Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes
Abstract
1. Introduction: Benefits of Water Reuse
- (i)
- Current regulations pertaining to microbiological water quality
- (ii)
- Monitoring and surveillance associated with water reuse
- (iii)
- Risk assessment of ARB and ARGs
- (iv)
- Mitigation approaches to safeguard water quality for reuse
2. Current Regulations Pertaining to Microbiological Water Quality
3. Monitoring and Surveillance of Antibiotic Resistance
4. Advancing Risk Assessment Frameworks for ARB and ARGs in Recycled Water
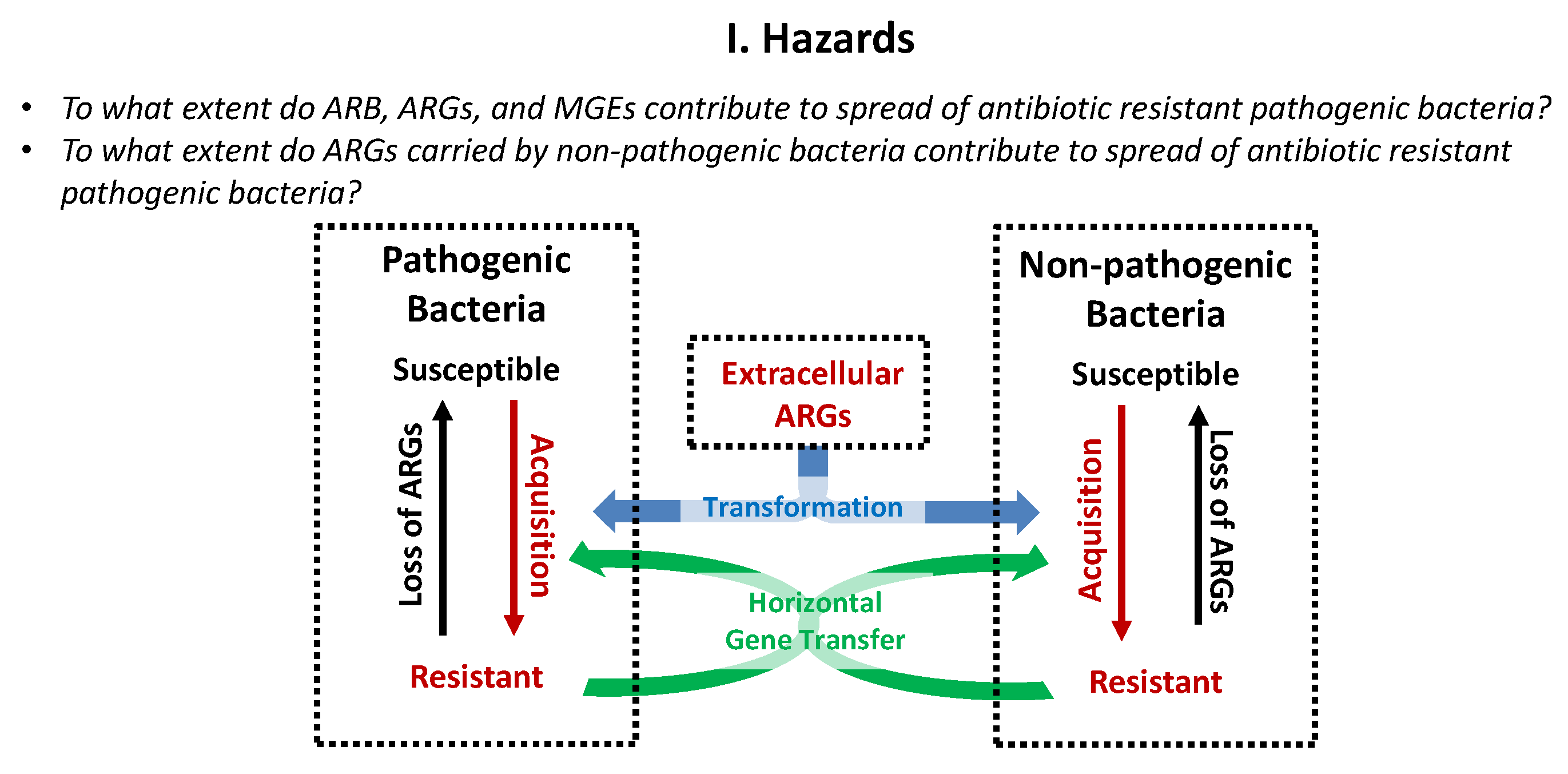
4.1. Hazards
4.1.1. Bacteria
4.1.2. Genes
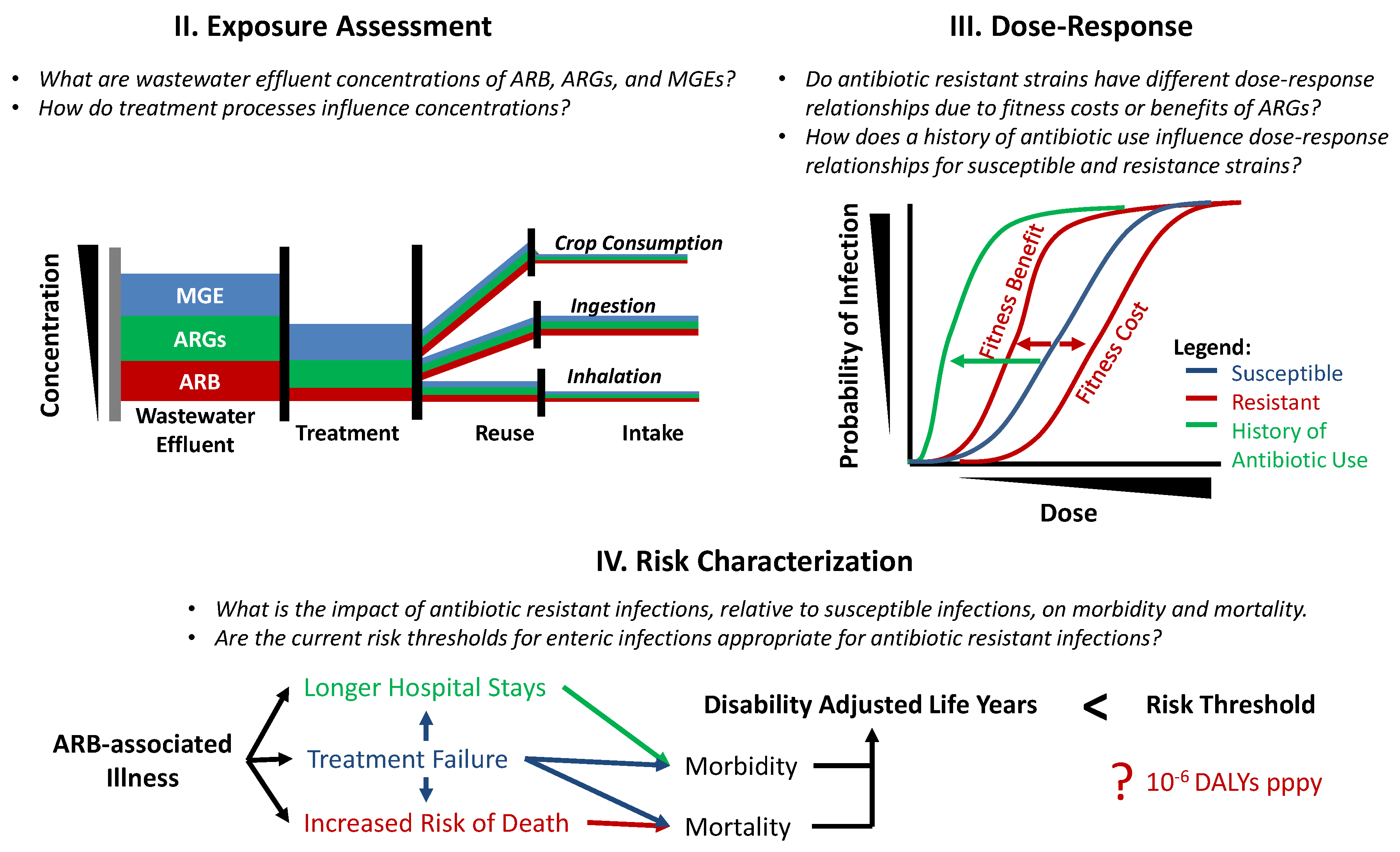
4.2. Exposures
4.3. Dose-Response Relationships
4.4. Risk Characterization and Management
5. Mitigation Strategies for Antimicrobial Resistance in Recycled Water
5.1. Source Prevention as a Barrier to Downstream Proliferation of ARB and ARGs
5.2. Treatment Technologies to Remove ARB and ARGs
6. Concluding Statement
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Jimenez, B.; Asano, T. Water Reuse: An International Survey of Current Practice, Issues and Needs (Scientific and Technical Report); IWA Publishing: London, UK, 2008. [Google Scholar]
- AQUASTAT. FAO’s Global Water Information System. Available online: http://www.fao.org/nr/water/aquastat/infographics/Infographics_all_eng.pdf (accessed on 5 October 2017).
- Thebo, A.L.; Drechsel, P.; Lambin, E.F. Global assessment of urban and peri-urban agriculture: Irrigated and rainfed croplands. Environ. Res. Lett. 2014, 9, 114002. [Google Scholar] [CrossRef]
- Pescod, M.B. Wastewater Treatment and Use in Agriculture. Fao Irrigation and Drainage Paper 47; FAO of the United Nations: Rome, Italy, 1992; p. 125. [Google Scholar]
- Thebo, A.L.; Drechsel, P.; Lambin, E.F.; Nelson, K.L. A global, spatially-explicit assessment of irrigated croplands influenced by urban wastewater flows. Environ. Res. Lett. 2017, 12, 074008. [Google Scholar] [CrossRef]
- Nuclear Regulatory Commission (NRC). Water Reuse: Potential for Expanding the Nation’s Water Supply through Reuse of Municipal Wastewater. Available online: https://www.nap.edu/catalog/13303/water-reuse-potential-for-expanding-the-nations-water-supply-through (accessed on 9 November 2017).
- International Water Association (IWA). History of Activated Sludge. Available online: http://www.iwa100as.org/history.php (accessed on 5 October 2017).
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473–474, 619–641. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.O.; Moreira, N.F.; Ribeiro, A.R.; Pereira, M.F.; Silva, A.M. Occurrence and removal of organic micropollutants: An overview of the watch list of eu decision 2015/495. Water Res. 2016, 94, 257–279. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Burgmann, H.; Sorum, H.; Norstrom, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Poirel, L.; Bessa, L.J.; Barbosa-Vasconcelos, A.; da Costa, P.M.; Nordmann, P. VIM-1, VIM-34, and IMP-8 carbapenemase-producing escherichia coli strains recovered from a Portuguese river. Antimicrob. Agents Chemother. 2016, 60, 2585–2586. [Google Scholar] [CrossRef] [PubMed]
- Mahon, B.M.; Brehony, C.; McGrath, E.; Killeen, J.; Cormican, M.; Hickey, P.; Keane, S.; Hanahoe, B.; Dolan, A.; Morris, D. Indistinguishable NDM-producing escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Eurosurveillance 2017, 22, 30513. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J.; Amezquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M. Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-direct proportionality between abundance and risk. Trends Microbiol. 2017, 25, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhao, Z.; Hernandez, D.; Jiang, S. Near real-time flow cytometry monitoring of bacterial and viral removal efficiencies during water reclamation processes. Water 2016, 8, 464. [Google Scholar] [CrossRef]
- Kokjohn, T.A.; Sayler, G.S. Attachment and replication of pseudomonas aeruginosa bacteriophages under conditions simulating aquatic environments. J. Gen. Microbiol. 1991, 137, 661–666. [Google Scholar] [CrossRef]
- Muniesa, M.; Jofre, J. Factors influencing the replication of somatic coliphages in the water environment. Antonie Van Leeuwenhoek 2004, 86, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.V.; Ripp, S.; Replicon, J.; Ogunseitan, O.A.; Kokjohn, T.A. Virus-mediated gene transfer in freshwater environments. In Gene Transfers and Environment: Proceedings of the Third European Meeting on Bacterial Genetics and Ecology (bageco-3), Villefranche-sur-mer, France, 20–22 November 1991; Gauthier, M.J., Ed.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 51–62. [Google Scholar]
- European Economic Community (EEC). Concerning Urban Wastewater Treatment, Council Directive 91/271/EEC, 1991.
- Becerra-Castro, C.; Lopes, A.R.; Vaz-Moreira, I.; Silva, E.F.; Manaia, C.M.; Nunes, O.C. Wastewater reuse in irrigation: A microbiological perspective on implications in soil fertility and human and environmental health. Environ. Int. 2015, 75, 117–135. [Google Scholar] [CrossRef] [PubMed]
- European Economic Community (EEC). Amending council directive 91/271/EEC, Commission directive 98/15/EC, 1998.
- European Economic Community (EEC). Establishing a framework for community action in the field of water policy–EU water framework, Directive (2000/60/EC), 2000.
- European Economic Community (EEC). Amending directives 2000/60/EC and 2008/105/EC as regards priority substances in the field of water policy, Directive 2013/39/EU, 2013.
- European Economic Community (EEC). On the quality of water intended for human consumption, Council directive 98/83/EC, 1998.
- World Health Organization (WHO). Quantitative Microbial Risk Assessment: Application for Water Safety Management; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization (WHO). Guidelines for Drinking Water Quality: Fourth Edition Incorporating the First Addendum. Available online: http://www.who.int/water_sanitation_health/publications/drinking-water-quality-guidelines-4-including-1st-addendum/en/ (accessed on 17 December 2017).
- World Health Organization (WHO). Who Guidelines for the Safe Use of Wastewater, Excreta and Greywater; WHO: Geneva, Switzerland, 2006. [Google Scholar]
- Manaia, C.M.; Macedo, G.; Fatta-Kassinos, D.; Nunes, O.C. Antibiotic resistance in urban aquatic environments: Can it be controlled? Appl. Microbiol. Biotechnol. 2016, 100, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Fontaneto, D.; Doppelbauer, J.; Corno, G. Fitness and recovery of bacterial communities and antibiotic resistance genes in urban wastewaters exposed to classical disinfection treatments. Environ. Sci. Technol. 2016, 50, 10153–10161. [Google Scholar] [CrossRef] [PubMed]
- McKinney, C.W.; Pruden, A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef] [PubMed]
- Washington, J.A. Principles of diagnosis. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Garner, E.; Wallace, J.S.; Argoty, G.A.; Wilkinson, C.; Fahrenfeld, N.; Heath, L.S.; Zhang, L.; Arabi, M.; Aga, D.S.; Pruden, A. Metagenomic profiling of historic Colorado front range flood impact on distribution of riverine antibiotic resistance genes. Sci. Rep. 2016, 6, 38432. [Google Scholar] [CrossRef] [PubMed]
- Gatica, J.; Tripathi, V.; Green, S.; Manaia, C.M.; Berendonk, T.; Cacace, D.; Merlin, C.; Kreuzinger, N.; Schwartz, T.; Fatta-Kassinos, D.; et al. High throughput analysis of integron gene cassettes in wastewater environments. Environ. Sci. Technol. 2016, 50, 11825–11836. [Google Scholar] [CrossRef] [PubMed]
- Balcazar, J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014, 10, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.Y.; Al-Jassim, N.; Ansari, M.I.; Mackie, R.I. Environmental and public health implications of water reuse: Antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes. Antibiotics 2013, 2, 367–399. [Google Scholar] [CrossRef] [PubMed]
- Harb, M.; Hong, P.-Y. Anaerobic membrane bioreactor effluent reuse: A review of microbial safety concerns. Fermentation 2017, 3, 39. [Google Scholar] [CrossRef]
- Garmendia, L.; Hernandez, A.; Sanchez, M.B.; Martinez, J.L. Metagenomics and antibiotics. Clin. Microbiol. Infect. 2012, 18 Suppl 4, 27–31. [Google Scholar] [CrossRef]
- Martinez, J.L.; Coque, T.M.; Baquero, F. Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat. Rev. Microbiol. 2015, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [PubMed]
- Wang, F.H.; Qiao, M.; Su, J.Q.; Chen, Z.; Zhou, X.; Zhu, Y.G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Singh, G.; Heath, L.S.; Pruden, A.; Xiao, W.; Zhang, L. Metastorm: A public resource for customizable metagenomics annotation. PLoS ONE 2016, 11, e0162442. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. Card 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jiang, X.; Chai, B.; Ma, L.; Li, B.; Zhang, A.; Cole, J.R.; Tiedje, J.M.; Zhang, T. ARGS-OAP: Online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured arg-database. Bioinformatics 2016, 32, 2346–2351. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.G.; Balkwill, D.L. Antibiotic resistance in bacteria isolated from the deep terrestrial subsurface. Microb. Ecol. 2009, 57, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.; Boucher, Y.; Labbate, M.; Holmes, A.; Krishnan, S.; Holley, M.; Stokes, H.W. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008, 190, 5095–5100. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Who Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 5 October 2017).
- Haas, C.N.; Rose, J.B.; Gerba, C.P. Quantitative Microbial Risk Assessment, 2nd ed.; World Health Organization (WHO): Geneva, Switzerland, 2014; p. 440. [Google Scholar]
- Pruden, A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 2014, 48, 5–14. [Google Scholar] [CrossRef] [PubMed]
- ECDC/EMEA. The Bacterial Challenge: Time to React; ECDC/EMEA: Solna, Sweden, 2009; ISBN 9789291931934. [Google Scholar]
- Al-Jassim, N.; Mantilla-Calderon, D.; Wang, T.; Hong, P.Y. Inactivation and gene expression of a virulent wastewater escherichia coli strain and the nonvirulent commensal escherichia coli DSM1103 strain upon solar irradiation. Environ. Sci. Technol. 2017, 51, 3649–3659. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.M.; Ailes, E.; Roberts, V.A.; Hill, V.; Hilborn, E.D.; Craun, G.F.; Rajasingham, A.; Kahler, A.; Garrison, L.; Hicks, L.; et al. Surveillance for waterborne disease outbreaks associated with drinking water―United States, 2007–2008. MMWR Surveill. Summ. 2011, 60, 38–68. [Google Scholar] [PubMed]
- Parsley, L.C.; Consuegra, E.J.; Kakirde, K.S.; Land, A.M.; Harper, W.F., Jr.; Liles, M.R. Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl. Environ. Microbiol. 2010, 76, 3753–3757. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Imamovic, L.; Jofre, J.; Schmidt, H.; Serra-Moreno, R.; Muniesa, M. Phage-mediated Shiga toxin 2 gene transfer in food and water. Appl. Environ. Microbiol. 2009, 75, 1764–1768. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Paul, J.H. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 1998, 64, 2780–2787. [Google Scholar] [PubMed]
- Ogunseitan, O.A. Genetic transduction in freshwater ecosystems. Freshwater Biol. 2008, 53, 1228–1239. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Calero-Caceres, W.; Muniesa, M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 2015, 79, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Penades, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Davison, J. Genetic exchange between bacteria in the environment. Plasmid 1999, 91, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Keen, E.C.; Bliskovsky, V.V.; Malagon, F.; Baker, J.D.; Prince, J.S.; Klaus, J.S.; Adhya, S.L. Novel “superspreader” bacteriophages promote horizontal gene transfer by transformation. MBio 2017, 8, e02115–e02116. [Google Scholar] [CrossRef] [PubMed]
- Al-Jassim, N.; Ansari, M.I.; Harb, M.; Hong, P.Y. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: Is the treated wastewater safe to reuse for agricultural irrigation? Water Res. 2015, 73, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Jumat, M.R.; Hasan, N.A.; Subramanian, P.; Heberling, C.; Colwell, R.R.; Hong, P.Y. Membrane bioreactor-based wastewater treatment plant in Saudi Arabia: Reduction of viral diversity, load, and infectious capacity. Water 2017, 9, 534. [Google Scholar] [CrossRef]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Vogwill, T.; MacLean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Mantilla-Calderon, D.; Hong, P.Y. Fate and persistence of a pathogenic ndm-1-positive escherichia coli strain in anaerobic and aerobic sludge microcosms. Appl. Environ. Microbiol. 2017, 83, e00640-17. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Danilchanka, O.; Guillard, T.; Cattoir, V.; Aschard, H.; Fu, Y.; Angoulvant, F.; Messika, J.; Ricard, J.D.; Mekalanos, J.J.; et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci. Transl. Med. 2015, 7, 297ra114. [Google Scholar] [CrossRef] [PubMed]
- Donskey, C.J. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin. Infect Dis. 2006, 43 (Suppl. 2), S62–S69. [Google Scholar] [CrossRef]
- Nordgard, L.; Brusetti, L.; Raddadi, N.; Traavik, T.; Averhoff, B.; Nielsen, K.M. An investigation of horizontal transfer of feed introduced DNA to the aerobic microbiota of the gastrointestinal tract of rats. BMC Res. Notes 2012, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Nordgard, L.; Nguyen, T.; Midtvedt, T.; Benno, Y.; Traavik, T.; Nielsen, K.M. Lack of detectable DNA uptake by bacterial gut isolates grown in vitro and by Acinetobacter baylyi colonizing rodents in vivo. Environ. Biosafety Res. 2007, 6, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, A.; Brusetti, L.; Arioli, S.; Nielsen, K.M.; Tamagnini, I.; Tamburini, A.; Sorlini, C.; Daffonchio, D. Detection of feed-derived maize DNA in goat milk and evaluation of the potential of horizontal transfer to bacteria. Eur. Food Res. Technol. 2008, 227, 1699–1709. [Google Scholar] [CrossRef]
- Wilcks, A.; Jacobsen, B.B. Lack of detectable DNA uptake by transformation of selected recipients in mono-associated rats. BMC Res. Notes 2010, 3, 49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Enault, F.; Briet, A.; Bouteille, L.; Roux, S.; Sullivan, M.B.; Petit, M.A. Phages rarely encode antibiotic resistance genes: A cautionary tale for virome analyses. ISME J. 2017, 11, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Hamilton, A.J.; Jiang, S.C. Assessment of public health risk associated with viral contamination in harvested urban stormwater for domestic applications. Sci. Total Environ. 2015, 523, 95–108. [Google Scholar] [CrossRef] [PubMed]
- CDC. One Health. Available online: https://www.cdc.gov/onehealth/index.html (accessed on 9 November 2017).
- USWaterAlliance. One Water Roadmap: The Sustainable Management of Life’s Most Essential Resource. Available online: http://uswateralliance.org//one-water/roadmap (accessed on 9 November 2017).
- Negreanu, Y.; Pasternak, Z.; Jurkevitch, E.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Environ. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef] [PubMed]
- Fahrenfeld, N.; Ma, Y.; O’Brien, M.; Pruden, A. Reclaimed water as a reservoir of antibiotic resistance genes: Distribution system and irrigation implications. Front. Microbiol. 2013, 4, 130. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Hu, H.W.; Shi, X.Z.; Wang, J.T.; Han, L.L.; Chen, D.; He, J.Z. Impacts of reclaimed water irrigation on soil antibiotic resistome in urban parks of victoria, australia. Environ. Pollut. 2016, 211, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Larsson, D.G.J. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Burgmann, H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Kotlarska, E.; Luczkiewicz, A.; Pisowacka, M.; Burzynski, A. Antibiotic resistance and prevalence of class 1 and 2 integrons in escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. Int. 2015, 22, 2018–2030. [Google Scholar] [CrossRef] [PubMed]
- Narciso-da-Rocha, C.; Varela, A.R.; Schwartz, T.; Nunes, O.C.; Manaia, C.M. BlaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital–urban wastewater treatment plant system. J. Glob. Antimicrob. Resist. 2014, 2, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Timraz, K.; Xiong, Y.H.; Al Qarni, H.; Hong, P.Y. Removal of bacterial cells, antibiotic resistance genes and integrase genes by on-site hospital wastewater treatment plants: Surveillance of treated hospital effluent quality. Environ. Sci.-Wat. Res. 2017, 3, 293–303. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Boulund, F.; Fick, J.; Kristiansson, E.; Larsson, D.G. Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front. Microbiol. 2014, 5, 648. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T. Strategies to assess and minimize the biological risk of antibiotic resistance in the environment. In Antimicrobial Resistance in the Environment; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp. 251–264. [Google Scholar]
- Garner, E.; Zhu, N.; Strom, L.; Edwards, M.; Pruden, A. A human exposome framework for guiding risk management and holistic assessment of recycled water quality. Environ. Sci. Water Res. Technol. 2016, 2, 580–598. [Google Scholar] [CrossRef]
- Ahammad, Z.S.; Sreekrishnan, T.R.; Hands, C.L.; Knapp, C.W.; Graham, D.W. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the upper ganges river. Environ. Sci. Technol. 2014, 48, 3014–3020. [Google Scholar] [CrossRef] [PubMed]
- Mantilla-Calderon, D.; Jumat, M.R.; Wang, T.; Ganesan, P.; Al-Jassim, N.; Hong, P.Y. Isolation and characterization of NDM-positive escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 2016, 60, 5223–5231. [Google Scholar] [CrossRef] [PubMed]
- Novo, A.; Manaia, C.M. Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2010, 87, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zeng, S.; He, M.; Gu, A.Z. Water disinfection byproducts induce antibiotic resistance-role of environmental pollutants in resistance phenomena. Environ. Sci. Technol. 2016, 50, 3193–3201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, A.Z.; He, M.; Li, D.; Chen, J. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 2017, 51, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Larsson, D.G.; Amezquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R.; et al. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.Q.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.X.; Li, F.X.; Hou, J.; Mu, Q.H.; Alvarez, P.J.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.B.; Guo, M.T.; Wei, W.J.; Yang, J. Reductions of bacterial antibiotic resistance through five biological treatment processes treated municipal wastewater. Environ. Sci. Pollut. Res. 2016, 23, 19495–19503. [Google Scholar] [CrossRef] [PubMed]
- Marcinek, H.; Wirth, R.; Muscholl-Silberhorn, A.; Gauer, M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 1998, 64, 626–632. [Google Scholar] [PubMed]
- Ferreira da Silva, M.; Tiago, I.; Verissimo, A.; Boaventura, R.A.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2006, 55, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jensen, J.N.; Aga, D.S.; Weber, A.S. Tetracycline as a selector for resistant bacteria in activated sludge. Chemosphere 2007, 66, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Książek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef] [PubMed]
- Neyestani, M.; Dickenson, E.; McLain, J.; Robleto, E.; Rock, C.; Gerrity, D. Impacts of solids retention time on trace organic compound attenuation and bacterial resistance to trimethoprim and sulfamethoxazole. Chemosphere 2017, 182, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Munck, C.; Albertsen, M.; Telke, A.; Ellabaan, M.; Nielsen, P.H.; Sommer, M.O. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 2015, 6, 8452. [Google Scholar] [CrossRef] [PubMed]
- WaterSecure. Chlorine disinfection. In WaterVal Validation Protocol; Australian WaterSecure Innovations Ltd.: Brisbane, Australia, 2017. [Google Scholar]
- USEPA. Disinfection Profiling and Benchmarking Guidance Manual; Agency, U.S.E.P., Ed.; U.S.EPA: Washington, DC, USA, 1999.
- Guo, M.-T.; Yuan, Q.-B.; Yang, J. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.E.; Tobin, R.S.; Junkins, B.; Kushner, D.J. Effect of chlorination on antibiotic resistance profiles of sewage related bacteria. Appl. Environ. Microbiol. 1984, 48, 73–77. [Google Scholar] [PubMed]
- Oncu, N.B.; Menceloglu, Y.Z.; Balcioglu, I.A. Comparison of the effectiveness of chlorine, ozone, and photocatalytic disinfection in reducing the risk of antibiotic resistance pollution. J. Adv. Oxid. Technol. 2011, 14, 196–203. [Google Scholar]
- Suquet, C.; Warren, J.J.; Seth, N.; Hurst, J.K. Comparative study of HOCl-inflicted damage to bacterial DNA ex vivo and within cells. Arch. Biochem. Biophys. 2010, 493, 135–142. [Google Scholar] [CrossRef] [PubMed]
- USEPA. Ultraviolet disinfection guidance manual for the final long term 2 enhanced surface water treatment rule. In EPA 815-R-06-007; United States Environmental Protection Agency: Washington, DC, USA, 2006; p. 436. [Google Scholar]
- Luddeke, F.; Hess, S.; Gallert, C.; Winter, J.; Gude, H.; Loffler, H. Removal of total and antibiotic resistant bacteria in advanced wastewater treatment by ozonation in combination with different filtering techniques. Water Res. 2015, 69, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Sigmon, C.; Shin, G.A.; Mieog, J.; Linden, K.G. Establishing surrogate-virus relationships for ozone disinfection of wastewater. Environ. Eng. Sci. 2015, 32, 451–460. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512–513, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Chung, H.J.; Wen Di, D.Y.; Dodd, M.C.; Hur, H.G.; Lee, Y. Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Res. 2017, 123, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Grare, M.; Duval, R.E.; Dague, E. Nanoscale effects of antibiotics on p. aeruginosa. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Grare, M.; Jauvert, E.; Coutable, A.; Regnouf-de-Vains, J.B.; Mourer, M.; Duval, R.E.; Dague, E. Nanoscale analysis of the effects of antibiotics and CX1 on a pseudomonas aeruginosa multidrug-resistant strain. Sci. Rep. 2012, 2, 575. [Google Scholar] [CrossRef] [PubMed]
- Breazeal, M.V.; Novak, J.T.; Vikesland, P.J.; Pruden, A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2013, 47, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hong, P.Y. Removal of antibiotic-resistant bacteria and antibiotic resistance genes affected by varying degrees of fouling on anaerobic microfiltration membranes. Environ. Sci. Technol. 2017, 51, 12200–12209. [Google Scholar] [CrossRef] [PubMed]
- Shoener, B.D.; Zhong, C.; Greiner, A.D.; O. Khunjar, W.; Hong, P.-Y.; Guest, J.S. Design of anaerobic membrane bioreactors for the valorization of dilute organic carbon waste streams. Energy Environ. Sci. 2016, 9, 1102–1112. [Google Scholar] [CrossRef]
- Harb, M.; Hong, P.Y. Molecular-based detection of potentially pathogenic bacteria in membrane bioreactor (MBR) systems treating municipal wastewater: A case study. Environ. Sci. Pollut. Res. Int. 2017, 24, 5370–5380. [Google Scholar] [CrossRef] [PubMed]
- Harb, M.; Wei, C.H.; Wang, N.; Amy, G.; Hong, P.Y. Organic micropollutants in aerobic and anaerobic membrane bioreactors: Changes in microbial communities and gene expression. Bioresour. Technol. 2016, 218, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Christgen, B.; Yang, Y.; Ahammad, S.Z.; Li, B.; Rodriquez, D.C.; Zhang, T.; Graham, D.W. Metagenomics shows that low-energy anaerobic-aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater. Environ. Sci. Technol. 2015, 49, 2577–2584. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.W.; Crook, J.; Anderson, M.A.; Bull, R.J.; Drewes, J.E.; Haas, C.N.; Jakubowski, W.; McCarty, P.L.; Nelson, K.L.; Rose, J.B.; et al. Expert Panel Final Report: Evaluation of the Feasibility of Developing Uniform Water Recycling Criteria for Direct Potable Reuse; National Water Research Institute for the State Water Resources Control Board: Sacramento, CA, USA, 2016. [Google Scholar]
| Type of Water | Type of Parameters | References |
|---|---|---|
| Wastewater | Chemical oxygen demand, COD | [23,25] |
| Biochemical oxygen demand, BOD—(can be replaced by total organic carbon, TOC, or total oxygen demand, TOD) | ||
| (BOD5 at 20 °C) | ||
| Total suspended solids | ||
| Total phosphorus | ||
| Total nitrogen | ||
| Water for reuse | pH | See [24] for examples of guidelines for Italy, Spain, Portugal, France. |
| Electrical conductivity, EC | ||
| Sodium absorption rate, SAR | ||
| Total suspended solids, TSS | ||
| Biological oxygen demand, BOD | ||
| Chemical oxygen demand, COD | ||
| Total nitrogen, TN or Nitrate nitrogen, or N–NO3 | ||
| Phosphate | ||
| Sulphate | ||
| Faecal coliforms or Escherichia coli | ||
| Nematode eggs | ||
| Surface water | 45 priority substances or groups of substances, 21 of which classified as priority hazardous substance. | [26,27] |
| Includes plant protection products, biocides, metals and other groups like Polyaromatic Hydrocarbons (PAH) that are mainly incineration by-products and Polybrominated Biphenylethers (PBDE) that are used as flame retardants | ||
| Drinking water | A total of 48 microbiological, chemical and indicator parameters must be monitored and tested regularly | [28] |
| Microbiological: | ||
| Escherichia coli | ||
| Enterococci | ||
| Pseudomonas aeruginosa | ||
| Clostridium perfringens including spores | ||
| Other heterotrophic bacteria |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, P.-Y.; Julian, T.R.; Pype, M.-L.; Jiang, S.C.; Nelson, K.L.; Graham, D.; Pruden, A.; Manaia, C.M. Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. Water 2018, 10, 244. https://doi.org/10.3390/w10030244
Hong P-Y, Julian TR, Pype M-L, Jiang SC, Nelson KL, Graham D, Pruden A, Manaia CM. Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. Water. 2018; 10(3):244. https://doi.org/10.3390/w10030244
Chicago/Turabian StyleHong, Pei-Ying, Timothy R. Julian, Marie-Laure Pype, Sunny C. Jiang, Kara L. Nelson, David Graham, Amy Pruden, and Célia M. Manaia. 2018. "Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes" Water 10, no. 3: 244. https://doi.org/10.3390/w10030244
APA StyleHong, P.-Y., Julian, T. R., Pype, M.-L., Jiang, S. C., Nelson, K. L., Graham, D., Pruden, A., & Manaia, C. M. (2018). Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. Water, 10(3), 244. https://doi.org/10.3390/w10030244








