Modified U-Tube for Ruling out Naked DNA Transfer during Conjugation and Application in Antibiotic Resistance Genes Transfer Research
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
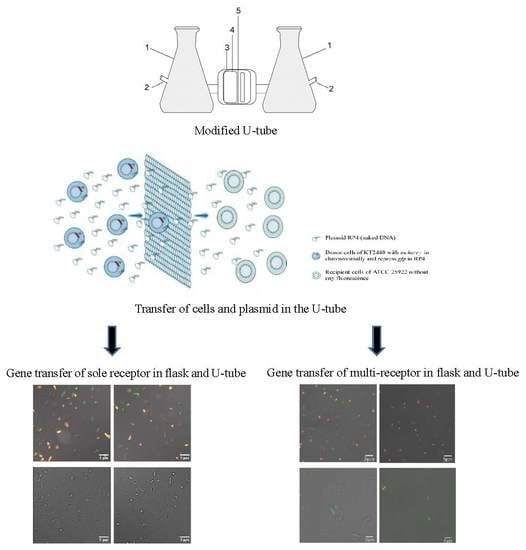
2.2. Modified U-Tube Device
2.3. Verification of U-Tube Performance
2.4. Gene Transfer Experiment
2.5. Sampling and Detection of the Gene Transfer
3. Results and Discussion
3.1. Transfer of Plasmid and Donor Strain in the U-Tube
3.2. Detection of Transconjugants in Gene Transfer Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J., Jr. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Chen, P.; Ding, R.; Zhang, P.; Li, X. Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in Beijing and Tianjin, China. Environ. Pollut. 2014, 193, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Knapp, C.W.; Christensen, B.T.; McCluskey, S.; Dolfing, J. Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 2016, 6, 21550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Chen, Y.; Zheng, X.; Su, Y.; Wan, R.; Yang, S. Distribution of tetracycline resistance genes in anaerobic treatment of waste sludge: The role of pH in regulating tetracycline resistant bacteria and horizontal gene transfer. Bioresour. Technol. 2016, 218, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Managaki, S.; Pärnänen, K.; Karkman, A.; Lyra, C.; Tamminen, M.; Suzuki, S.; Virta, M. Sulphonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS ONE. 2014, 9, e92702. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Taviani, E.; Ceccarelli, D.; Lazaro, N.; Bani, S.; Cappuccinelli, P.; Colwell, R.R.; Colombo, M.M. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Mivrobiol. Ecol. 2008, 64, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Mispagel, H.; Gray, J.T. Antibiotic resistance from wastewater oxidation ponds. Water Environ. Res. 2005, 77, 2996–3002. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Wolffs, P.F. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Nancharaiah, Y.V.; Wattiau, P.; Wuertz, S.; Bathe, S.; Mohan, S.V.; Wilderer, P.A.; Hausner, M. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl. Environ. Microb. 2003, 69, 4846–4852. [Google Scholar] [CrossRef]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.H.; Sørensen, S.J.; Smets, B.F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015, 9, 934–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakibaie, M.R.; Jalilzadeh, K.A.; Yamakanamardi, S.M. Horizontal transfer of antibiotic resistance genes among gram negative bacteria in sewage and lake water and influence of some physico-chemical parameters of water on conjugation process. J. Environ. Biol. 2009, 30, 45–49. [Google Scholar] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Bocer, T.; Mackiewicz, P.; Janska, H. A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots to mitochondrial genome of Phaseolus. Plant Mol. Biol. 2004, 56, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Chancey, S.T.; Agrawal, S.; Schroeder, M.R.; Farley, M.M.; Tettelin, H.; Stephens, D.S. Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumonia. Front. Microbiol. 2015, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, A.; Dai, T.; Li, F.; Xie, H.; Chen, L.; Wen, D. Cell-free DNA: a neglected source for antibiotic resistance genes spreading from WWTPs. Environ. Sci. Technol. 2018, 52, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Luo, Y.; Mathieu, J.; Wang, Q.; Feng, L.; Mu, Q.; Alvarez, P.J.J. Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ. Sci. Technol. 2013, 48, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fertil. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Norman, A.; Hansen, L.H.; Sørensen, S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yun, Z.; Ha, U.; Lee, S.; Park, H.; Kwon, E.E.; Chandran, K. Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations. Sci. Total Environ. 2014, 468, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mao, D.; Luo, Y. Ionic liquid facilitates the conjugative transfer of antibiotic resistance genes mediated by plasmid RP4. Environ. Sci. Technol. 2015, 49, 8731–8740. [Google Scholar] [CrossRef] [PubMed]
- Tejerizo, G.T.; Bañuelos, L.A.; Cervantes, L.; Gaytán, P.; Pistorio, M.; Romero, D.; Brom, S. Development of molecular tools to monitor conjugative transfer in rhizobia. J. Microbiol. Method 2015, 117, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Gulez, G.; Altıntaş, A.; Fazli, M.; Dechesne, A.; Workman, C.T.; Tolker-Nielsen, T.; Smets, B.F. Colony morphology and transcriptome profiling of Pseudomonas putida KT2440 and its mutants deficient in alginate or all EPS synthesis under controlled matric potentials. Microbiol. Open 2014, 3, 457–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klümper, U. Permissiveness of Soil Microbial Communities towards Broad Host Range Plasmids. Ph.D. Thesis, Technical University of Denmark, Lyngby, Demark, June 2015. [Google Scholar]
- Luo, Y.; Wang, Q.; Lu, Q.; Mu, Q.; Mao, D. An ionic liquid facilitates the proliferation of antibiotic resistance genes mediated by class І integrons. Environ. Sci. Technol. Lett. 2014, 1, 266–270. [Google Scholar] [CrossRef]
- Davis, B.D. Studies on nutritionally deficient bacterial mutants isolated by means of penicillin. Experientia 1950, 6, 41–50. [Google Scholar] [CrossRef]
- Cervantes, L.; Bustos, P.; Girard, L.; Santamaría, R.I.; Dávila, G.; Vinuesa, P.; Romero, D.; Brom, S. The conjugative plasmid of a bean-nodulating Sinorhizobium fredii strain is assembled from sequences of two Rhizobium plasmids and the chromosome of a Sinorhizobium strain. BMC Microbiol. 2011, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Jutkina, J.; Rutgersson, C.; Flach, C.F.; Larsson, D.J. An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci. Total Environ. 2016, 548, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, J.; Li, B.; Wen, X.; Liang, P.; Huang, X. A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 2018, 642, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, Q.; Mao, D.; Cui, Y.; Luo, Y. The horizontal transfer of antibiotic resistance genes is enhanced by ionic liquid with different structure of varying alkyl chain length. Front. Microbiol. 2015, 6, 864. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Yankelevich, T.; Dechesne, A.; Merkey, B.; Sternberg, C.; Smets, B.F. An individual-based approach to explain plasmid invasion in bacterial populations. FEMS Microbiol. Ecol. 2010, 75, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellanger, X.; Guilloteau, H.; Bonot, S.; Merlin, C. Demonstrating plasmid-based horizontal gene transfer in complex environmental matrices: A practical approach for a critical review. Sci. Total Environ. 2014, 493, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Musovic, S.; Dechesne, A.; Sørensen, J.; Smets, B.F. Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microbiol. 2010, 76, 4813–4818. [Google Scholar] [CrossRef] [PubMed]
- Dahlberg, C.; Bergström, M.; Hermansson, M. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 1998, 64, 2670–2675. [Google Scholar] [PubMed]
- Van Elsas, J.D.; Fry, J.; Hirsch, P.; Molin, S.; Thomas, C.M. Ecology of plasmid transfer and spread. In The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread, 1st ed.; CRC Press: Boca Raton, FL, USA, 2000; pp. 175–206. [Google Scholar]
- Yang, D.; Wang, J.; Qiu, Z.; Jin, M.; Shen, Z.; Chen, Z.; Li, J.W. Horizontal transfer of antibiotic resistance genes in a membrane bioreactor. J. Biotechnol. 2013, 167, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchrieser, C.; Charpentier, X. Induction of competence for natural transformation in Legionella pneumophila and exploitation for mutant construction. Methods Mol. Biol. 2013, 954, 183–195. [Google Scholar] [PubMed]
- Charpentier, X.; Kay, E.; Schneider, D.; Shuman, H.A. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 2011, 193, 1114–1121. [Google Scholar] [CrossRef] [PubMed]





| No. | Group | Device | Donor | Recipient | Sample Time (min) | Sampling Point | Detection |
|---|---|---|---|---|---|---|---|
| 1# | Verification | U-tube | RP4 * | Sterile LB | 15, 60, 180 | Recipient flask | PCR |
| 2# | P. putida (RP4) | E. coli | 15, 60, 180 | Recipient flask | PCR/plate counting | ||
| 3# | Flask | RP4 | Sterile LB | 15, 60, 180 | PCR | ||
| 4# | P. putida (RP4) | E. coli | 15, 60, 180 | PCR/plate counting | |||
| 5# | Gene Transfer | U-tube | P. putida (RP4) | E. coli | 180 | Recipient flask | FCM/CLSM |
| 6# | P. putida (RP4) | microcosms | 180 | Recipient flask | FCM/CLSM | ||
| 7# | Flask | P. putida (RP4) | E.coli | 180 | FCM/CLSM | ||
| 8# | P. putida (RP4) | microcosms | 180 | FCM/CLSM | |||
| 9# | Control | Flask | P. putida (RP4) | 180 | FCM/CLSM | ||
| 10# | E. coli | 180 | FCM/CLSM | ||||
| 11# | microcosms | 180 | FCM/CLSM | ||||
| Sampling Time and Results | Treatment Settings | ||||||
|---|---|---|---|---|---|---|---|
| Donor | 2# | 4# | |||||
| Sampling time (min) | 180 | 15 | 60 | 180 | 15 | 60 | 180 |
| Numbers of colony | 587 ± 6.1 | No colony | 358 ± 14.4 | 854 ± 13.4 | 1371 ± 28.9 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Liu, X.; Li, B.; Han, L.; Ma, X.; Meng, F.; Li, M. Modified U-Tube for Ruling out Naked DNA Transfer during Conjugation and Application in Antibiotic Resistance Genes Transfer Research. Water 2018, 10, 1313. https://doi.org/10.3390/w10101313
Zhang N, Liu X, Li B, Han L, Ma X, Meng F, Li M. Modified U-Tube for Ruling out Naked DNA Transfer during Conjugation and Application in Antibiotic Resistance Genes Transfer Research. Water. 2018; 10(10):1313. https://doi.org/10.3390/w10101313
Chicago/Turabian StyleZhang, Ning, Xiang Liu, Bing Li, Limei Han, Xuejiao Ma, Fanbin Meng, and Miao Li. 2018. "Modified U-Tube for Ruling out Naked DNA Transfer during Conjugation and Application in Antibiotic Resistance Genes Transfer Research" Water 10, no. 10: 1313. https://doi.org/10.3390/w10101313
APA StyleZhang, N., Liu, X., Li, B., Han, L., Ma, X., Meng, F., & Li, M. (2018). Modified U-Tube for Ruling out Naked DNA Transfer during Conjugation and Application in Antibiotic Resistance Genes Transfer Research. Water, 10(10), 1313. https://doi.org/10.3390/w10101313