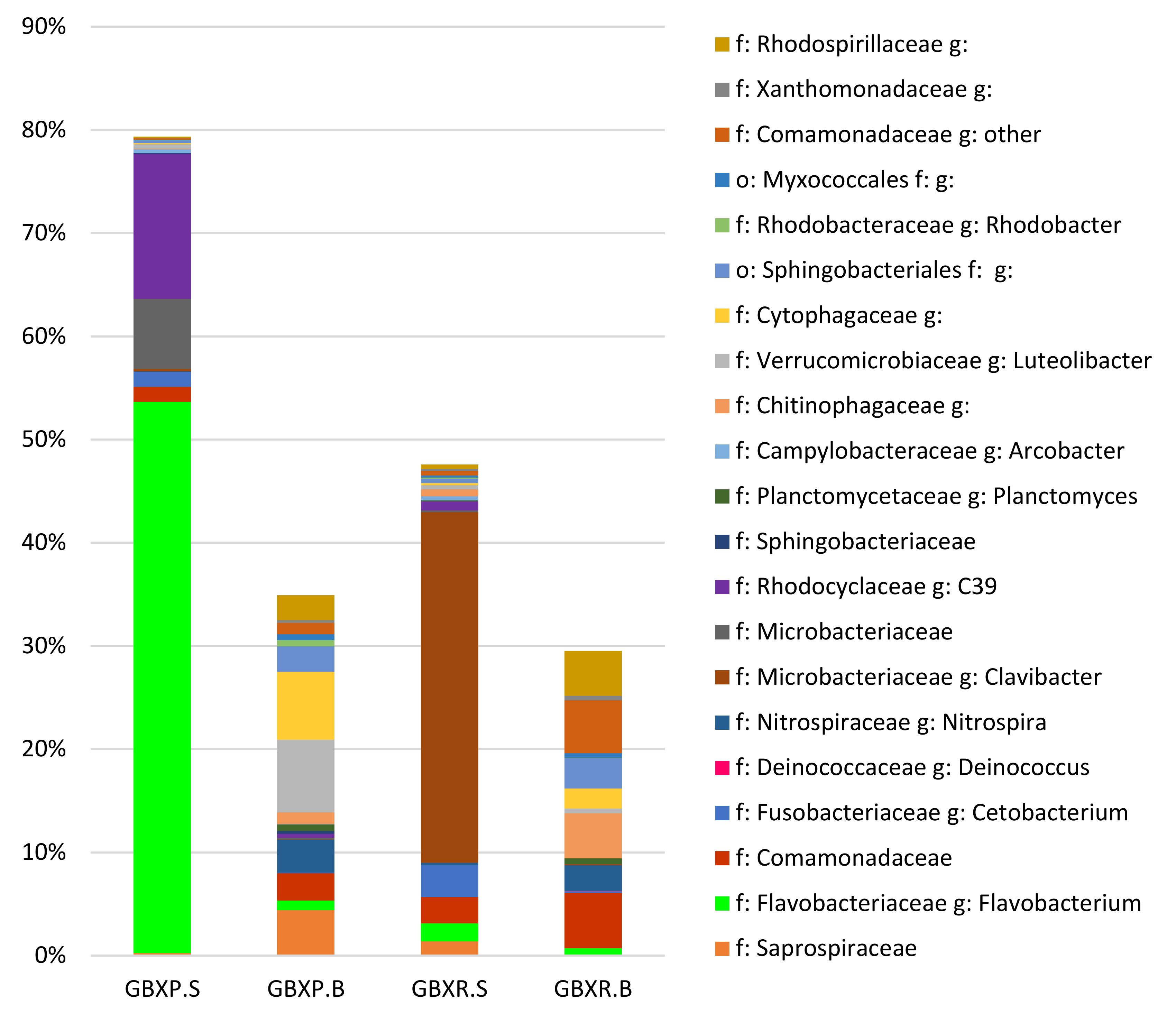
3.5. Detailed Analysis of the Microbial Communities in the Coupled and Decoupled Gembloux Systems
4. Discussion
4.1. Predominant Taxa
4.2. Potential Roles/Functions of the Identified Taxa
4.3. Core Microbiomes
5. Conclusion and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Delaide, B.; Goddek, S.; Gott, J.; Soyeurt, H.; Haissam Jijakli, M.; Lalman, J.; Junge, R. Lettuce (Lactuca sativa L. var. Sucrine) growth performance in complemented aquaponic solution outperforms hydroponics. Water 2016, 8, 467. [Google Scholar] [CrossRef]
- Rakocy, J.E. Aquaponics-Integrating Fish and Plant Culture. In Aquaculture Production Systems; Tidwell, J.H., Ed.; John Wiley and Sons: Hoboken, NJ, USA, 2012; pp. 343–386. [Google Scholar]
- Rakocy, J.E.; Masser, M.P.; Losordo, T.M. Recirculating aquaculture tank production systems: Aquaponics-integrating fish and plant culture. SRAC Publ. South Reg. Aquac. Cent. 2006, 16, 454. [Google Scholar]
- Delaide, B.; Delhaye, G.; Dermience, M.; Gott, J.; Soyeurt, H.; Jijakli, M.H. Plant and fish production performance, nutrient mass balances, energy and water use of the PAFF Box, a small-scale aquaponic system. Aquac. Eng. 2017, 78, 130–139. [Google Scholar] [CrossRef]
- Buzby, K.M.; Lin, L.S. Scaling aquaponic systems: Balancing plant uptake with fish output. Aquac. Eng. 2014, 63, 39–44. [Google Scholar] [CrossRef]
- Schmautz, Z.; Loeu, F.; Liebisch, F.; Graber, A.; Mathis, A.; Bulc, T.G.; Junge, R. Tomato productivity and quality in aquaponics: Comparison of three hydroponic methods. Water 2016, 8, 533. [Google Scholar] [CrossRef]
- Schmautz, Z.; Graber, A.; Jaenicke, S.; Goesmann, A.; Junge, R.; Smits, T.H.M. Microbial diversity in different compartments of an aquaponics system. Arch. Microbiol. 2017, 199, 613–620. [Google Scholar] [CrossRef]
- Graber, A.; Junge, R. Aquaponic Systems: Nutrient recycling from fish wastewater by vegetable production. DES 2009, 246, 147–156. [Google Scholar] [CrossRef]
- Timmons, M.B.; Ebeling, J.M. Recirculating Aquaculture, 3rd ed.; Ithaca Publishing: New York, NY, USA, 2013; ISBN 978-0-9712646. [Google Scholar]
- Resh, H.M. Hydroponic Food Production: A Definitive Guidebook for the Advanced Home Gardener and the Commercial Hydroponic Grower; CRC Press: Boca Raton, FL, USA, 2013; ISBN 1439878692. [Google Scholar]
- Rurangwa, E.; Verdegem, M.C.J. Microorganisms in recirculating aquaculture systems and their management. Rev. Aquac. 2015, 7, 117–130. [Google Scholar] [CrossRef]
- Itoi, S.; Ebihara, N.; Washio, S.; Sugita, H. Nitrite-oxidizing bacteria, Nitrospira, distribution in the outer layer of the biofilm from filter materials of a recirculating water system for the goldfish Carassius auratus. Aquaculture 2007, 264, 297–308. [Google Scholar] [CrossRef]
- Bartelme, R.P.; McLellan, S.L.; Newton, R.J. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528. [Google Scholar] [CrossRef]
- Hu, B.; Shen, L.; Xu, X.; Zheng, P. Anaerobic ammonium oxidation (anammox) in different natural ecosystems: Table 1. Biochem. Soc. Trans. 2011, 39, 1811–1816. [Google Scholar] [CrossRef]
- Goddek, S.; Schmautz, Z.; Scott, B.; Delaide, B.; Keesman, K.; Wuertz, S.; Junge, R. The Effect of Anaerobic and Aerobic Fish Sludge Supernatant on Hydroponic Lettuce. Agronomy 2016, 6, 37. [Google Scholar] [CrossRef]
- Yogev, U.; Sowers, K.R.; Mozes, N.; Gross, A. Nitrogen and carbon balance in a novel near-zero water exchange saline recirculating aquaculture system. Aquaculture 2017, 467, 118–126. [Google Scholar] [CrossRef]
- Schneider, O.; Sereti, V.; Eding, E.H.; Verreth, J.A.J. Analysis of nutrient flows in integrated intensive aquaculture systems. Aquac. Eng. 2004, 32, 379–401. [Google Scholar] [CrossRef]
- Jorquera, M.; Martínez, O.; Maruyama, F.; Marschner, P.; de la Luz Mora, M. Current and Future Biotechnological Applications of Bacterial Phytases and Phytase-Producing Bacteria. Microb. Environ. 2008, 23, 182–191. [Google Scholar] [CrossRef]
- Gravel, V.; Dorais, M.; Dey, D.; Vandenberg, G. Fish effluents promote root growth and suppress fungal diseases in tomato transplants. Can. J. Plant Sci. 2015, 95, 427–436. [Google Scholar] [CrossRef]
- Sirakov, I.; Lutz, M.; Graber, A.; Mathis, A.; Staykov, Y.; Smits, T.; Junge, R. Potential for Combined Biocontrol Activity against Fungal Fish and Plant Pathogens by Bacterial Isolates from a Model Aquaponic System. Water 2016, 8, 518. [Google Scholar] [CrossRef]
- Yildiz, H.Y.; Keskin, E.; Bekcan, S.; Kaynar, S.; Parisi, G. Molecular Identification of Dominant Bacterial Taxa in the Aquaponic System with Co-Culture of Tilapia (Oreochromis Aureus) and Tomato (Solanum lycopersicum ) Using Environmental DNA. Water 2017, 9, 1–8. [Google Scholar]
- Munguia-Fragozo, P.; Alatorre-Jacome, O.; Rico-Garcia, E.; Torres-Pacheco, I.; Cruz-Hernandez, A.; Ocampo-Velazquez, R.V.; Garcia-Trejo, J.F.; Guevara-Gonzalez, R.G. Perspective for Aquaponic Systems: “omic” Technologies for Microbial Community Analysis. BioMed Res. Int. 2015. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E. The family Deinococcaceae. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 613–615. [Google Scholar]
- Tsuchiya, C.; Sakata, T.; Sugita, H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2008, 46, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Baldani, J.I.; Rouws, L.; Magalhaes Cruz, L.; Lopes Olivares, F.; Schmid, M.; Hartmann, A. The Family Oxalobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–1012. ISBN 9783642301971. [Google Scholar]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Davis, M.J.; Graves Gillaspie, A.; Vidaver, A.K.; Harris’, R.W. Clavibacter: A New Genus Containing Some Phytopathogenic Coryneform Bacteria Pathogens That Cause Ratoon Stunting Disease of Sugarcane and Bermudagrass Stunting Disease? Int. J. Syst. Bacteriol. 1984, 84, 107–117. [Google Scholar] [CrossRef]
- Bittsanszky, A.; Uzinger, N.; Gyulai, G.; Mathis, A.; Junge, R.; Villarroel, M.; Kotzen, B.; Komives, T. Nutrient supply of plants in aquaponic systems. Ecocycles 2016, 2, 17–20. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Ye, Y.; Zhou, Y.; Fodor, A.A. A Core Human Microbiome as Viewed through 16S rRNA Sequence Clusters. PLoS ONE 2012, 7, e34242. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef]
- Dennert, F.; Imperiali, N.; Staub, C.; Schneider, J.; Laessle, T.; Zhang, T.; Wittwer, R.; van der Heijden, M.G.A.; Smits, T.H.M.; Schlaeppi, K.; et al. Conservation tillage and organic farming induce minor variations in Pseudomonas abundance, their antimicrobial function and soil disease resistance. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Evtushenko, L.; Takeuchi, M. The family Microbacteriaceae. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 1020–1098. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microb. 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Hynes, R.K.; Leung, G.C.Y.; Hirkala, D.L.M.; Nelson, L.M. Isolation, selection, and characterization of beneficial rhizobacteria from pea, lentil, and chickpea grown in western Canada. Can. J. Microbiol. 2008, 54, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Kolton, M.; Erlacher, A.; Berg, G.; Cytryn, E. The Flavobacterium Genus in the Plant Holobiont: Ecological, Physiological, and Applicative Insights. In Microbial Models: From Environmental to Industrial Sustainability; Springer: Singapore city, Singapore, 2016; pp. 189–207. [Google Scholar]
- Liu, J.; Lu, Z.; Yang, J.; Xing, M.; Yu, F.; Guo, M. Effect of earthworms on the performance and microbial communities of excess sludge treatment process in vermifilter. Bioresour. Technol. 2012, 117, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Soltani, A.-A.; Khavazi, K.; Asadi-Rahmani, H.; Omidvari, M.; Abaszadeh Dahaji, P.; Mirhoseyni, H. Plant Growth Promoting Characteristics in Some Flavobacterium spp. Isolated from Soils of Iran. J. Agric. Sci. 2010, 2, 106–115. [Google Scholar] [CrossRef]
- Kim, J.T.; Han, K.S.; Kim, B.R.; Kim, J.S.; Lee, B.C.; Yang, E.S.; Kwon, K.H.; Son, J.R.; Kim, H.G.I.; Yu, S.H. Biological Control of Plant Diseases Using Flavobacterium Hercynium Epb-C313; Uliège Library: Liège, Belgium, 2010. [Google Scholar]
- Reichenbach, H. The Lysobacter Genus. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 939–957. [Google Scholar]
- Lee, K.-J.; Oh, B.-T.; Seralathan, K.-K. Advances in Plant Growth Promoting Rhizobacteria for Biological Control of Plant Diseases. In Bacteria in Agrobiology: Disease Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; p. 495. [Google Scholar]
- Folman, L.B.; Postma, J.; van Veen, J.A. Characterisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 2003, 158, 107–115. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Nielsen, P.H. 70 The Family Saprospiraceae. In The Prokaryotes -Other Major Linages of Bacteria and the Archea; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Xia, Y.; Kong, Y.; Thomsen, T.R.; Nielsen, P.H. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (“Candidatus epiflobacter” spp.) in activated sludge. Appl. Environ. Microbiol. 2008, 74, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, W.; Huang, J.; He, S.; Yan, Y.; Zhu, W.; Wu, S.; Zhang, X. Nitrogen removal by the enhanced floating treatment wetlands from the secondary effluent. Bioresour. Technol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Monsees, H.; Keitel, J.; Paul, M.; Kloas, W.; Wuertz, S. Potential of aquacultural sludge treatment for aquaponics: Evaluation of nutrient mobilization under aerobic and anaerobic conditions. Aquac. Environ. Interact. 2017, 9, 9–18. [Google Scholar] [CrossRef]
- Wongkiew, S.; Hu, Z.; Chandran, K.; Lee, J.W.; Khanal, S.K. Nitrogen transformations in aquaponic systems: A review. Aquac. Eng. 2017, 76, 9–19. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Huang, W.; Wang, Y.; Zhou, J. Bacterial community structure in simultaneous nitrification, denitrification and organic matter removal process treating saline mustard tuber wastewater as revealed by 16S rRNA sequencing. Bioresour. Technol. 2017, 228, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, M.A.H.J.; Harhangi, H.R.; Van De Pas-Schoonen, K.; Van De Vossenberg, J.; Flik, G.; Jetten, M.S.M.; Klaren, P.H.M.; Op Den Camp, H.J.M. Biodiversity of N-cycle bacteria in nitrogen removing moving bed biofilters for freshwater recirculating aquaculture systems. Aquaculture 2010, 306, 177–184. [Google Scholar] [CrossRef]
| Code | Operator | Location | Design | Fish Species | Feed Type | Biochips Type | Plant Type | Sampling Date | Sump Samples | Biofilter Samples | Extra Samples |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCG | Provincial Trial Centre for Vegetable Production | Kruishoutem, Belgium | Aquaponics—open loop | Scrotum barco | Vegetarian | Eco Pondchips | Tomatoes | 29/03/2017 | 1 sump 60 (low density) 1 sump 100 (high density) | 1 biofilter 60 (low density) 1 biofilter 100 (high density) | |
| INA | Inagro | Rumbeke-Beitem, Belgium | Aquaponics—open loop | Sander lucioperca | Omnivorous | Kaldnes media | Tomatoes | 18/04/2017 | 1 sump fish 1 sump hydroponics | 1 biofilter | |
| UF | UrbanFarmers | The Hague, Netherlands | Aquaponics—open loop | Oreochromis niloticus | Omnivorous | Kaldnes media | Microgreens Leafy greens Fruity vegetables | 23/03/2017 | 1 sump | 1 biofilter | 1 biofilm |
| IGB | Leibnitz-Institute of freshwater ecology and inland fisheries | Berlin, Germany | Aquaponics—open loop | Oreochromis niloticus | Omnivorous | Kaldnes media | Tomatoes | 07/04/2017 | 1 sump | 1 biofilter | |
| GBXP | Gembloux Agro Bio Tech, PAFF Box system | Gembloux, Belgium | Aquaponics—closed loop | Oreochromis niloticus | Vegetarian | Microbeads | Leafy greens Fruity vegetables | 27/04/2017 | 4 sump | 4 biofilter | |
| GBXR | Gembloux Agro Bio Tech, RAS system | Gembloux, Belgium | Aquaculture | Oreochromis niloticus | Vegetarian | Biocerapond | N.R. | 03/04/2017 | 1 sump | 1 biofilter | |
| BQF | Belgian Quality Fish | Dottignies, Belgium | Aquaculture | Acipenser spp. Huso sp. | Omnivorous | Kaldnes media | N.R. | 29/03/2017 | 1 sump | 1 biofilter | 1 biofilter denitrification |
| WU | Wageningen University | Wageningen, Netherlands | Aquaculture | Eel, catfish | Omnivorous | Kaldnes media | N.R. | 12/04/2017 | 1 sump eel 1 sump catfish | 1 biofilter eel |
| Code | Sampling Zone | Number of Reads before Filtering | Chimeric Reads | Chloroplast and Mitochondrial Reads | Singleton Reads | Number of Reads after Filtering | % of Unassigned Reads | Shannon Index (after Filtering) | EquitabilityIndex (after Filtering) |
|---|---|---|---|---|---|---|---|---|---|
| PCG.S.60 | sump low density | 75,840 | 83 | 9 | 1016 | 74,732 | 7.6% | 5.65 | 0.52 |
| PCG.S.100 | sump high density | 131,231 | 443 | 100 | 3822 | 127,166 | 13.9% | 6.55 | 0.56 |
| PCG.B.60 | biofilter low density | 104,241 | 77 | 10 | 2661 | 101,493 | 9.5% | 7.88 | 0.69 |
| PCG.B.100 | biofilter high density | 78,392 | 66 | 7 | 1635 | 76,684 | 10.6% | 7.73 | 0.70 |
| INA.S.fi | sump fish loop | 107,998 | 146 | 117 | 2634 | 105,101 | 19.0% | 7.68 | 0.66 |
| INA.S.pl | sump before plant compartment | 92,790 | 708 | 2 | 3581 | 87,985 | 6.8% | 8.63 | 0.71 |
| INA.B | biofilter | 100,948 | 154 | 124 | 2831 | 97,839 | 19.2% | 7.37 | 0.65 |
| UF.S | sump | 117,695 | 1122 | 22 | 3175 | 113,376 | 14.6% | 6.77 | 0.57 |
| UF.B | biofilter | 134,730 | 223 | 5 | 7336 | 126,866 | 20.3% | 8.30 | 0.67 |
| UF.b | biofilm | 99,487 | 260 | 56 | 2407 | 96,764 | 11.4% | 6.94 | 0.59 |
| IGB.S | sump | 63,482 | 186 | 10 | 2657 | 57,612 | 12.9% | 7.50 | 0.62 |
| IGB.B | biofilter | 59,923 | 74 | 45 | 2192 | 58,091 | 13.1% | 8.44 | 0.75 |
| GBXP.S | sump | 97,905 | 340 | 864 | 4 | 96,697 | 4.0% | 3.79 | 0.34 |
| GBXP.B | biofilter | 69,831 | 52 | 0 | 2233 | 67,546 | 14.7% | 8.31 | 0.74 |
| GBXR.S | sump | 11,096 | 1037 | 100 | 2318 | 112,641 | 4.8% | 5.91 | 0.51 |
| GBXR.B | biofilter | 124,569 | 378 | 81 | 3937 | 120,173 | 5.7% | 7.65 | 0.65 |
| BQF.S | sump | 81,204 | 50 | 10 | 1087 | 80,057 | 3.3% | 2.83 | 0.26 |
| BQF.B | biofilter | 56,448 | 85 | 13 | 3090 | 53,260 | 19.7% | 8.42 | 0.74 |
| BQF.deni | denitrification biofilter | 65,693 | 14 | 1 | 1966 | 63,712 | 29.5% | 6.72 | 0.63 |
| WU.S.cat | sump catfish system | 44,743 | 119 | 4 | 1142 | 43,478 | 7.9% | 6.20 | 0.58 |
| WU.S.eel | sump eel system | 75,055 | 32 | 1 | 671 | 74,351 | 4.2% | 3.22 | 0.32 |
| WU.B.eel | biofilter eel system | 101,169 | 177 | 8 | 2059 | 98,925 | 7.8% | 6.43 | 0.60 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eck, M.; Sare, A.R.; Massart, S.; Schmautz, Z.; Junge, R.; Smits, T.H.M.; Jijakli, M.H. Exploring Bacterial Communities in Aquaponic Systems. Water 2019, 11, 260. https://doi.org/10.3390/w11020260
Eck M, Sare AR, Massart S, Schmautz Z, Junge R, Smits THM, Jijakli MH. Exploring Bacterial Communities in Aquaponic Systems. Water. 2019; 11(2):260. https://doi.org/10.3390/w11020260
Chicago/Turabian StyleEck, Mathilde, Abdoul Razack Sare, Sébastien Massart, Zala Schmautz, Ranka Junge, Theo H. M. Smits, and M. Haïssam Jijakli. 2019. "Exploring Bacterial Communities in Aquaponic Systems" Water 11, no. 2: 260. https://doi.org/10.3390/w11020260
APA StyleEck, M., Sare, A. R., Massart, S., Schmautz, Z., Junge, R., Smits, T. H. M., & Jijakli, M. H. (2019). Exploring Bacterial Communities in Aquaponic Systems. Water, 11(2), 260. https://doi.org/10.3390/w11020260









