Abstract
Aquaponics is a production system based on the dynamic equilibrium between fish, plants, and microorganisms. In order to better understand the role of microorganisms in this tripartite relationship, we studied the bacterial communities hosted in eight aquaponic and aquaculture systems. The bacterial communities were analyzed by 16S rRNA gene deep sequencing. At the phylum level, the bacterial communities from all systems were relatively similar with a predominance of Proteobacteria and Bacteroidetes. At the genus level, however, the communities present in the sampled systems were more heterogeneous. The biofilter samples harbored more diverse communities than the corresponding sump samples. The core microbiomes from the coupled and decoupled systems shared more common operational taxonomic units than with the aquaculture systems. Eventually, some of the taxa identified in the systems could have beneficial functions for plant growth and health, but a deeper analysis would be required to identify the precise functions involved in aquaponics.
1. Introduction
Aquaponics is a combination of hydroponic and recirculating aquaculture technologies [1,2]. It offers the possibility of recycling nutrient-rich waste water from fish into organic fertilizers for the plants grown in the system [3], thus reducing the use of fertilizers of mineral origin and the environmental impact of both fish and plant production [4,5]. The use of the aquaculture wastewater to fertilize the plants can avoid the discharge of phosphorus- and nitrogen-enriched water into already nitrogen-loaded surface- and groundwater [5,6].
Along with plants and fish, microorganisms are present in aquaponics. Bacteria are key players in processes which are central for the functioning and equilibrium of an aquaponic system [7]. The best studied process is nitrification, during which ammonia (the main nitrogen form excreted by the fish) is transformed via nitrite to nitrate, which is less toxic for the fish [8] and preferred by plants [9,10]. The main bacteria involved in this transformation are the ammonia oxidizing bacteria (AOB), such as Nitrosococcus, Nitrosospira, and Nitrosomonas, and the nitrite oxidizing bacteria (NOB), such as Nitrobacter, Nitrospira [11], Nitrococcus, and Nitrospina [12]. Some Nitrospira populations are also able to perform the complete ammonia to nitrate transformation—and are known as complete ammonia oxidizers (COMAMMOX)—by themselves [13,14]. Archaea, such as the Thaumarchaeota, can also be involved in the ammonia-oxidizing process [13]. Finally, the anaerobic ammonium oxidation (ANAMMOX) group, members of the Planctomycetes responsible for the anaerobic transformation of ammonium and nitrite into nitrous oxide and N2 [15] may play a role as well where oxygen levels are low.
In addition to nitrification, microorganisms are involved in other important processes. They can contribute to extracting the various macro- and micronutrients from the feed leftovers and solid feces and make them available for plant uptake [16]. Depending on the aquaculture compartment, design, fish species, and feed type, 15–60% of the consumed feed is actually converted into fish biomass and used for fish metabolism. The rest is excreted and is available for the bacteria to decompose [9,17,18]. Besides this, bacteria could also play a role in the solubilization of nutrients encompassed in solid compounds, such as phytates [19]. Additionally, microorganisms in aquaponics are also involved in various plant growth promotion and protection pathways, such as biocontrol or the enhancement of root growth [7,20,21]. However, these pathways are not sufficiently elucidated yet.
Here, we compared a set of aquaculture (AQ) and aquaponic (AP) systems, which differ in terms of plant and fish species and/or feed type. AP designs included both “coupled” or closed loop AP systems (one loop containing fish and plants) and “decoupled” or open loop AP systems (two separate loops for fish and plants). The aim of this study was to gain insight into the diversity of the bacterial communities in these systems and, if possible, to link their potential functions to plant growth and plant health. For this, the bacterial communities present in biofilter and sump samples, which were the two common units in all systems, were characterized using 16S rRNA gene deep sequencing.
2. Materials and Methods
2.1. Samples Collected in This Study and Samples Preparation
Sump and biofilter samples were collected from eight different systems (Table 1). Three were operated as aquaculture and five as aquaponics. Samples were collected in the period between March and April 2017 as described below. A detailed description of the systems is given in Supplementary Material (Description of the visited aquaponic and aquaculture systems, Figure S1 and Table S1). For comparison to previously published data [7], the dataset of the aquaponic system of the Zürich University of Applied Sciences (ZHAW) was downloaded from the European Bioinformatics Institute database (EBI) and analyzed similarly to the data generated in this study.

Table 1.
Comparison of the sampled aquaculture and aquaponics systems.
2.1.1. Sump Samples
For each sample, two liters of water were collected in sterile Pyrex bottles. In order to concentrate the bacteria, the samples were filtered through 0.2 μm filters (Supor®—with a vacuum pump. The filters were then placed in a 50 mL sterile Falcon tube containing 30 mL of sterile water. After vortexing the Falcon tube for 4 min, the filters where removed and the tube centrifuged at 7607× g for 10 min [22]. The pellet was then directly used for DNA extraction.
2.1.2. Biofilter Samples
The biochips used in the different systems varied in shape and size. Therefore, a constant number of biochips per sample could not be taken, as this would not always fit in a 50 mL Falcon tube. Instead, as many biochips as possible (between 10 and 30 depending on the size) were inserted in a 50 mL sterile Falcon tube containing 30 mL of sterile water in order to ensure the harvest of a maximum quantity of bacteria. The Falcon tubes were placed on a vortex for 2 min before placing them for 5 min in an ultrasonic bath (Ultrasonic cleaner, model USC600T, VWR, Leuven, Belgium). The Falcon tubes were then centrifuged at 7607× g for 10 min and the pellet was collected for DNA extraction.
2.2. DNA Extraction
The Fast DNA Spin Kit using Cell Lysis Solution TC (MP Biomedicals, Santa Ana, CA, USA) was used for the DNA extractions. The manufacturer’s protocol was modified as follows: Samples were homogenised with a Power-Mix Model L46 (Labinco, Breda, The Netherlands) at speed setting 7 for 40 s, then incubated on ice for 2 min and again homogenized for 40 s. Subsequently, to remove cell debris, tubes were centrifuged at 14,000× g for 10 min. All DNA extracts were stored unopened at 4 °C until further analyses.
2.3. Sequencing
Library preparation and sequencing were carried out by DNA Vision S.A. (company, Gosselies, Belgium). The library preparation and indexing steps were done using the Nextera Index kit v2 (Illumina, San Diego, CA, USA), while sequencing was conducted on an Illumina Miseq (2 × 250 bp) (Illumina Inc., San Diego, CA, USA) with the Miseq reagent kit v3 (600-cycles, Illumina, San Diego, CA, USA). Sequencing primers were chosen to cover the hypervariable regions V1-V3 of the 16S ribosomal RNA gene as recommended by Munguia-Fragozo et al. [23] and Schmautz et al. [7]. The following primers were used:
- Forward V1-V3
- 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCCTGGCTCAG-3’
- Reverse V1-V3
- 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATTACCGCGGCTGCTGG-3’
Data are available under the accession PRJNA513832 on the Sequence Read Archive databse (SRA) of the National Center for Biotechnology Information (NCBI).
2.4. Bioinformatics
The analysis of the sequencing data was conducted with the QIIME pipeline v1.9.1 (http://qiime.org/) [24]. Forward and reverse sequences were merged in one file per sample with multiple_join_paired_ends.py. Paired fastq files were converted into fasta files with convert_fastaqual_fastq.py. Sequences were assigned to Operational Taxonomic Units (OTUs) with a cut-off of 97% sequence identity to the reference database Greengenes 13_8 with pick_de_novo_otus.py. Chimeric sequences were identified with the Chimera Slayer tool and then removed with filter_fasta.py. Singletons and sequences originating from chloroplasts and mitochondria were discarded.
For further analysis of the bacteria communities’ composition in the 22 samples, the samples were rarefied at 40,000 sequences with single_rarefaction.py (rarefaction curves available in the Supplementary Material; Figure S2). Bar charts representing the relative abundances of the various OTUs were obtained using summarize_taxa_through_plots.py. Core microbiomes were generated using compute_core_microbiome.py.
2.5. Statistics
The Shannon and equitability indices were calculated via the alpha_diversity.py. The Shannon and equitability indices of biofilter versus sump were compared via the global core_diversity_analyses.py workflow with a nonparametric t-test (using Monte Carlo permutations). Principal coordinates analyses were carried out with the beta_diversity_through_plots.py script in order to compare the communities.
3. Results
3.1. Metagenome Sequencing
Whereas the previous study on the Wädenswil Aquaponics System [7] gave a first impression on the bacterial communities in a single setting, a comparison to other systems was not performed. For this reason, we collected 22 samples from various aquaponic and aquaculture systems in Western Europe (Table 1). Eleven samples were collected from sumps, nine from biofilters, one from an additional denitrification biofilter, and one from the periphyton present on tank walls (Table 2). The bacterial communities thereof were analyzed in this study using 16S rRNA gene deep sequencing. The average Q30 of the sequence in the samples was 80%, which indicated samples of good DNA quality. Of the total reads, 11.8% were not assigned at the phylum level.

Table 2.
Summary of metagenomics data.
3.2. Taxonomic Assignment of Reads
Based on the taxonomic assignment of the reads, it can be observed that the bacterial communities in the different systems were highly variable. Of all filtered reads in all samples, an average 11.8% ± 6.7% could not be assigned to any OTU. In general, two major phyla were found throughout the samples (Figure 1): Proteobacteria, representing 34.6% ± 10.1% of the total reads, and Bacteroidetes, representing 25.5% ± 14.0%. Other phyla were found in lower quantities in the samples. However, some samples held exceptionally high amounts of individual phyla. An example here is the presence of 73.1% reads representing the Thermi phylum, mainly represented by a single OTU (Deinococcus) in the sump sample of the Belgian Quality Fish (BQF) system (Figure 1). Except for the biofilter of the same system, this phylum was only present at very low levels in the other systems.
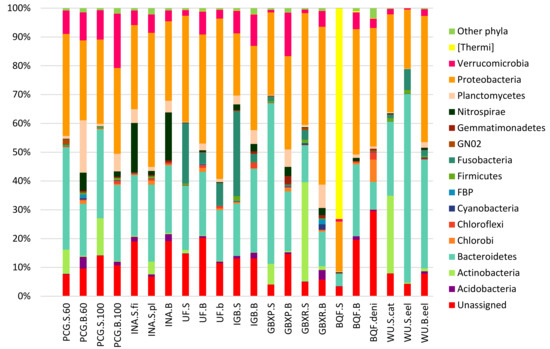
Figure 1.
Bar charts representing the relative abundances of the phyla in each sample. Phyla which represented less than 0.2% of the total reads are gathered under “other phyla” BHI 80139, BRC1, Chlamydia, Elusimicrobia, Fibrobacteras, GN04, GOUTA4, Lentispaerae, NKB19, OP11, OP3, OP8, PAUC34f, SBR1093, SR1, Spirochaetes, Synergistetes, TM6, TM7, Tenericutes, WPS2, WS1, WS2, WS3, WWE1, and Caldithrix).
It could be noted that in almost all systems, the biofilter sample harbored a more diverse community as the Shannon indices of the biofilter samples were significantly higher (p < 0.05) than the Shannon indices of the sump group (Table 2). The equitability was also significantly higher in the biofilter samples than is the sump samples (p < 0.05). The only exception was the INA system. A more thorough exploration of this system would be required to explain this difference.
At the genus level, reads were assigned to more than 700 different OTUs. To allow for a more in-depth analysis of the genera present in aquaponic systems, it was decided to focus on the OTUs representing more than 1% of the total reads per sample (Figure 2). For some OTUs, the identification process was only possible at the family level.
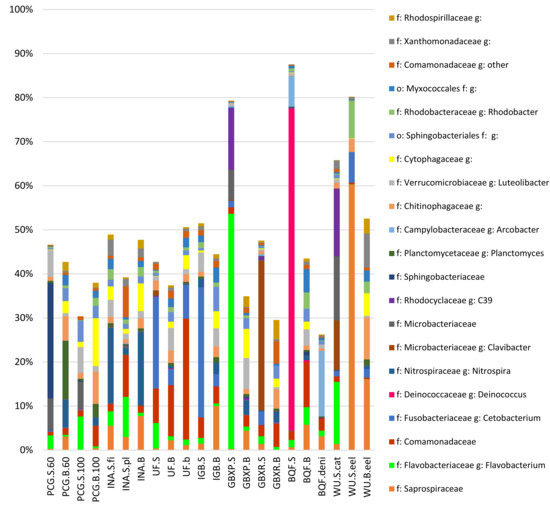
Figure 2.
Bar charts representing the relative abundances of the families and genera representing more than 1% of the total reads for each sample.
3.3. Potential Influence of the System Design on Microbial Communities
One OTU assigned to the genus Deinoccocus was mainly present in the BQF system and represented 73% of the BQF sump sample. Members of the genus Deinoccoccus are heterotrophic organisms resistant to UV radiation [25] and to our knowledge, there isno link between the rearing of sturgeons and the presence of this genus. As the BQF system was implemented with an ozone plus UV light disinfection treatment (Supplementary Material, Description of the visited aquaponic and aquaculture systems), it could be expected that this organism was dominating the community based on its resistance to such treatment. However, most aquaponic systems, such as the Wädenswil Aquaponics system [7], use UV light to prevent the proliferation of undesirable microorganisms and to keep the water clean and clear [9], without observing such a development of Deinoccocus. It could thus be that the dominance of Deinococcus spp. in the sump was due to a higher strength of the UV light in combination with the ozone treatment. It was also observed in both biofilter samples, but at levels below 0.5% of the community. This indicated that the Deinococcus spp. were more planktonic, while we observed a broader diversity in the biofilms on the carrier in the biofilters.
The species Cetobacterium somerae belongs to the Fusobacteriaceae family and has been commonly found in guts of freshwater fish [7,12,26]. The discovery of Cetobacterium in some of the samples might indicate that the system design in these cases was not sufficiently adapted to remove sufficient amounts of fish feces from the water of the fish tank. On the day when the Urban Farmers (UF) (Supplementary Material, Description of the visited aquaponic and aquaculture systems) samples were collected, pipes were clogged with fish sludge in the drum filter compartment and thus sludge may have passed the drum filter towards the biofilter and sump compartments. This might explain the large amounts of reads assigned to the genus Cetobacterium in the UF samples (Figure 2). Additionally, it was observed that Cetobacterium were more often detected in the sump of systems than in biofilter samples. It was assumed that the sump in these systems could offer sufficiently anaerobic zones whereas the moving bed biofilters were fully aerobic [3]. However, if the system hosted an important quantity of Cetobacterium in the sump, one could also observe their presence in the biofilter albeit at a smaller proportion (Figure 2).
Conversion of nitrogen compounds is of utmost importance for recirculating systems to avoid toxicity problems of the different nitrogen forms for each species. Of the known nitrifying bacteria, the Nitrosomonadaceae family was present in most samples. Even though the relative abundance of this family was quite low (between 0% and 1.7% of reads; average = 0.3%; stdev = 0.5%), the order of magnitude observed in most samples in this study was similar to the one observed in the study of the Wädenswil Aquaponics system [7]. The most abundant nitrifying bacteria were those of the genus Nitrospira. This is also in accordance with the study conducted on the Wädenswil Aquaponics system [7], and may indicatethat the COMAMMOX process is more common to aquatic culture systems.
3.4. Core Microbiomes
3.4.1. General Core Microbiome
Generally, a large diversity of bacteria was observed and all systems hosted different bacterial communities (Figure 1). However, in spite of this diversity and the specificities of each system, common bacterial groups were found in all aquaponic and aquaculture systems. A core microbiome containing only the OTUs present in all samples was extracted from the data set and, regardless of the system and sample location, four OTUs were identified. OTUs representing unidentified genera from the Oxalobacteraceae family and the Comamonadaceae family were identified as being present in all samples. The Oxalobacteraceae family harbors several heterotrophic bacteria that can be found in water, soil and also in association with plants [27]. Regarding the Comamonadaceae family, it is also found in aquaculture or aquaponic systems in other studies [7,12]. In their review, Munguia-Fragoso et al. [23] reported that Comamonas sp. were identified in several bacterial communities of freshwater recirculating aquaculture systems (RAS). At the genus level, OTUs assigned to the genera Cetobacterium were as well part of the general core microbiome. Although Cetobacterium is rather an anaerobe, its presence in all systems could be explained by its common presence in fish guts [26,28].
3.4.2. System-Specific Core Microbiomes
On the basis of the three basic setups that were sampled in this study, we also calculated core microbiomes for each of these setups (Figure 3). The AQ group contained nine common OTUs, and the decoupled AP group harbored a core microbiome of 34 OTUs. The coupled AP group contained only the plant and fish farming (PAFF) Box samples and, therefore, harbored a core microbiome of 636 OTUs. Whilst only one and five OTUs were common for aquaculture and the two aquaponics systems, the two aquaponic groups share more OTUs. A total of 17 OTUs at different levels were found, indicating that there are some not yet identified conditions that are specific to aquaponic systems, independently of the setup.
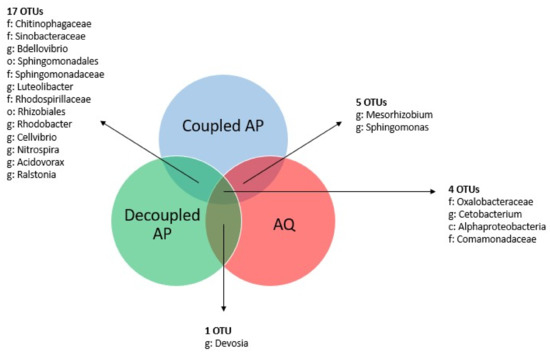
Figure 3.
Venn diagram representing the intersection of different core microbiomes obtained through the grouping of samples based on the system setup (i.e., coupled aquaponics, decoupled aquaponics, and aquaculture).
3.4.3. Sampling Site-Specific Core Microbiomes
Based on the sampling strategy, sampling site-specific core microbiomes were generated from biofilter and sump samples. The sump core microbiome was composed of 22 OTUs, while the biofilter core microbiome was composed of 28 OTUs. The larger numbers of OTUs in site-specific core microbiomes indicated that both zones had site-specific bacterial communities. This was confirmed by principal coordinates analysis (PcoA), indicating the presence of two sample clusters: a narrow cluster grouping biofilter samples and a wider cluster grouping the sump samples (Figure 4). Between these two site-specific core microbiomes, ten OTUs were common (four of them belonging to the global core microbiome). The six additional OTUs found in all biofilters and sumps were Sphingomonas, Devosia, Novosphingobium, Acidovorax, Ralstonia, and an unidentified OTU from the Rhizobiaceae family. Nitrospira could be found in all biofilter samples. However, this was not the case for the genera Nitrosomonas and Nitrobacter.
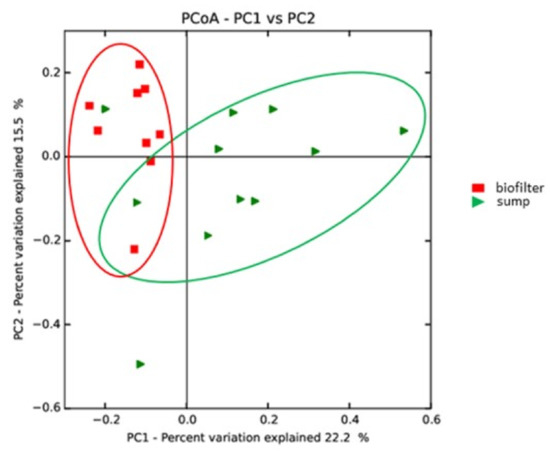
Figure 4.
Weighted UniFrac principal coordinates analysis presenting the separation between the sumps and the biofilter samples. Axis 1 and 2 explain 37.8% of the total variability.
3.5. Detailed Analysis of the Microbial Communities in the Coupled and Decoupled Gembloux Systems
Up to now, we have compared systems that were highly heterogeneous in their design, size, and operational strategies. This study also included two systems that were highly comparable: the recirculating aquaculture system (RAS) and the Plant and Fish Farming (PAFF) Box of Gembloux Agro–Bio Tech, as both systems shared the same size, fish, feed type and incoming water quality. The main difference was that the PAFF Box is operated as closed circular system, whereas the RAS is an aquaculture system. In order to observe the impact of plants in the system on the composition of the bacterial community, we chose to compare the communities in these two systems in more detail.
The RAS sump sample was dominated by the two genera Clavibacter and Cetobacterium, whereas the PAFF Box sample contained a majority of the genus Flavobacterium and C39, an OTU belonging to the Rhodocyclaceae family (Figure 5). The presence of such large numbers of reads assigned to Clavibacter, a genus that mainly contains plant pathogenic species [29], needs to be examined in more detail to confirm the assignment of all reads to this genus. The RAS biofilter sample was clearly dominated by members of the genus Lysobacter and also hosted Nitrospira and Novosphingobium, while the PAFF Box biofilter contained C39, Nitrospira, Flavobacterium, members of the Microbacteriaceae family, and Cetobacterium (Figure 5). This indicated that, despite similarities in the global setup (fish species, feed type, incoming water quality, and size of the fish tanks), each system developed its own specific community. The presence of plants in the aquaculture loop thus had a large influence on the composition of the bacterial community.
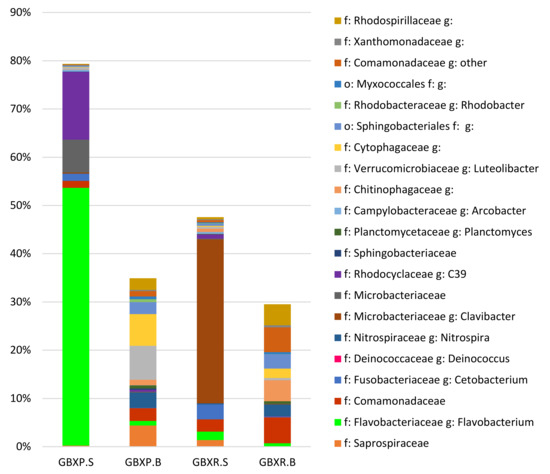
Figure 5.
Relative abundances of the families and genera representing more than 1% of the total reads within each sample, for the biofilter and sump samples of the recirculating aquaculture system (RAS) and plant and fish farming (PAFF) Box systems of Gembloux Agro-Bio. Tech.
4. Discussion
4.1. Predominant Taxa
In order to work towards better understanding of how bacterial communities function in aquaponics, we decided to focus on the potential impact of the bacteria in the aquaponic solution on plant growth. A few studies have reported that, with aquaponics, they obtained plant yields as good as in hydroponics despite the aquaponic solution containing lower nutrient concentrations [1,8,30]. A first step to take towards the elucidation of this increased growth would be to check for similarities in the bacterial communities in different aquaponic systems. In order to do so, it was decided to study the core microbiomes of our samples. The concept of core microbiomes has been used and described in several other research fields, such as the plant holobiont [31], humans [32], or milk microbiomes [33]. The core microbiome has been defined by Lemanceau et al. [31] (p. 1) as “the microbial community that is systematically associated with a given host”. Until now, the concept of core microbiome has been focused on the taxonomic composition of a community. However, it could also be argued that a core microbiome should have specific functionalities responding to the needs of their associated host [31].
At the phylum level, Proteobacteria and Bacteroidetes were the two major groups representing, respectively, 35% and 26% of the reads obtained from the different aquaponics and aquaculture systems. This was in accordance with the observation from Schmautz et al. [7], who also found that Proteobacteria (approximately 50%) and Bacteroidetes (15–20%) were the major phyla in their root zone, biofilter, and periphyton samples. This also corroborated observations made in freshwater aquaculture [23]. Other phyla common to the observations by Schmautz et al. [7], the freshwater data cited by Munguia-Fragoso et al. [23], and the present study were the phyla Actinobacteria, Planctomycetes and Nitrospirae.
Few reads assigned to the genus Pseudomonas were detected in the visited systems. The number of assigned reads to the pseudomonads was in the same order of magnitude as previously observed by Schmautz et al. [7]. Pseudomonas are usually found in close proximity to roots and are less prevalent in bulk soil [34]. As, in this study, we did not investigate the root zone of the aquaponics systems, this genus may thus rather be represented by planktonic species in the samples taken.
4.2. Potential Roles/Functions of the Identified Taxa
In aquaponics, we are interested in the bacteria, which could help us ensure fish welfare and plant care. When it comes to plants, bacteria could help their growth and health through growth stimulation and biocontrol. Taxa, such as the Microbacteriaceae family, are known to be able to form associations with plants [35] and have been detected in the microbiome of barley roots along with members of the Comamonadaceae family [36]. In the latter family, several species have been detected to possess skills for siderophore production and protection against Fusarium and Rhizoctonia [37]. The Microbacteriaceae family contains species which have aminocyclopropane-1-carboxylate (ACC) deaminase activity, siderophore, and indole production [37]. The Flavobacterium genus is widely present in nature and mostly known for its capacity to degrade complex organic molecules [38,39]. Flavobacterium spp. are often found in association with plant roots and plant leaves and are believed to be involved in plant growth and protection [38,39,40]. Several strains were detected to be able to participate in the solubilization of insoluble phosphate, the production of auxin, and the production of siderophore [37,40]. Flavobacterium are also used to fight against plant pathogens (Phytophtora infestans) in biological control formulation [41]. In aquaculture and aquaponics, Flavobacterium have also been detected and can be considered as a common genus found in such systems [7,12,23]. The Lysobacter genus has also been identified as plant growth promoting bacteria (PGPB) and can help fight against plant disease through the production of antibiotics [42,43,44]. The presence of all of those species in most of the samples in this study confirmed the data from the samples from the Wädenswill aquaponics system [7] and strengthens the statement that the microbiome in aquaponics or aquaculture systems may be able to secure the health and growth of the plants in the first-named type of systems.
A crucial function of the bacteria communities in aquaponics would be the solubilization of the fish dejections and fish feed leftovers into macro- and micronutrients, which the plants can absorb. The members of the genera Flavobacterium and Sphingobacterium could participate in the decomposition of organic matter [39]. The Saprospiraceae family is typically found in aquatic environment, such as wastewater treatment plants [39,45,46], and could be involved in the degradation of complex carbon molecules, such as proteins [45,46]. Many other detected genera include heterotrophic organisms able to degrade biomass in the system, as well.
Several of the observed OTUs were related to a role in the nitrogen cycle. The genus Nitrospira was detected in all biofilter samples. Nitrospira is commonly known as a NOB [11,12,14,47]. Daims et al. [14] showed that certain strains of the Nitrospira genus could actually be complete nitrifiers, i.e., able to oxidize ammonia to nitrate without the help of AOB, a process now known under the name COMAMMOX. Denitrification has also been often observed in aquaculture and aquaponics [48,49]. Members of the genus Arcobacter are known to perform denitrification [50] and have been particularly found in the denitrifying biofilter of the BQF system (Figure 2). The phylum Planctomycetes has already been observed in recirculating aquaculture [51] and contains ANAMMOX bacteria [39,51]. However, with the current database used for assignment to genus level, it could not be confirmed that ANAMMOX bacteria were present in the examined systems.
4.3. Core Microbiomes
Bacterial populations in aquaponic systems were highly diverse whether between systems or between the different compartments therein. In each sample, all the taxa representing less than 1% of the total number of reads were discarded, and this represented at least 50% of the reads (Figure 2). Despite this diversity, a core microbiome common to all samples could be identified. Moreover, the core microbiomes were composed of 28 (7.6% of the total reads common to all biofilters) and 22 OTUs (6.1% of the total reads are common to all sumps) for the biofilter and sump samples, respectively. This brought forth the hypothesis that a common bacterial base may exist between all aquaponic systems despite differences in fish species, system layout, or fish feed.
Despite the differences in the bacterial communities due to system specificities, there were still similarities between all examined systems. A principal coordinate analysis combining the data from the tested systems with the data collected by Schmautz et al. [7] (Supplementary Material; Figure S3) showed that the samples collected from the plant roots, periphyton, and biofilter compartments of the Wädenswil Aquaponic system (ZHAW) clustered closely with the other samples. This showed that there was a common pattern concerning the composition of the bacteria community in diversified aquaponic systems located in Western Europe. It would then be interesting to broaden the study to systems situated worldwide and also on a longer period of time.
5. Conclusion and Perspectives
This study was one of the first investigations into the diversity of bacterial communities present in a variety of aquaponic and aquaculture systems. It offered a global overview of the microbial taxa therein and of the potential roles that microorganisms could play in plant care. Nevertheless, it was shown that the different system setups had a large influence on the bacterial communities, and it needs to be investigated in more detail which species performs what role in such systems.
As the currently available datasets were from a single time point and only limited compartments within single systems were sampled, a more comprehensive sampling of single systems over time would be required to study the influence of sample time and location within a system. This may explain the currently obtained data better within the frame of the operational differences, but helps us also to understand the biological processes taking place in a single system.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4441/11/2/260/s1, Supplemental material containing a description of the visited aquaponic and aquaculture systems, Table S1: Water quality parameters, Figure S1: Recirculating aquaculture system of the Integrated and Urban Plant Pathology Laboratory, Figure S2: Rarefaction curves of every samples indicating the number of observed OTUs, according to sequencing depth, Figure S3: Weighted UniFrac Principal Coordinates Analysis, including the eight visited systems and the Wädenswill Aquaponic System.
Author Contributions
M.E., S.M., and M.H.J. conceived and designed the experiments; M.E. performed the experiments; M.E., A.R.S., Z.S., and T.H.M.S. analyzed data; and M.E., A.R.S., S.M., Z.S., R.J., T.H.M.S. and M.H.J. wrote and edited the manuscript.
Funding
This research was funded by F.R.S.-F.N.R.S. M.E. is a F.R.S.-FNRS PhD research fellow (grant reference 30206153).
Acknowledgments
The authors would like to thank the different partners that agreed to let us visit and sample their systems: UrbanFarmers, The Hague, The Netherlands; Provincial Trial Centre for Vegetable Production, Kruishoutem, Belgium; Belgian Quality Fish, Dottignies, Belgium; Leibnitz–Institute of Freshwater Ecology and Inland Fisheries, Berlin, Germany; University of Wageningen, Wageningen, The Netherlands; and Inagro, Rumbeke–Beitem, Belgium. The authors would like to acknowledge the FRS–FNRS for financial support. THMS acknowledges the support by the Swiss National Science Foundation (SNF project 310030_169665) and by the Department of Life Sciences and Facility Management of the Zurich University of Applied Sciences (ZHAW) in Wädenswil, Switzerland.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
References
- Delaide, B.; Goddek, S.; Gott, J.; Soyeurt, H.; Haissam Jijakli, M.; Lalman, J.; Junge, R. Lettuce (Lactuca sativa L. var. Sucrine) growth performance in complemented aquaponic solution outperforms hydroponics. Water 2016, 8, 467. [Google Scholar] [CrossRef]
- Rakocy, J.E. Aquaponics-Integrating Fish and Plant Culture. In Aquaculture Production Systems; Tidwell, J.H., Ed.; John Wiley and Sons: Hoboken, NJ, USA, 2012; pp. 343–386. [Google Scholar]
- Rakocy, J.E.; Masser, M.P.; Losordo, T.M. Recirculating aquaculture tank production systems: Aquaponics-integrating fish and plant culture. SRAC Publ. South Reg. Aquac. Cent. 2006, 16, 454. [Google Scholar]
- Delaide, B.; Delhaye, G.; Dermience, M.; Gott, J.; Soyeurt, H.; Jijakli, M.H. Plant and fish production performance, nutrient mass balances, energy and water use of the PAFF Box, a small-scale aquaponic system. Aquac. Eng. 2017, 78, 130–139. [Google Scholar] [CrossRef]
- Buzby, K.M.; Lin, L.S. Scaling aquaponic systems: Balancing plant uptake with fish output. Aquac. Eng. 2014, 63, 39–44. [Google Scholar] [CrossRef]
- Schmautz, Z.; Loeu, F.; Liebisch, F.; Graber, A.; Mathis, A.; Bulc, T.G.; Junge, R. Tomato productivity and quality in aquaponics: Comparison of three hydroponic methods. Water 2016, 8, 533. [Google Scholar] [CrossRef]
- Schmautz, Z.; Graber, A.; Jaenicke, S.; Goesmann, A.; Junge, R.; Smits, T.H.M. Microbial diversity in different compartments of an aquaponics system. Arch. Microbiol. 2017, 199, 613–620. [Google Scholar] [CrossRef]
- Graber, A.; Junge, R. Aquaponic Systems: Nutrient recycling from fish wastewater by vegetable production. DES 2009, 246, 147–156. [Google Scholar] [CrossRef]
- Timmons, M.B.; Ebeling, J.M. Recirculating Aquaculture, 3rd ed.; Ithaca Publishing: New York, NY, USA, 2013; ISBN 978-0-9712646. [Google Scholar]
- Resh, H.M. Hydroponic Food Production: A Definitive Guidebook for the Advanced Home Gardener and the Commercial Hydroponic Grower; CRC Press: Boca Raton, FL, USA, 2013; ISBN 1439878692. [Google Scholar]
- Rurangwa, E.; Verdegem, M.C.J. Microorganisms in recirculating aquaculture systems and their management. Rev. Aquac. 2015, 7, 117–130. [Google Scholar] [CrossRef]
- Itoi, S.; Ebihara, N.; Washio, S.; Sugita, H. Nitrite-oxidizing bacteria, Nitrospira, distribution in the outer layer of the biofilm from filter materials of a recirculating water system for the goldfish Carassius auratus. Aquaculture 2007, 264, 297–308. [Google Scholar] [CrossRef]
- Bartelme, R.P.; McLellan, S.L.; Newton, R.J. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528. [Google Scholar] [CrossRef]
- Hu, B.; Shen, L.; Xu, X.; Zheng, P. Anaerobic ammonium oxidation (anammox) in different natural ecosystems: Table 1. Biochem. Soc. Trans. 2011, 39, 1811–1816. [Google Scholar] [CrossRef]
- Goddek, S.; Schmautz, Z.; Scott, B.; Delaide, B.; Keesman, K.; Wuertz, S.; Junge, R. The Effect of Anaerobic and Aerobic Fish Sludge Supernatant on Hydroponic Lettuce. Agronomy 2016, 6, 37. [Google Scholar] [CrossRef]
- Yogev, U.; Sowers, K.R.; Mozes, N.; Gross, A. Nitrogen and carbon balance in a novel near-zero water exchange saline recirculating aquaculture system. Aquaculture 2017, 467, 118–126. [Google Scholar] [CrossRef]
- Schneider, O.; Sereti, V.; Eding, E.H.; Verreth, J.A.J. Analysis of nutrient flows in integrated intensive aquaculture systems. Aquac. Eng. 2004, 32, 379–401. [Google Scholar] [CrossRef]
- Jorquera, M.; Martínez, O.; Maruyama, F.; Marschner, P.; de la Luz Mora, M. Current and Future Biotechnological Applications of Bacterial Phytases and Phytase-Producing Bacteria. Microb. Environ. 2008, 23, 182–191. [Google Scholar] [CrossRef]
- Gravel, V.; Dorais, M.; Dey, D.; Vandenberg, G. Fish effluents promote root growth and suppress fungal diseases in tomato transplants. Can. J. Plant Sci. 2015, 95, 427–436. [Google Scholar] [CrossRef]
- Sirakov, I.; Lutz, M.; Graber, A.; Mathis, A.; Staykov, Y.; Smits, T.; Junge, R. Potential for Combined Biocontrol Activity against Fungal Fish and Plant Pathogens by Bacterial Isolates from a Model Aquaponic System. Water 2016, 8, 518. [Google Scholar] [CrossRef]
- Yildiz, H.Y.; Keskin, E.; Bekcan, S.; Kaynar, S.; Parisi, G. Molecular Identification of Dominant Bacterial Taxa in the Aquaponic System with Co-Culture of Tilapia (Oreochromis Aureus) and Tomato (Solanum lycopersicum ) Using Environmental DNA. Water 2017, 9, 1–8. [Google Scholar]
- Munguia-Fragozo, P.; Alatorre-Jacome, O.; Rico-Garcia, E.; Torres-Pacheco, I.; Cruz-Hernandez, A.; Ocampo-Velazquez, R.V.; Garcia-Trejo, J.F.; Guevara-Gonzalez, R.G. Perspective for Aquaponic Systems: “omic” Technologies for Microbial Community Analysis. BioMed Res. Int. 2015. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E. The family Deinococcaceae. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 613–615. [Google Scholar]
- Tsuchiya, C.; Sakata, T.; Sugita, H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2008, 46, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Baldani, J.I.; Rouws, L.; Magalhaes Cruz, L.; Lopes Olivares, F.; Schmid, M.; Hartmann, A. The Family Oxalobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–1012. ISBN 9783642301971. [Google Scholar]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Davis, M.J.; Graves Gillaspie, A.; Vidaver, A.K.; Harris’, R.W. Clavibacter: A New Genus Containing Some Phytopathogenic Coryneform Bacteria Pathogens That Cause Ratoon Stunting Disease of Sugarcane and Bermudagrass Stunting Disease? Int. J. Syst. Bacteriol. 1984, 84, 107–117. [Google Scholar] [CrossRef]
- Bittsanszky, A.; Uzinger, N.; Gyulai, G.; Mathis, A.; Junge, R.; Villarroel, M.; Kotzen, B.; Komives, T. Nutrient supply of plants in aquaponic systems. Ecocycles 2016, 2, 17–20. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Ye, Y.; Zhou, Y.; Fodor, A.A. A Core Human Microbiome as Viewed through 16S rRNA Sequence Clusters. PLoS ONE 2012, 7, e34242. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef]
- Dennert, F.; Imperiali, N.; Staub, C.; Schneider, J.; Laessle, T.; Zhang, T.; Wittwer, R.; van der Heijden, M.G.A.; Smits, T.H.M.; Schlaeppi, K.; et al. Conservation tillage and organic farming induce minor variations in Pseudomonas abundance, their antimicrobial function and soil disease resistance. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Evtushenko, L.; Takeuchi, M. The family Microbacteriaceae. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 1020–1098. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microb. 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Hynes, R.K.; Leung, G.C.Y.; Hirkala, D.L.M.; Nelson, L.M. Isolation, selection, and characterization of beneficial rhizobacteria from pea, lentil, and chickpea grown in western Canada. Can. J. Microbiol. 2008, 54, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Kolton, M.; Erlacher, A.; Berg, G.; Cytryn, E. The Flavobacterium Genus in the Plant Holobiont: Ecological, Physiological, and Applicative Insights. In Microbial Models: From Environmental to Industrial Sustainability; Springer: Singapore city, Singapore, 2016; pp. 189–207. [Google Scholar]
- Liu, J.; Lu, Z.; Yang, J.; Xing, M.; Yu, F.; Guo, M. Effect of earthworms on the performance and microbial communities of excess sludge treatment process in vermifilter. Bioresour. Technol. 2012, 117, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Soltani, A.-A.; Khavazi, K.; Asadi-Rahmani, H.; Omidvari, M.; Abaszadeh Dahaji, P.; Mirhoseyni, H. Plant Growth Promoting Characteristics in Some Flavobacterium spp. Isolated from Soils of Iran. J. Agric. Sci. 2010, 2, 106–115. [Google Scholar] [CrossRef]
- Kim, J.T.; Han, K.S.; Kim, B.R.; Kim, J.S.; Lee, B.C.; Yang, E.S.; Kwon, K.H.; Son, J.R.; Kim, H.G.I.; Yu, S.H. Biological Control of Plant Diseases Using Flavobacterium Hercynium Epb-C313; Uliège Library: Liège, Belgium, 2010. [Google Scholar]
- Reichenbach, H. The Lysobacter Genus. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 939–957. [Google Scholar]
- Lee, K.-J.; Oh, B.-T.; Seralathan, K.-K. Advances in Plant Growth Promoting Rhizobacteria for Biological Control of Plant Diseases. In Bacteria in Agrobiology: Disease Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; p. 495. [Google Scholar]
- Folman, L.B.; Postma, J.; van Veen, J.A. Characterisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 2003, 158, 107–115. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Nielsen, P.H. 70 The Family Saprospiraceae. In The Prokaryotes -Other Major Linages of Bacteria and the Archea; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Xia, Y.; Kong, Y.; Thomsen, T.R.; Nielsen, P.H. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (“Candidatus epiflobacter” spp.) in activated sludge. Appl. Environ. Microbiol. 2008, 74, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, W.; Huang, J.; He, S.; Yan, Y.; Zhu, W.; Wu, S.; Zhang, X. Nitrogen removal by the enhanced floating treatment wetlands from the secondary effluent. Bioresour. Technol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Monsees, H.; Keitel, J.; Paul, M.; Kloas, W.; Wuertz, S. Potential of aquacultural sludge treatment for aquaponics: Evaluation of nutrient mobilization under aerobic and anaerobic conditions. Aquac. Environ. Interact. 2017, 9, 9–18. [Google Scholar] [CrossRef]
- Wongkiew, S.; Hu, Z.; Chandran, K.; Lee, J.W.; Khanal, S.K. Nitrogen transformations in aquaponic systems: A review. Aquac. Eng. 2017, 76, 9–19. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Huang, W.; Wang, Y.; Zhou, J. Bacterial community structure in simultaneous nitrification, denitrification and organic matter removal process treating saline mustard tuber wastewater as revealed by 16S rRNA sequencing. Bioresour. Technol. 2017, 228, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, M.A.H.J.; Harhangi, H.R.; Van De Pas-Schoonen, K.; Van De Vossenberg, J.; Flik, G.; Jetten, M.S.M.; Klaren, P.H.M.; Op Den Camp, H.J.M. Biodiversity of N-cycle bacteria in nitrogen removing moving bed biofilters for freshwater recirculating aquaculture systems. Aquaculture 2010, 306, 177–184. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).