Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants
Abstract
1. Introduction
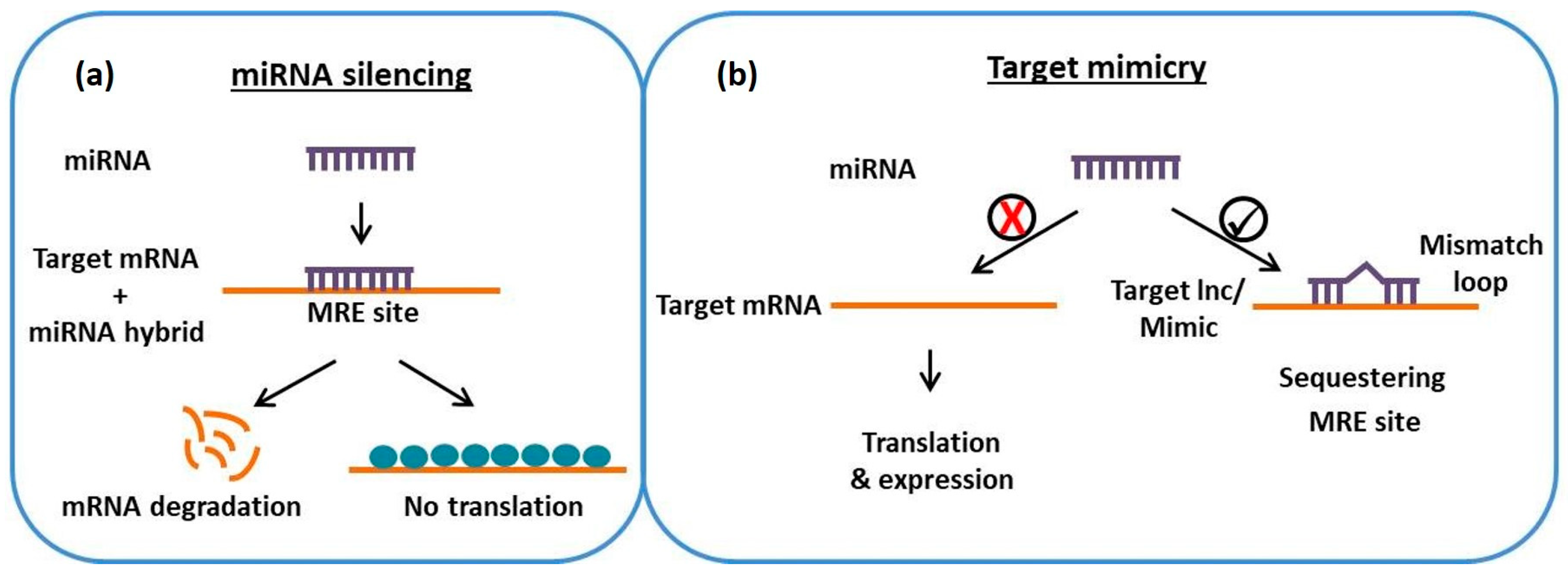
2. Role of lncRNAs as Endogenous Target Mimics for MicroRNAs
3. Long non-coding RNAs Expressed under Nitrogen and Phosphorus Deprivation
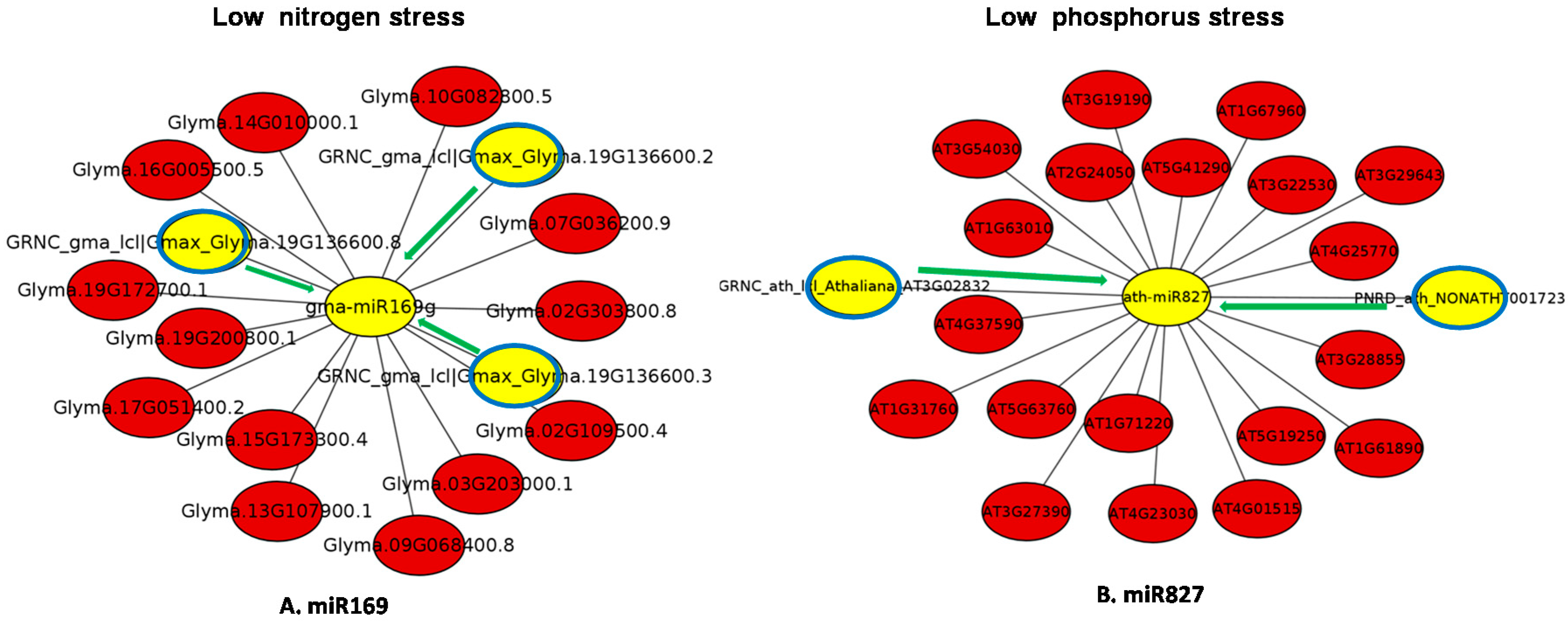
4. Putative Endogenous Target Mimics under Low Nitrogen and Phosphate Stress
5. Conclusions and Future Prospects
Funding
Conflicts of Interest
References
- Erdmann, V.A.; Barciszewska, M.Z.; Szymanski, M.; Hochberg, A.; de Groot, N.; Barciszewski, J. The non-coding RNAs as riboregulators. Nucleic Acids Res. 2001, 29, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and their roles in stress response in plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinform. 2015, 3, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Ravasi, T.; Suzuki, H.; Pang, K.C.; Katayama, S.; Furuno, M.; Okunishi, R.; Fukuda, S.; Ru, K.; Frith, M.C.; Gongora, M.M.; et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006, 16, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Lafreniere, R.G.; Powers, V.E.; Sebastio, G.; Ballabio, A.; Pettigrew, A.L.; Ledbetter, D.H.; Levy, E.; Craig, I.W.; Willard, H.F. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 1991, 349, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Allis, C.D. RNA meets chromatin. Gene. Dev. 2005, 19, 1635–1655. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, J.; Le Baccon, P.; Wutz, A.; Heard, E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006, 20, 2223–2237. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Chureau, C.; Samain, S.; Weissenbach, J.; Avner, P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 2006, 312, 1653–1655. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Li, G.; Son, J.; Xu, C.F.; Margueron, R.; Neubert, T.A.; Reinberg, D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long noncoding RNA expression. Nucleic Acids Res. 2009, 37, D122–D126. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.K.; Yang, Y.T.; Ma, X.; Han, B.; Wang, Z.S.; Zhao, Q.Y.; Wu, L.Q.; Qu, Z.Q. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Disease 2016, e2203. [Google Scholar] [CrossRef] [PubMed]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Okada, T.; Fukushima, T.; Tsudzuki, T.; Sugiura, M.; Yukawa, Y. A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biol. 2012, 9, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.C.; Frithand, M.C.; Mattick, J.S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006, 22, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Hao, Z.; Yan, J.; Li, G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genomics 2015, 16, 793. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-J.; Wang, Z.-M.; Wang, M.; Wang, X.-J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Sun, H.-X.; Park, B.S.; Huang, C.-H.; Yeh, S.-D.; Jung, C.; Chua, N.-H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Gendall, A.R.; Levy, Y.Y.; Wilson, A.; Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001, 107, 525–535. [Google Scholar] [CrossRef]
- Wood, C.C.; Robertson, M.; Tanner, G.; Peacock, W.J.; Dennis, E.S.; Helliwell, C.A. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE3. Proc. Natl. Acad. Sci. USA 2006, 103, 14631–14636. [Google Scholar] [CrossRef] [PubMed]
- Nischal, L.; Mohsin, M.; Khan, I.; Kardam, H.; Wadhwa, A.; Abrol, Y.P.; Iqbal, M.; Ahmad, A. Identification and comparative analysis of microRNAs associated with low-N tolerance in rice genotypes. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Elanchezhian, R.; Krishnapriya, V.; Pandey, R.; Rao, A.S.; Abrol, Y.P. Physiological and molecular approaches for improving phosphorus uptake efficiency of crops. Curr. Sci. India 2015, 108, 1271–1279. [Google Scholar]
- Frink, C.R.; Waggoner, P.E.; Ausubel, J.H. Nitrogen fertilizer: retrospect and prospect. Proc. Natl. Acad. Sci. USA 1999, 96, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genomics 2016, 291, 1663–1680. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Liang, Z.; Ge, M.; Qi, W.; Zhang, T.; Lin, F.; Peng, Z.; Zhao, H. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genomics 2016, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Amaral, N.S.D.; Melo, N.C.; Maia, B.d.M.; Rocha, R.M. Noncoding RNA profiles in tobacco- and alcohol-associated diseases. Genes 2017, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Steinkraus, B.R.; Toegel, M.; Fulga, T.A. Tiny giants of gene regulation: Experimental strategies for microRNA functional studies. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 311–362. [Google Scholar] [CrossRef] [PubMed]
- Dogini, D.B.; Pascoal, V.D.B.; Avansini, S.H.; Vieira, A.S.; Pereira, T.C.; Lopes-Cendes, I. The new world of RNAs. Genet. Mol. Biol. 2014, 37, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Lozada-Chávez, I.; Stadler, P.F.; Prohaska, S.J. Hypothesis for the modern RNA world: A pervasive non-coding RNA-based genetic regulation is a prerequisite for the emergence of multicellular complexity. Orig. Life Evol. Biosph. 2011, 41, 587–607. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of MicroRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Li, Y.; Li, J.; Millar, A.A. Inhibiting plant microRNA activity: Molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 2015, 13, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, F.; Axtell, M.J. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 2014, 26, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Strachan, T.; Read, A.P. Human Molecular Genetics; Garland Science, Taylor & Francis Group: New York, NY, USA, 2011. [Google Scholar]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.V.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2014, 43, D168–D173. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K. MicroRNAs and target mimics for crop improvement. Curr. Sci. India 2015, 108, 1624–1633. [Google Scholar]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef] [PubMed]
- Karakulah, G.; Yucebilgili-Kurtoglu, K.; Unver, T. PeTMbase: A Database of Plant Endogenous Target Mimics (eTMs). PLoS ONE 2016, 11, e0167698. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhi, H.; Zhang, Y.; Liu, Y.; Zhang, J.; Gao, Y.; Guo, M.; Ning, S.; Li, X. miRSponge: A manually curated database for experimentally supported miRNA sponges and ceRNAs. Database (Oxford) 2015, bav0982015. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Meng, X.; Li, X.; Illing, N.; Ingle, R.A.; Wang, J.; Chen, M. PceRBase: A database of plant competing endogenous RNA. Nucleic Acids Res. 2017, 45, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, MicroRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.L.; Leyva-Gonzalez, M.A.; Gonzalez-Morales, S.I.; Lopez-Bucio, J.; Herrera-Estrella, L. Phosphatenutrition: Improvinglow-phosphatetolerance in crops. Annu. Rev. Plant Biol. 2014, 65. [Google Scholar] [CrossRef]
- Vidal, E.A.; Moyano, T.C.; Canales, J.; Gutiérrez, R.A. Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5611–5618. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; LejayChandra, L.; et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite profiling of low-P tolerant and low-P sensitive maize genotypes under phosphorus starvation and restoration conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef] [PubMed]
- Nazir, M.; Pandey, R.; Siddiqi, T.O.; Ibrahim, M.M.; Qureshi, I.M.; Vengavasi, K.; Abraham, G.; Ahmad, A. Nitrogen-deficiency stress induces protein expression differentially in low-N tolerant and low-N sensitive maize genotypes. Front. Plant Sci. 2016, 7, 298. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Bhosale, R.; Huang, G.; Pandey, B.K.; Parker, H.; Zappala, S.; Yang, J.; Dievart, A.; Bureau, C.; Ljung, K.; et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018, 9, 1–7. [Google Scholar] [CrossRef]
- Gutierrez, R.A. Systems biology for enhanced plant nitrogen nutrition. Science 2012, 336, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.W.; Frommer, W.B. New Technologies for 21st Century Plant Science. Plant Cell 2012, 24, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.A.S.; Zakaria, A.G.; Said, F.B.; Tuinstra, M. Identification of quantitative trait loci for nitrogen use efficiency in maize. Mol. Breeding 1999, 5, 187–195. [Google Scholar] [CrossRef]
- Garnett, T.; Conn, V.; Kaiser, B.N. Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ. 2009, 32, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; AieseCigliano, R.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genomics 2014, 15, 179. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Amitha Mithra, S.V.; Chaudhary, S.; Tyagi, P.; Venkadesan, S.; Rani, M.; Mandal, P.K. Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic N starvation reveals differences in chloroplast and starch metabolism-related genes. Genes 2018. [Google Scholar] [CrossRef] [PubMed]
- Marschner, H.; Marschner, P. Mineral Nutrition of Higher Plants, 3rd ed.; Elsevier/Academic Press: London, UK; Waltham, MA, USA, 2012. [Google Scholar]
- Pandey, R.; Zinta, G.; AbdElgawad, H.; Ahmad, A.; Jain, V.; Janssens, I.A. Physiological and molecular alterations in plants exposed to high [CO2] under phosphorus stress. Biotechnol. Adv. 2015, 33, 303–316. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Herńandez-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L. Phosphate availability alters architecture and causes changes hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Nacry, P.; Canivenc, G.; Muller, B.; Azmi, A.; Van Onckelen, V.; Rossignol, M.; Doumas, P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005, 138, 2061–2074. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Calderón, L.; López-Bucio, J.; Chacón-López, A.; Cruz-Ramírez, A.; Nieto-Jacobo, F.; Dubrovsky, J.G.; Herrera-Estrella, J. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2006, 46, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Svistoonoff, S.; Creff, A.; Reymond, M.; Sigoillot-Claude, C.; Ricaud, L.; Blanchet, A.; Nussaume, L.; Desnos, T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007, 39, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-L.; Almvik, M.; Clarke, N.; Eich-Greatorex, S.; Øgaard, A.F.; Krogstad, T.; Lambers, H.; Clarke, J.L. Contrasting responses of root morphology and root-exuded organic acids to low phosphorus availability in three important food crops with divergent root traits. AoB Plants 2015, 7, plv097. [Google Scholar] [CrossRef] [PubMed]
- Vengavasi, K.; Pandey, R. Root exudation index: Screening organic acid exudation and phosphorus acquisition efficiency in soybean genotypes. Crop Pasture Sci. 2016, 67, 1096–1109. [Google Scholar] [CrossRef]
- Vengavasi, K.; Pandey, R.; Abraham, G.; Yadav, R.K. Comparative analysis of soybean root proteome reveals molecular basis of differential carboxylate efflux under low phosphorus stress. Genes 2017, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Krogstad, T.; Clarke, N.; Øgaard, A.F.; Clarke, J.L. Impact of phosphorus on rhizosphere organic anions of wheat at different growth stages under field conditions. AoB Plants 2017, 9, plx008. [Google Scholar] [CrossRef]
- Vengavasi, K.; Pandey, R. Root exudation potential in contrasting soybean genotypes in response to low soil phosphorus availability is determined by photo-biochemical processes. Plant Physiol. Biochem. 2018, 124, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Palma, D.A.; Blumwald, E.; Plaxton, W.C. Upregulation of vascular H+-translocating pyrophosphatase by phosphate starvation of Brassica napus (rapeseed) suspension cell cultures. FEBS Lett. 2000, 486, 155–158. [Google Scholar] [CrossRef]
- Theodorou, M.E.; Plaxton, W.C. Purification and characterization of pyrophosphate dependent phosphofructokinase from phosphate-starved Brassica nigra suspension cells. Plant Physiol. 1996, 112, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Vengavasi, K.; Kumar, A.; Pandey, R. Transcript abundance, enzyme activity and metabolite concentration regulates differential carboxylate efflux in soybean under low phosphorus stress. Indian J. Plant Physi. 2016, 21, 179–188. [Google Scholar] [CrossRef]
- Chiou, T.J.; Aung, K.; Lin, S.-I.; Wu, C.-C.; Chiang, S.-F.; Su, C.-L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Doerner, P. Phosphate starvation signaling: A threesome controls systemic Pi homeostasis. Curr. Opin. Plant Biol. 2008, 11, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Lin, S.I.; Chiou, T.J. Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 2009, 60, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Shin, H.-S.; Chen, R.; Harrison, M.J. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006, 45, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.F.; Chiou, T.J. The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 2011, 156, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen, J.; Mysore, K.S.; Zhang, W.H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Muchhal, U.S.; Raghothama, K.G. Differential expression of TPSI1, a phosphate starvation-induced gene in tomato. Plant Mol. Biol. 1997, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Burleigh, S.M.; Harrison, M.J. Characterization of the Mt4 gene from Medicago truncatula. Gene 1998, 216, 47–53. [Google Scholar] [CrossRef]
- Burleigh, S.H.; Harrison, M.J. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999, 119, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhang, Y.; Dong, J.; Sun, Y.; Lim, B.L.; Liu, D.; Lu, Z.J. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 2016, 17, 655. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.H.C.; Sun, H.; Bowler, C.; Chua, N. Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 2016, 210, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRbase: microRNA sequences, targets and gene nomenclature. Nucleic Acid Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Datta, S.K.; Datta, K. miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 2015, 6, 232. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.N.; Rothstein, S.J.; Spangenberg, G.; Kant, S. Role of microRNAs involved in plant response to nitrogen and phosphorous limiting conditions. Front. Plant Sci. 2015, 6, 629. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A MicroRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ding, H.; Zhu, J.K.; Zhang, F.; Li, W.X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ying, S.; Huang, H.; Li, K.; Wu, P.; Shou, H. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009, 57, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, W.; Ying, Y.; Li, S.; Secco, D.; Tyerman, S.; Whelan, J.; Shou, H. Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 2012, 196, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Yamaguchi, M.; Shen, H.; Sasaki, T.; Yamamoto, Y.; Matsumoto, H. Phosphorus deficiency enhances plasma membrane HC-ATPase activity and citrate exudation in greater purple lupin (Lupinuspilosus). Funct. Plant Biol. 2004, 31, 1075–1083. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Yu, D. Identification of nitrogen starvation-responsive MicroRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.Q.; Zhu, Y.Y.; SQ, H.; Yang, Z.M. Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J. Plant Physiol. 2010, 167, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, X.; Guo, C.; Gu, J.; Xiao, K. Identification and characterization of micro RNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J. Plant Biochem. Biot. 2013, 22, 113–123. [Google Scholar] [CrossRef]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, S.; Nonis, A.; Begheldo, M.; Manoli, A.; Palme, K.; Caporale, G.; Ruperti, B.; Quaggiotti, S. Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 2012, 35, 1137–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Hao, Q.; Sha, A.; Zhou, R.; Zhou, X.; Yuan, L. Elucidation of miRNAs-mediated responses to low nitrogen stress by deep sequencing of two soybean genotypes. PLoS ONE 2013, 8, e67423. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, H.; Hamera, H.; Chen, X.; Fang, R. miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.D.; Jackson, J.; Moussaieff, A.; Aharoni, A.; Benfey, P.N. Cell types specific transcriptional profiling: Implications for metabolite profiling. Plant J. 2012, 70, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Liao, J.; Li, Z.; Yu, Y.; Zhang, J.; Li, Q.; Qu, L.; Shu, W.; Chen, Y. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [PubMed]
| Name | Features | Links | Reference |
|---|---|---|---|
| TAPIR | Only tool for the TM prediction in plants; applies Franco-Zorrilla rule for target mimicry; contains data for 10 plant species; RNA hybrid and miRBase are the data sources | http://bioinformatics.psb.ugent.be/webtools/tapir/ | [55] |
| miRSponge | 1.6% data is from plant and others are of non-plant; experimentally validated; literature mining is the data source | http://www.bio-bigdata.net/miRSponge/ | [57] |
| PeTMbase | Contains 2728 TMs for 11 species; uses Wu et al. (2013) target mimicry rule; GreeNC, PNRD, miRBase, NCBI SRA are the data sources | http://petmbase.org | [56] |
| PceRBase | First database for plant TMs; 167608 TMs from 26 plant species; Phytozomev10, TAIR10, MSU RGGP & miRBase and literature are the data sources | http://bis.zju.edu.cn/pcernadb/ | [58] |
| miRNA ID | eTM ID | lncRNA ID | Low N | Low P | References |
|---|---|---|---|---|---|
| ath-miR156a-5p | ath_eTM_miR156a-5p-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (P↑) | [112] | |
| ath_eTM_miR156a-5p-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156b-5p | ath_eTM_miR156b-5p-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156b-5p-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156c-5p | ath_eTM_miR156c-5p-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156c-5p-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156d-5p | ath_eTM_miR156d-5p-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156d-5p-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156e | ath_eTM_miR156e-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156e-1 | PNRD_ath_NONATHT000580 | ||||
| gma-miR156e | gma_eTM_miR156e-3 | GRNC_gma_lcl|Gmax_Glyma.18G293400.2 | Soybean (P↑) | [114] | |
| gma_eTM_miR156e-2 | GRNC_gma_lcl|Gmax_Glyma.18G293400.1 | ||||
| gma_eTM_miR156e-1 | GRNC_gma_lcl|Gmax_Glyma.05G242200.1 | ||||
| ath-miR156f-5p | ath_eTM_miR156f-5p-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156f-5p-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156g | ath_eTM_miR156g-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156g-1 | PNRD_ath_NONATHT000580 | ||||
| ath-miR156h | ath_eTM_miR156h-3 | GRNC_ath_lcl|Athaliana_AT3G18217.1 gene = AT3G18217 | A. thaliana (N↑) | [113] | |
| ath_eTM_miR156h-2 | GRNC_ath_lcl|Athaliana_AT1G52347.1 gene = AT1G52347 | ||||
| ath_eTM_miR156h-1 | PNRD_ath_NONATHT000580 | ||||
| gma-miR159a-3p | gma_eTM_miR159a-3p-1 | gma_TCONS_00088249 | Soybean (P↑) | [114] | |
| tae-miR159b | tae_eTM_miR159b-2 | GRNC_tae_lcl|Taestivum_Traes_2DS_9A9CAF0B0.1 | Wheat(P↑) | [115] | |
| tae_eTM_miR159b-1 | GRNC_tae_lcl|Taestivum_Traes_1AL_8C8E43898.1 | ||||
| zma-miR160a-3p | zma_eTM_miR160a-3p-1 | GRNC_zma_lcl|Zmays_AC211588.3_FGT002 | Maize (N↑) | [116] | |
| zma-miR160a-5p | zma_eTM_miR160a-5p-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160a-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160a-5p-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160a-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160a-5p-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR160b-5p | zma_eTM_miR160b-5p-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160b-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160b-5p-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160b-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160b-5p-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR160c-5p | zma_eTM_miR160c-5p-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160c-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160c-5p-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160c-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160c-5p-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR160d-3p | zma_eTM_miR160d-3p-3 | GRNC_zma_lcl|Zmays_GRMZM2G064666_T02 | Maize (N↑) | [113] | |
| zma_eTM_miR160d-3p-2 | GRNC_zma_lcl|Zmays_GRMZM2G054392_T01 | ||||
| zma_eTM_miR160d-3p-1 | GRNC_zma_lcl|Zmays_GRMZM2G052412_T01 | ||||
| zma-miR160d-5p | zma_eTM_miR160d-5p-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160d-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160d-5p-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160d-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160d-5p-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR160e | zma_eTM_miR160e-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160e-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160e-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160e-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160e-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR160g-5p | zma_eTM_miR160g-5p-5 | GRNC_zma_lcl|Zmays_GRMZM5G849473_T01 | Maize (N↑) | [116] | |
| zma_eTM_miR160g-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G531719_T01 | ||||
| zma_eTM_miR160g-5p-3 | GRNC_zma_lcl|Zmays_GRMZM2G149698_T05 | ||||
| zma_eTM_miR160g-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G011007_T01 | ||||
| zma_eTM_miR160g-5p-1 | PNRD_zma_GRMZM5G849473_T01 | ||||
| zma-miR164f-5p | zma_eTM_miR164f-5p-1 | GRNC_zma_lcl|Zmays_GRMZM2G008252_T01 | Maize (N↑) | [116] | |
| zma-miR164f-3p | zma_eTM_miR164f-3p-1 | GRNC_zma_lcl|Zmays_GRMZM5G837428_T01 | Maize (N↑) | [113] | |
| zma-miR166j-3p | zma_eTM_miR166j-3p-4 | GRNC_zma_lcl|Zmays_GRMZM2G134604_T01 | Maize (N↓) | [117] | |
| zma_eTM_miR166j-3p-3 | zma_eTM_miR166j-3p-3 | ||||
| zma_eTM_miR166j-3p-2 | zma_eTM_miR166j-3p-2 | ||||
| zma_eTM_miR166j-3p-1 | zma_TCONS_00089106 | ||||
| zma-miR166k-3p | zma_eTM_miR166k-3p-4 | GRNC_zma_lcl|Zmays_GRMZM2G134604_T01 | Maize (N↓) | [117] | |
| zma_eTM_miR166k-3p-3 | GRNC_zma_lcl|Zmays_GRMZM2G110279_T02 | ||||
| zma_eTM_miR166k-3p-2 | GRNC_zma_lcl|Zmays_GRMZM2G110279_T01 | ||||
| zma_eTM_miR166k-3p-1 | zma_TCONS_00089106 | ||||
| zma-miR166n-3p | zma_eTM_miR166n-3p-4 | GRNC_zma_lcl|Zmays_GRMZM2G134604_T01 | Maize (N↓) | [117] | |
| zma_eTM_miR166n-3p-3 | GRNC_zma_lcl|Zmays_GRMZM2G110279_T02 | ||||
| zma_eTM_miR166n-3p-2 | GRNC_zma_lcl|Zmays_GRMZM2G110279_T01 | ||||
| zma_eTM_miR166n-3p-1 | zma_TCONS_00089106 | ||||
| zma-miR167g-3p | zma_eTM_miR167g-3p-1 | zma_TCONS_00081049 | Maize (N↓) | [116] | |
| zma-miR167g-5p | zma_eTM_miR167g-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G174168_T02 | Maize (N↓) | [116] | |
| zma_eTM_miR167g-5p-1 | GRNC_zma_lcl|Zmays_GRMZM2G174168_T01 | ||||
| zma-miR167h-3p | zma_eTM_miR167h-3p-7 | GRNC_zma_lcl|Zmays_GRMZM2G326635_T01 | Maize (N↓) | [116] | |
| zma_eTM_miR167h-3p-6 | GRNC_zma_lcl|Zmays_GRMZM2G175272_T03 | ||||
| zma_eTM_miR167h-3p-5 | GRNC_zma_lcl|Zmays_GRMZM2G159741_T04 | ||||
| zma_eTM_miR167h-3p-4 | GRNC_zma_lcl|Zmays_GRMZM2G158766_T04 | ||||
| zma_eTM_miR167h-3p-3 | GRNC_zma_lcl|Zmays_GRMZM2G125239_T04 | ||||
| zma_eTM_miR167h-3p-2 | PNRD_zma_TCONS_00034773 | ||||
| zma_eTM_miR167h-3p-1 | zma_TCONS_00012947 | ||||
| zma-miR167h-5p | zma_eTM_miR167h-5p-2 | GRNC_zma_lcl|Zmays_GRMZM2G174168_T02 | Maize (N↓) | [116] | |
| zma_eTM_miR167h-5p-1 | GRNC_zma_lcl|Zmays_GRMZM2G174168_T01 | ||||
| gma-miR169f | gma_eTM_miR169f-3 | GRNC_gma_lcl|Gmax_Glyma.19G136600.8 | Soybean (N↑) | [118] | |
| gma_eTM_miR169f-2 | GRNC_gma_lcl|Gmax_Glyma.19G136600.3 | ||||
| gma_eTM_miR169f-1 | GRNC_gma_lcl|Gmax_Glyma.19G136600.2 | ||||
| gma-miR169g | gma_eTM_miR169g-3 | GRNC_gma_lcl|Gmax_Glyma.19G136600.8 | Soybean (N↑) | [118] | |
| gma_eTM_miR169g-2 | GRNC_gma_lcl|Gmax_Glyma.19G136600.3 | ||||
| gma_eTM_miR169g-1 | GRNC_gma_lcl|Gmax_Glyma.19G136600.2 | ||||
| ath-miR169a-3p | ath_eTM_miR169a-3p-2 | GRNC_ath_lcl|Athaliana_AT1G44940.2 | A. thaliana(P↓) | [113] | |
| ath_eTM_miR169a-3p-1 | GRNC_ath_lcl|Athaliana_AT1G44940.1 | ||||
| zma-miR319a-5p | zma_eTM_miR319a-5p-4 | GRNC_zma_lcl|Zmays_GRMZM2G438722_T03 | Maize (N↑) | [116] | |
| zma_eTM_miR319a-5p-3 | zma_TCONS_00089764 | ||||
| zma_eTM_miR319a-5p-2 | zma_TCONS_00089763 | ||||
| zma_eTM_miR319a-5p-1 | zma_TCONS_00024738 | ||||
| zma-miR395d-5p | zma_eTM_miR395d-5p-1 | zma_TCONS_00091080 | Maize (N↓) | [116] | |
| zma-miR395g-5p | zma_eTM_miR395g-5p-1 | zma_TCONS_00091080 | |||
| gma-miR398b | gma_eTM_miR398b-1 | GRNC_gma_lcl|Gmax_Glyma.12G204100.1 | Soybean (P↓) | [114] | |
| ath-miR399f | ath_eTM_miR399f-5 | GRNC_ath_lcl|Athaliana_AT5G03545.1 | A. thaliana (N↑↓) | A. thaliana (P↑) | [113] |
| ath_eTM_miR399f-4 | GRNC_ath_lcl|Athaliana_AT3G09922.1 | ||||
| ath_eTM_miR399f-3 | PNRD_ath_At4-2 | ||||
| ath-miR399e | ath_eTM_miR399f-2 | PNRD_ath_At4 | |||
| ath_eTM_miR399f-1 | PNRD_ath_AtIPS1 | ||||
| ath_eTM_miR399e-1 | GRNC_ath_lcl|Athaliana_AT1G53708.1 | A. thaliana (N↑↓) | A. thaliana (P↑) | [113] | |
| ath-miR399d | ath_eTM_miR399d-5 | GRNC_ath_lcl|Athaliana_AT5G03545.1 | A. thaliana (N↑↓) | A. thaliana (P↑) | [113] |
| ath_eTM_miR399d-4 | GRNC_ath_lcl|Athaliana_AT3G09922.1 | ||||
| ath_eTM_miR399d-3 | PNRD_ath_At4-2 | ||||
| ath-miR399b | ath_eTM_miR399d-2 | PNRD_ath_At4 | |||
| ath_eTM_miR399d-1 | PNRD_ath_AtIPS1 | ||||
| ath_eTM_miR399b-6 | GRNC_ath_lcl|Athaliana_AT5G03545.1 | A. thaliana (N↑↓) | A. thaliana (P↑) | [113] | |
| ath_eTM_miR399b-5 | GRNC_ath_lcl|Athaliana_AT3G09922.1 | ||||
| ath_eTM_miR399b-4 | GRNC_ath_lcl|Athaliana_AT1G53708.1 | ||||
| ath-miR399a | ath_eTM_miR399b-3 | PNRD_ath_At4-2 | |||
| ath_eTM_miR399b-2 | PNRD_ath_At4 | ||||
| ath_eTM_miR399b-1 | PNRD_ath_AtIPS1 | ||||
| ath_eTM_miR399a-6 | GRNC_ath_lcl|Athaliana_AT5G03545.1 | A. thaliana (N↑↓) | A. thaliana (P↑) | [113] | |
| ath_eTM_miR399a-5 | GRNC_ath_lcl|Athaliana_AT3G09922.1 | ||||
| ath_eTM_miR399a-4 | GRNC_ath_lcl|Athaliana_AT1G53708.1 | ||||
| ath_eTM_miR399a-3 | PNRD_ath_At4-2 | ||||
| ath_eTM_miR399a-2 | PNRD_ath_At4 | ||||
| ath_eTM_miR399a-1 | PNRD_ath_AtIPS1 | ||||
| tae-miR408 | tae_eTM_miR408-2 | GRNC_tae_lcl|Taestivum_Traes_5BL_EED36D3B9.35 | Wheat (P↓) | [115] | |
| tae_eTM_miR408-1 | tae_TCONS_00103503 | ||||
| osa-miR444a-3p.2 | osa_eTM_miR444a-3p.2-1 | GRNC_osa_lcl|Osativa_LOC_Os12g19080.1 | Rice((N↑) | [119] | |
| ath-miR827 | ath_eTM_miR827-2 | GRNC_ath_lcl|Athaliana_AT3G02832.1 gene = AT3G02832 | A. thaliana (P↑) | [112] | |
| ath_eTM_miR827-1 | PNRD_ath_NONATHT001723 | ||||
| ath-miR828 | ath_eTM_miR828-1 | PNRD_ath_NONATHT000094 | A. thaliana (P↑) | [112] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borah, P.; Das, A.; Milner, M.J.; Ali, A.; Bentley, A.R.; Pandey, R. Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants. Genes 2018, 9, 459. https://doi.org/10.3390/genes9090459
Borah P, Das A, Milner MJ, Ali A, Bentley AR, Pandey R. Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants. Genes. 2018; 9(9):459. https://doi.org/10.3390/genes9090459
Chicago/Turabian StyleBorah, Priyanka, Antara Das, Matthew J. Milner, Arif Ali, Alison R. Bentley, and Renu Pandey. 2018. "Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants" Genes 9, no. 9: 459. https://doi.org/10.3390/genes9090459
APA StyleBorah, P., Das, A., Milner, M. J., Ali, A., Bentley, A. R., & Pandey, R. (2018). Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants. Genes, 9(9), 459. https://doi.org/10.3390/genes9090459






