Disease Resistance Mechanisms in Plants
Abstract
:1. Introduction
1.1. Plant Disease Resistance Has Emerged as a Complex, Multicomponent System
1.2. Plant Defense Relies on Detection and Response
1.3. Pathogen Virulence Relies on Attack and Evasion
2. Pathogen Detection
2.1. PRRs and WAKs Detect PAMPs and DAMPs
2.2. NLRs Detect Pathogen Effectors
3. Signal Transduction
3.1. Resistance Involves Multiple Signaling Mechanisms
3.2. Plant Hormones Systematically Initiate and Repress Resistance
3.3. TFs Play an Essential Role in Transcriptional Reprogramming
3.4. Epigenetic Regulation and PDR Provide Additional Regulatory Mechanisms
4. Defense Response
4.1. HR, ROS, and Cell Wall Modification Inhibit Pathogen Infection
4.2. Enzymes and Enzyme Inhibitors Counter Pathogenic Effectors and Facilitate Detection
4.3. Defensins and Thaumatin-Like Proteins Offensively Inhibit Pathogen Infection
4.4. Phytoalexins and Beneficial Symbionts Are Chemical and Biological Plant Weapons
5. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Whetzel, H.H. An Outline of the History of Phytopathology; WB Saunders: Philadelphia, PA, USA, 1918. [Google Scholar]
- Moutinho-Pereira, J.; Magalhaes, N.; Torres de Castro, L.; Manuela Chaves, M.; Torres-Pereira, J. Physiological responses of grapevine leaves to bordeaux mixture under light. Vitis 2001, 40, 117–121. [Google Scholar]
- Kutschera, U.; Hossfeld, U. Physiological phytopathology: Origin and evolution of a scientific discipline. J. Appl. Bot. Food Qual. 2012, 85, 1–5. [Google Scholar]
- Rhoades, V.H. The location of a gene for disease resistance in maize. Proc. Natl. Acad. Sci. USA 1935, 21, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, W. The Cereal Rusts: Origins, Specificity, Structure, and Physiology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 1. [Google Scholar]
- Flor, H.H. Inheritance of pathogenicity in melampsora lini. Phytopathology 1942, 32, 653–669. [Google Scholar]
- Hammond-Kosack, K.E.; Jones, J. Resistance gene-dependent plant defense responses. Plant Cell 1996, 8, 1773–1791. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Jones, J.D. Plant disease resistance genes. Ann. Rev. Plant Biol. 1997, 48, 575–607. [Google Scholar] [CrossRef] [PubMed]
- Antolín-Llovera, M.; Ried, M.K.; Binder, A.; Parniske, M. Receptor kinase signaling pathways in plant-microbe interactions. Ann. Rev. Phytopathol. 2012, 50, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Decreux, A.; Messiaen, J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005, 46, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Ann. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, Z.; Zheng, X.; Zhang, Z. The role of G-proteins in plant immunity. Plant Signal. Behav. 2012, 7, 1284–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Peeters, N.; Rivas, S. Ubiquitination during plant immune signaling. Plant Physiol. 2012, 160, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Ann. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Ann. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Upadhyaya, N.M.; Stiller, J.; Ellis, J.G.; Dodds, P.N.; Kazan, K.; Manners, J.M. Genomic analysis of Xanthomonas translucens pathogenic on wheat and barley reveals cross-kingdom gene transfer events and diverse protein delivery systems. PLoS ONE 2014, 9, e84995. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Stukenbrock, E.H.; Liu, Z.; Meinhardt, S.; Ling, H.; Faris, J.D.; Rasmussen, J.B.; Solomon, P.S.; McDonald, B.A.; Oliver, R.P. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 2006, 38, 953–956. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Ann. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van der Krol, S.; Shibuya, N.; Joosten, M.H.; Thomma, B.P. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.-M.; Chai, J. Structural basis for flg22-induced activation of the arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Prince, D.C.; Drurey, C.; Zipfel, C.; Hogenhout, S.A. The leucine-rich repeat receptor-like kinase brassinosteroid insensitive1-associated kinase1 and the cytochrome p450 phytoalexin deficient3 contribute to innate immunity to aphids in arabidopsis. Plant Physiol. 2014, 164, 2207–2219. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-J.; Ronald, P.C. Cleavage and nuclear localization of the rice xa21 immune receptor. Nat. Commun. 2012, 3, 920. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Biswas, R.; Lenz, H.D.; Rauhut, T.; Ranf, S.; Kemmerling, B.; Götz, F.; Glawischnig, E.; Lee, J.; Felix, G. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 2007, 282, 32338–32348. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef] [PubMed]
- Fliegmann, J.; Mithöfer, A.; Wanner, G.; Ebel, J. An ancient enzyme domain hidden in the putative β-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 2004, 279, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Verzaux, E.; Chaparro-Garcia, A.; Bijsterbosch, G.; Keizer, L.P.; Zhou, J.; Liebrand, T.W.; Xie, C.; Govers, F.; Robatzek, S. Elicitin recognition confers enhanced resistance to phytophthora infestans in potato. Nat. Plants 2015, 1, 15034. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 1997, 11, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, S.; Deng, Z.; Wang, X.; Chen, T.; Zhang, J.; Chen, S.; Ling, H.; Zhang, A.; Wang, D. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 2007, 52, 420–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Ge, S.; Xing, L.; Han, D.; Kang, Z.; Zhang, G.; Wang, X.; Wang, X.; Chen, P.; Cao, A. RLP1.1, a novel wheat receptor-like protein gene, is involved in the defence response against Puccinia striiformis f. sp. tritici. J. Exp. Bot. 2013, 64, 3735–3746. [Google Scholar] [CrossRef] [PubMed]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Pruitt, R.; Nürnberger, T. Sensing danger: Key to activating plant immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Ann. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- van den Burg, H.A.; Harrison, S.J.; Joosten, M.H.; Vervoort, J.; de Wit, P.J. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 2006, 19, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Takken, F.L.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Christopoulou, M.; Caldwell, K.S. Impacts of resistance gene genetics, function, and evolution on a durable future. Ann. Rev. Phytopathol. 2013, 51, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Ann. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Vernaldi, S.; Maekawa, T. Evolution and conservation of plant NLR functions. Front. Immunol. 2013, 4, 297. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.H.; Belhaj, K.; Bozkurt, T.O.; Birk, M.S.; Kamoun, S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for PTO-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 2016, 209, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Baggs, E.; Dagdas, G.; Krasileva, K. NLR diversity, helpers and integrated domains: Making sense of the NLR identity. Curr. Opin. Plant Biol. 2017, 38, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.T.; Chung, E.-H.; Habring-Müller, A.; Demar, M.; Schwab, R.; Dangl, J.L.; Weigel, D.; Chae, E. Activation of a plant NLR complex through heteromeric association with an autoimmune risk variant of another NLR. Curr. Biol. 2017, 27, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Kito, M.; Nishiyama, M.; Furuya, K.; Hong, S.-K.; Miyake, K.; Beppu, T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3 (2). Gene 1990, 95, 49–56. [Google Scholar] [CrossRef]
- Hengartner, M.O.; Horvitz, H.R. Programmed cell death in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 1994, 4, 581–586. [Google Scholar] [CrossRef]
- Zou, H.; Henzel, W.J.; Liu, X.; Lutschg, A.; Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c–dependent activation of caspase-3. Cell 1997, 90, 405–413. [Google Scholar] [CrossRef]
- Andersen, E.J.; Ali, S.; Reese, R.N.; Yen, Y.; Neupane, S.; Nepal, M.P. Diversity and evolution of disease resistance genes in barley (Hordeum vulgare L.). Evol. Bioinformat. Online 2016, 12, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Marone, D.; Russo, M.A.; Laidò, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int. J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Le, T.-N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.C.K.; Zhu, Q.-H.; Taylor, J.M.; Kazan, K.; Llewellyn, D.J.; Zhang, R. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidl, M.F.; Thomma, B.P. Transposable elements direct the coevolution between plants and microbes. Trends Genet. 2017, 33, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell Online 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.-M.; Teh, T.; Wang, C.-I.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Biezen, E.A.; Jones, J.D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- van der Hoorn, R.A.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P.N. A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front. Plant Sci. 2014, 5, 606. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Krasileva, K.V.; Banfield, M.J.; Terauchi, R.; Kamoun, S. The “sensor domains” of plant NLR proteins: More than decoys? Front. Plant Sci. 2015, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marathe, R.; Dinesh-Kumar, S. Plant defense: One post, multiple guards?! Mol. Cell 2003, 11, 284–286. [Google Scholar] [CrossRef]
- Axtell, M.J.; Staskawicz, B.J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 2003, 112, 369–377. [Google Scholar] [CrossRef]
- Holt, B.F.; Boyes, D.C.; Ellerström, M.; Siefers, N.; Wiig, A.; Kauffman, S.; Grant, M.R.; Dangl, J.L. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal arabidopsis development. Dev. Cell 2002, 2, 807–817. [Google Scholar] [CrossRef]
- Noutoshi, Y.; Ito, T.; Seki, M.; Nakashita, H.; Yoshida, S.; Marco, Y.; Shirasu, K.; Shinozaki, K. A single amino acid insertion in the WRKY domain of the arabidopsis TIR-NBS-LRR-WRKY -type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 2005, 43, 873–888. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, C.; Huet, G.; Jauneau, A.; Camborde, L.; Trémousaygue, D.; Kraut, A.; Zhou, B.; Levaillant, M.; Adachi, H.; Yoshioka, H. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015, 161, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef] [PubMed]
- Casey, L.W.; Lavrencic, P.; Bentham, A.R.; Cesari, S.; Ericsson, D.J.; Croll, T.; Turk, D.; Anderson, P.A.; Mark, A.E.; Dodds, P.N. The CC domain structure from the wheat stem rust resistance protein SR33 challenges paradigms for dimerization in plant NLR proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 12856–12861. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef] [PubMed]
- Knepper, C.; Savory, E.A.; Day, B. The role of NDR1 in pathogen perception and plant defense signaling. Plant Signal. Behav. 2011, 6, 1114–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 2005, 8, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, S.; Boevink, P.C.; Armstrong, M.R.; Ramos, M.B.; Hein, I.; Birch, P.R. Relocalization of late blight resistance protein R3A to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell 2012, 24, 5142–5158. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.-H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ülker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Schornack, S.; Win, J.; Shindo, T.; Ilyas, M.; Oliva, R.; Cano, L.M.; Jones, A.M.; Huitema, E.; van der Hoorn, R.A. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 2011, 108, 20832–20837. [Google Scholar] [CrossRef] [PubMed]
- Nomura, K.; DebRoy, S.; Lee, Y.H.; Pumplin, N.; Jones, J.; He, S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 2006, 313, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Chaparro-Garcia, A.; Schwizer, S.; Sklenar, J.; Yoshida, K.; Petre, B.; Bos, J.I.; Schornack, S.; Jones, A.M.; Bozkurt, T.O.; Kamoun, S. Phytophthora infestans RXRL-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PLoS ONE 2015, 10, e0137071. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Zhang, Z.; Friesen, T.L.; Raats, D.; Fahima, T.; Brueggeman, R.S.; Lu, S.; Trick, H.N.; Liu, Z.; Chao, W. The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2016, 2, e1600822. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, Y.; Kim, Y.M.; Faris, J.D.; Shelver, W.L.; Wit, P.J.; Xu, S.S.; Friesen, T.L. SnTox1, a Parastagonospora nodorum necrotrophic effector, is a dual-function protein that facilitates infection while protecting from wheat-produced chitinases. New Phytol. 2016, 211, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Ciuffetti, L.M.; Manning, V.A.; Pandelova, I.; Betts, M.F.; Martinez, J.P. Host-selective toxins, Ptr toxA and Ptr toxB, as necrotrophic effectors in the Pyrenophora tritici-repentis–wheat interaction. New Phytol. 2010, 187, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zurn, J.D.; Kariyawasam, G.; Faris, J.D.; Shi, G.; Hansen, J.; Rasmussen, J.B.; Acevedo, M. Inverse gene-for-gene interactions contribute additively to tan spot susceptibility in wheat. Theor. Appl. Genet. 2017, 130, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning, V.A.; Chu, A.L.; Steeves, J.E.; Wolpert, T.J.; Ciuffetti, L.M. A host-selective toxin of Pyrenophora tritici-repentis, Ptr toxA, induces photosystem changes and reactive oxygen species accumulation in sensitive wheat. Mol. Plant Microbe Interact. 2009, 22, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-TU by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galletti, R.; Ferrari, S.; De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide-or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011, 157, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D.; Kohorn, S.L.; Todorova, T.; Baptiste, G.; Stansky, K.; McCullough, M. A dominant allele of Arabidopsis pectin-binding wall-associated kinase induces a stress response suppressed by MPK6 but not MPK3 mutations. Mol. Plant 2012, 5, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Melech-Bonfil, S.; Sessa, G. Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 2010, 64, 379–391. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, O.; Pedley, K.F.; Martin, G.B. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004, 23, 3072–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, C.-S.; Martin, G.B. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011, 16, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Frye, C.A.; Tang, D.; Innes, R.W. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 2001, 98, 373–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Temple, B.R.; Jones, A.M. The plant heterotrimeric G-protein complex. Annu. Rev. Plant Biol. 2007, 58, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Botella, J.R. Plant G-proteins come of age: Breaking the bond with animal models. Front. Chem. 2016, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Temple, B.R.; Jones, C.D.; Jones, A.M. Evolution of a signaling nexus constrained by protein interfaces and conformational states. PLoS Comput. Biol. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Assmann, S.M. Plants: The latest model system for G-protein research. EMBO Rep. 2004, 5, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, W.-Y.; Park, H.C.; Lee, S.Y.; Bohnert, H.J.; Yun, D.-J. Sumo and sumoylation in plants. Mol. Cells 2011, 32, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, B.; Du, L.; Wang, H.; Yang, T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 2013, 163, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Mizoguchi, T.; Yoshida, R.; Ichimura, K.; Shinozaki, K. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in arabidopsis. Mol. Cell 2011, 41, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, X.; Lin, W.; Chen, S.; Lu, D.; Niu, Y.; Li, L.; Cheng, C.; McCormack, M.; Sheen, J. Bifurcation of arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013, 9, e1003127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuda, K.; Mine, A.; Bethke, G.; Igarashi, D.; Botanga, C.J.; Tsuda, Y.; Glazebrook, J.; Sato, M.; Katagiri, F. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 2013, 9, e1004015. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Ann. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Century, K.S.; Shapiro, A.D.; Repetti, P.P.; Dahlbeck, D.; Holub, E.; Staskawicz, B.J. NDR1, a pathogen-induced component required for arabidopsis disease resistance. Science 1997, 278, 1963–1965. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Lorang, J.; Kidarsa, T.; Bradford, C.; Gilbert, B.; Curtis, M.; Tzeng, S.-C.; Maier, C.; Wolpert, T. Tricking the guard: Exploiting plant defense for disease susceptibility. Science 2012, 338, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Geraats, B.P.; Bakker, P.A.; Lawrence, C.B.; Achuo, E.A.; Höfte, M.; Van Loon, L. Ethylene-insensitive tobacco shows differentially altered susceptibility to different pathogens. Phytopathology 2003, 93, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, P.; Shockey, J.; Lévesque, C.A.; Cook, R.J. A role for jasmonate in pathogen defense of arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7209–7214. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Penninckx, I.A.; Cammue, B.P.; Broekaert, W.F. The complexity of disease signaling in arabidopsis. Curr. Opin. Immunol. 2001, 13, 63–68. [Google Scholar] [CrossRef]
- McConn, M.; Creelman, R.A.; Bell, E.; Mullet, J.E. Jasmonate is essential for insect defense in arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Ruther, J.; Kleier, S. Plant–plant signaling: Ethylene synergizes volatile emission in Zea mays induced by exposure to (Z)-3-hexen-1-ol. J. Chem. Ecol. 2005, 31, 2217–2222. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Alborn, H.T.; Tumlinson, J.H. Synergistic interactions between volicitin, jasmonic acid and ethylene mediate insect-induced volatile emission in Zea mays. Physiol. Plant. 2003, 117, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-G.; Stork, W.; Mudgett, M.B. Xanthomonas type III effector XopD desumoylates tomato transcription factor SIERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 2013, 13, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asselbergh, B.; De Vleesschauwer, D.; Höfte, M. Global switches and fine-tuning—ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Lind, C.; Dreyer, I.; López-Sanjurjo, E.J.; von Meyer, K.; Ishizaki, K.; Kohchi, T.; Lang, D.; Zhao, Y.; Kreuzer, I.; Al-Rasheid, K.A. Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr. Biol. 2015, 25, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Barth, C.; Moeder, W.; Klessig, D.F.; Conklin, P.L. The timing of senescence and response to pathogens is altered in the ascorbate-deficient arabidopsis mutant vitamin C-1. Plant Physiol. 2004, 134, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Pavet, V.; Olmos, E.; Kiddle, G.; Mowla, S.; Kumar, S.; Antoniw, J.; Alvarez, M.E.; Foyer, C.H. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 2005, 139, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Cascone, P.; Madonna, V.; Lelio, I.; Esposito, F.; Avitabile, C.; Romanelli, A.; Guerrieri, E.; Vitiello, A.; Pennacchio, F. Plant-to-plant communication triggered by systemin primes anti-herbivore resistance in tomato. Sci. Rep. 2017, 7, 15522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.L.; Fiil, B.K.; Petersen, K.; Nielsen, H.B.; Botanga, C.J.; Thorgrimsen, S.; Palma, K.; Suarez-Rodriguez, M.C.; Sandbech-Clausen, S.; Lichota, J. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008, 27, 2214–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Ju, H.; Zhou, G.; Zhu, C.; Erb, M.; Wang, X.; Wang, P.; Lou, Y. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wei, T.; Yin, K.Q.; Chen, Z.; Gu, H.; Qu, L.J.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Moffat, C.S.; Ingle, R.A.; Wathugala, D.L.; Saunders, N.J.; Knight, H.; Knight, M.R. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Xu, J.; He, Y.; Yang, K.-Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, M.G.; Dinh, S.T.; Oh, Y.; Gaquerel, E.; Baldwin, I.T.; Galis, I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC Plant Biol. 2013, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Okada, K.; Miyamoto, K.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J. Biol. Chem. 2009, 284, 26510–26518. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lyngkjaer, M.F.; Collinge, D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Meng, X.; Wang, R.; Mao, G.; Han, L.; Liu, Y.; Zhang, S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in arabidopsis. PLoS Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKks drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Yu, D.; Jiao, J.; Jing, S.; Schulze-Lefert, P.; Shen, Q.-H. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 2013, 25, 1158–1173. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Xinqiong, L.; Nakayama, A.; Sugano, S.; Jiang, C.-J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Bartley, L.E.; Chen, X.; Dardick, C.; Chern, M.; Ruan, R.; Canlas, P.E.; Ronald, P.C. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 2008, 1, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife 2013, 2, e00983. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, F.; Tian, W.; Guo, Z.; Chen, X.; Xu, X.; Shang, H.; Hu, X. The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. PLoS ONE 2017, 12, e0181963. [Google Scholar] [CrossRef] [PubMed]
- McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Pritchard, L.; Gomez, S.; Morales, J.; Whisson, S.C.; Beynon, J.L.; Birch, P.R. An RXLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013, 9, e1003670. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Toruño, T.Y.; Elowsky, C.G.; Zhang, C.; Steinbrenner, J.; Beynon, J.; Alfano, J.R. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014, 201, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Goregaoker, S.P.; Culver, J.N. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. J. Virol. 2009, 83, 9720–9730. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; Moscou, M.J.; Ward, E.R.; Horvath, D.M. Engineering plant disease resistance based on tal effectors. Ann. Rev. Phytopathol. 2013, 51, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Wang, J.; Zeng, X.; Gu, K.; Qiu, C.; Yang, X.; Zhou, Z.; Goh, M.; Luo, Y.; Murata-Hori, M. The rice TAL effector–dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 2014, 26, 497–515. [Google Scholar] [CrossRef] [PubMed]
- Römer, P.; Hahn, S.; Jordan, T.; Strauß, T.; Bonas, U.; Lahaye, T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 2007, 318, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Conrath, U.; Peterhänsel, C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011, 12, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Holeski, L.M.; Jander, G.; Agrawal, A.A. Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 2012, 27, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- López Sánchez, A.; Stassen, J.H.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.M.; Song, H.R.; Han, S.K.; Han, M.; Kim, C.Y.; Park, J.; Lee, Y.H.; Jeon, J.S.; Noh, Y.S.; Noh, B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012, 71, 135–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.-C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Mosher, R.A.; Durrant, W.E.; Wang, D.; Song, J.; Dong, X. A comprehensive structure–function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 2006, 18, 1750–1765. [Google Scholar] [CrossRef] [PubMed]
- Burketova, L.; Trda, L.; Ott, P.G.; Valentova, O. Bio-based resistance inducers for sustainable plant protection against pathogens. Biotechnol. Adv. 2015, 33, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.R.; Paterson, L. Parents lend a helping hand to their offspring in plant defence. Biol. Lett. 2012, 8, 871–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ameye, M.; Audenaert, K.; De Zutter, N.; Steppe, K.; Van Meulebroek, L.; Vanhaecke, L.; De Vleesschauwer, D.; Haesaert, G.; Smagghe, G. Priming of wheat with the green leaf volatile Z-3-hexenyl acetate enhances defense against Fusarium graminearum but boosts deoxynivalenol production. Plant Physiol. 2015, 167, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Beachy, R.N. Mechanisms and applications of pathogen-derived resistance in transgenic plants. Curr. Opin. Biotechnol. 1997, 8, 215–220. [Google Scholar] [CrossRef]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Namba, S.; Ling, K.; Gonsalves, C.; Slightom, J.L.; Gonsalves, D. Protection of transgenic plants expressing the coat protein gene of watermelon mosaic virus ii or zucchini yellow mosaic virus against six potyviruses. Phytopathology 1992, 82, 940–946. [Google Scholar] [CrossRef]
- Navarro, L.; Jay, F.; Nomura, K.; He, S.Y.; Voinnet, O. Suppression of the microRNA pathway by bacterial effector proteins. Science 2008, 321, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.; Wang, X.; Liu, X.; Xiang, Q.; Jiang, S. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.F.; Rupp, J.L.S.; Trick, H.N.; Fellers, J.P. Stable resistance to Wheat streak mosaic virus in wheat mediated by RNAi. In Vitro Cell. Dev. Biol. Plant 2014, 50, 665–672. [Google Scholar] [CrossRef]
- Tian, B.; Li, J.; Oakley, T.R.; Todd, T.C.; Trick, H.N. Host-derived artificial microRNA as an alternative method to improve soybean resistance to soybean cyst nematode. Genes 2016, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhao, S.-L.; Li, J.-L.; Hu, X.-H.; Wang, H.; Cao, X.-L.; Xu, Y.-J.; Zhao, Z.-X.; Xiao, Z.-Y.; Yang, N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, F.; Zhu, H. Alternative splicing in plant immunity. Int. J. Mol. Sci. 2014, 15, 10424–10445. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, T.; Eulgem, T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 2013, 110, E3535–E3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sela, H.; Spiridon, L.N.; Petrescu, A.J.; Akerman, M.; Mandel-Gutfreund, Y.; Nevo, E.; Loutre, C.; Keller, B.; Schulman, A.H.; Fahima, T. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol. Plant Pathol. 2012, 13, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saintenac, C.; Zhang, W.; Salcedo, A.; Rouse, M.N.; Trick, H.N.; Akhunov, E.; Dubcovsky, J. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 2013, 341, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Halterman, D.A.; Wei, F.; Wise, R.P. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003, 131, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Halterman, D.; Zhou, F.; Wei, F.; Wise, R.P.; Schulze-Lefert, P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 2001, 25, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Dellota, E. Jr.; Yamane, D.; Jin, H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Shi, J.; Zhai, Y.; Hou, Y.; Ma, W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA 2015, 112, 5850–5855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, T.; Trifonova, E.A.; Kochetov, A.V.; Kanayama, Y. Expression of an extracellular ribonuclease gene increases resistance to cucumber mosaic virus in tobacco. BMC Plant Biol. 2016, 16, 246. [Google Scholar] [CrossRef] [PubMed]
- Galiana, E.; Bonnet, P.; Conrod, S.; Keller, H.; Panabières, F.; Ponchet, M.; Poupet, A.; Ricci, P. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol. 1997, 115, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Hugot, K.; Ponchet, M.; Marais, A.; Ricci, P.; Galiana, E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Mol. Plant Microbe Interact. 2002, 15, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Filipenko, E.; Kochetov, A.; Kanayama, Y.; Malinovsky, V.; Shumny, V. PR-proteins with ribonuclease activity and plant resistance against pathogenic fungi. Russian J. Genet. Appl. Res. 2013, 3, 474–480. [Google Scholar] [CrossRef]
- Caporale, C.; Di Berardino, I.; Leonardi, L.; Bertini, L.; Cascone, A.; Buonocore, V.; Caruso, C. Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett. 2004, 575, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Almagro, L.; Gómez Ros, L.; Belchi-Navarro, S.; Bru, R.; Ros Barceló, A.; Pedreno, M. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2008, 60, 377–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Mittler, R.; Herr, E.H.; Orvar, B.L.; Van Camp, W.; Willekens, H.; Inzé, D.; Ellis, B.E. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. USA 1999, 96, 14165–14170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Ann. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Kärkönen, A.; Kuchitsu, K. Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 2015, 112, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Cessna, S.G.; Sears, V.E.; Dickman, M.B.; Low, P.S. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 2000, 12, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Kabbage, M.; Kim, H.-J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, B.; Dunwell, J.M.; Ray, J.; Schmitt, M.; Cuming, A. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 1993, 268, 12239–12242. [Google Scholar] [PubMed]
- Dumas, B.; Sailland, A.; Cheviet, J.; Freyssinet, G.; Pallett, K. Identification of barley oxalate oxidase as a germin-like protein. C. R. Acad. Sci. Ser. III Sci. 1993, 316, 793–798. [Google Scholar]
- Dumas, B.; Freyssinet, G.; Pallett, K.E. Tissue-specific expression of germin-like oxalate oxidase during development and fungal infection of barley seedlings. Plant Physiol. 1995, 107, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Ji, R.; Guo, X.; Foster, S.J.; Chen, H.; Dong, C.; Liu, Y.; Hu, Q.; Liu, S. Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 2008, 228, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, P.A.; Anderson, T.; Lane, B.G.; Davidson, A.L.; Simmonds, D.H. Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol. Mol. Plant Pathol. 2001, 59, 297–307. [Google Scholar] [CrossRef]
- Liu, F.; Wang, M.; Wen, J.; Yi, B.; Shen, J.; Ma, C.; Tu, J.; Fu, T. Overexpression of barley oxalate oxidase gene induces partial leaf resistance to Sclerotinia sclerotiorum in transgenic oilseed rape. Plant Pathol. 2015, 64, 1407–1416. [Google Scholar] [CrossRef]
- Liang, H.; Maynard, C.A.; Allen, R.D.; Powell, W.A. Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol. Biol. 2001, 45, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Ramputh, A.; Arnason, J.; Cass, L.; Simmonds, J. Reduced herbivory of the European corn borer (Ostrinia nubilalis) on corn transformed with germin, a wheat oxalate oxidase gene. Plant Sci. 2002, 162, 431–440. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, Z.; Andersen, C.H.; Schmelzer, E.; Gregersen, P.L.; Collinge, D.B.; Smedegaard-Petersen, V.; Thordal-Christensen, H. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol. Biol. 1998, 36, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Bäumlein, H.; Mock, H.-P.; Himmelbach, A.; Schweizer, P. The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Buchenauer, H. Ultrastructural and cytochemical studies on cellulose, xylan and pectin degradation in wheat spikes infected by Fusarium culmorum. J. Phytopathol. 2000, 148, 263–275. [Google Scholar] [CrossRef]
- Wanjiru, W.M.; Zhensheng, K.; Buchenauer, H. Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 2002, 108, 803–810. [Google Scholar] [CrossRef]
- Kong, L.; Anderson, J.M.; Ohm, H.W. Induction of wheat defense and stress-related genes in response to Fusarium graminearum. Genome 2005, 48, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Anguelova-Merhar, V.; Westhuizen, A.; Pretorius, Z. β-1,3-glucanase and chitinase activities and the resistance response of wheat to leaf rust. J. Phytopathol. 2001, 149, 381–384. [Google Scholar] [CrossRef]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broekaert, W.; Van Parijs, J.; Allen, A.; Peumans, W. Comparison of some molecular, enzymatic and antifungal properties of chitinases from thorn-apple, tobacco and wheat. Physiol. Mol. Plant Pathol. 1988, 33, 319–331. [Google Scholar] [CrossRef]
- Singh, A.; Kirubakaran, S.I.; Sakthivel, N. Heterologous expression of new antifungal chitinase from wheat. Protein Express. Purif. 2007, 56, 100–109. [Google Scholar] [CrossRef] [PubMed]
- van Scheltinga, A.C.T.; Kalk, K.H.; Beintema, J.J.; Dijkstra, B.W. Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure 1994, 2, 1181–1189. [Google Scholar] [CrossRef] [Green Version]
- Nagano, N.; Orengo, C.A.; Thornton, J.M. One fold with many functions: The evolutionary relationships between tim barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002, 321, 741–765. [Google Scholar] [CrossRef]
- Leubner-Metzger, G.; Meins, F., Jr. 3 functions and regulation of plant β-(pr-2). In Pathogenesis-Related Proteins in Plants; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Doxey, A.C.; Yaish, M.W.; Moffatt, B.A.; Griffith, M.; McConkey, B.J. Functional divergence in the Arabidopsis β-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 2007, 24, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; De Wit, P.J. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.L.; van der Hoorn, R.A. Ten prominent host proteases in plant-pathogen interactions. Int. J. Mol. Sci. 2018, 19, 639. [Google Scholar] [CrossRef] [PubMed]
- Naumann, T.A.; Wicklow, D.T.; Price, N.P. Identification of a chitinase-modifying protein from Fusarium verticillioides truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 2011, 286, 35358–35366. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Naumann, T.A.; Price, N.P.; Rogozhin, E.A.; Andreev, Y.A.; Vassilevski, A.A.; Odintsova, T.I. Novel mode of action of plant defense peptides–hevein-like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 2014, 281, 4754–4764. [Google Scholar] [CrossRef] [PubMed]
- Pekkarinen, A.I.; Longstaff, C.; Jones, B.L. Kinetics of the inhibition of Fusarium serine proteinases by barley (Hordeum vulgare L.) inhibitors. J. Agric. Food Chem. 2007, 55, 2736–2742. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L.; Rigden, D.J.; R Melo, F.; Bloch, C.; Silva, C.P.; Grossi de Sá, M.F. Activity of wheat α-amylase inhibitors towards bruchid α-amylases and structural explanation of observed specificities. FEBS J. 2000, 267, 2166–2173. [Google Scholar] [CrossRef] [Green Version]
- Feng, G.H.; Richardson, M.; Chen, M.S.; Kramer, K.J.; Morgan, T.D.; Reeck, G.R. α-amylase inhibitors from wheat: Amino acid sequences and patterns of inhibition of insect and human α-amylases. Insect Biochem. Mol. Biol. 1996, 26, 419–426. [Google Scholar] [CrossRef]
- Mundy, J.; Svendsen, I.; Hejgaard, J. Barley α-amylase/subtilisin inhibitor. I. Isolation and characterization. Carlsberg Res. Commun. 1983, 48, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Petrucci, T.; Rab, A.; Tomasi, M.; Silano, V. Further characterization studies of the α-amylase protein inhibitor of gel electrophoretic mobility 0.19 from the wheat kernel. Biochim. Biophys. Acta (BBA)-Protein Struct. 1976, 420, 288–297. [Google Scholar] [CrossRef]
- Franco, O.L.; Rigden, D.J.; Melo, F.R.; Grossi-de-Sá, M.F. Plant α-amylase inhibitors and their interaction with insect α-amylases. FEBS J. 2002, 269, 397–412. [Google Scholar] [CrossRef] [Green Version]
- Steppuhn, A.; Baldwin, I.T. Resistance management in a native plant: Nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol. Lett. 2007, 10, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A. Fungal effector protein AVR2 targets diversifying defense-related CYS proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Rooney, H.C.; van’t Klooster, J.W.; van der Hoorn, R.A.; Joosten, M.H.; Jones, J.D.; de Wit, P.J. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 2005, 308, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Win, J.; Tian, M.; Schornack, S.; Kaschani, F.; Ilyas, M.; van der Hoorn, R.A.; Kamoun, S. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 2009, 106, 1654–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano-Torres, J.L.; Wilbers, R.H.; Gawronski, P.; Boshoven, J.C.; Finkers-Tomczak, A.; Cordewener, J.H.; America, A.H.; Overmars, H.A.; Van’t Klooster, J.W.; Baranowski, L. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA 2012, 109, 10119–10124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reisige, K.; Gorzelanny, C.; Daniels, U.; Moerschbacher, B.M. The C28 aldehyde octacosanal is a morphogenetically active component involved in host plant recognition and infection structure differentiation in the wheat stem rust fungus. Physiol. Mol. Plant Pathol. 2006, 68, 33–40. [Google Scholar] [CrossRef]
- Tsuba, M.; Katagiri, C.; Takeuchi, Y.; Takada, Y.; Yamaoka, N. Chemical factors of the leaf surface involved in the morphogenesis of Blumeria graminis. Physiol. Mol. Plant Pathol. 2002, 60, 51–57. [Google Scholar] [CrossRef]
- Woloshuk, C.P.; Kolattukudy, P. Mechanism by which contact with plant cuticle triggers cutinase gene expression in the spores of Fusarium solani f. sp. pisi. Proc. Natl. Acad. Sci. USA 1986, 83, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Gottwald, S.; Samans, B.; Lück, S.; Friedt, W. Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: Two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genom. 2012, 13, 369. [Google Scholar] [CrossRef] [PubMed]
- Finkina, E.; Melnikova, D.; Bogdanov, I. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar]
- Nimchuk, Z.; Marois, E.; Kjemtrup, S.; Leister, R.T.; Katagiri, F.; Dangl, J.L. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 2000, 101, 353–363. [Google Scholar] [CrossRef]
- Boyes, D.C.; Nam, J.; Dangl, J.L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 1998, 95, 15849–15854. [Google Scholar] [CrossRef] [PubMed]
- Leister, R.T.; Ausubel, F.M.; Katagiri, F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 1996, 93, 15497–15502. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Tanaka, T.; Duke, S.O.; Porter, J.K.; Wray, E.M.; Hodges, L.; Sessions, A.E.; Wang, E.; Merrill, A.H., Jr.; Riley, R.T. Fumonisin-and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 1994, 106, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Desaki, Y.; Miya, A.; Venkatesh, B.; Tsuyumu, S.; Yamane, H.; Kaku, H.; Minami, E.; Shibuya, N. Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 2006, 47, 1530–1540. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, D.; Zähringer, U.; Gerber, I.; Dubery, I.; Hartung, T.; Bors, W.; Hutzler, P.; Durner, J. Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15811–15816. [Google Scholar] [CrossRef] [PubMed]
- Canonne, J.; Froidure-Nicolas, S.; Rivas, S. Phospholipases in action during plant defense signaling. Plant Signal. Behav. 2011, 6, 13–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Phospholipase d and phosphatidic acid in plant defence response: From protein–protein and lipid–protein interactions to hormone signalling. J. Exp. Bot. 2015, 66, 1721–1736. [Google Scholar] [CrossRef] [PubMed]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L. Oxi1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef] [PubMed]
- de Zélicourt, A.; Letousey, P.; Thoiron, S.; Campion, C.; Simoneau, P.; Elmorjani, K.; Marion, D.; Simier, P.; Delavault, P. Ha-def1, a sunflower defensin, induces cell death in Orobanche parasitic plants. Planta 2007, 226, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Mendez, E.; Moreno, A.; Colilla, F.; Pelaez, F.; Limas, G.G.; Mendez, R.; Soriano, F.; Salinas, M.; Haro, C. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, γ-hordothionin, from barley endosperm. FEBS J. 1990, 194, 533–539. [Google Scholar] [CrossRef]
- Colilla, F.J.; Rocher, A.; Mendez, E. Γ-purothionins: Amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 1990, 270, 191–194. [Google Scholar] [CrossRef]
- Bruix, M.; Jimenez, M.; Santoro, J.; Gonzalez, C.; Colilla, F.; Mendez, E.; Rico, M. Solution structure of. Gamma. 1-H and gamma. 1-P thionins from barley and wheat endosperm determined by 1H-NMR: A structural motif common to toxic arthropod proteins. Biochemistry 1993, 32, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Terras, F.R.G.; Eggermont, K.; Kovaleva, V.; Raikhel, N.V.; Osborn, R.W.; Kester, A.; Rees, S.B.; Torrekens, S.; Van Leuven, F.; Vanderleyden, J. Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell 1995, 7, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Méndez, E.; Rocher, A.; Calero, M.; Girbés, T.; Citores, L.; Soriano, F. Primary structure of ω-hordothionin, a member of a novel family of thionins from barley endosperm, and its inhibition of protein synthesis in eukaryotic and prokaryotic cell-free systems. FEBS J. 1996, 239, 67–73. [Google Scholar] [CrossRef]
- Van Der Weerden, N.L.; Lay, F.T.; Anderson, M.A. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 2008, 283, 14445–14452. [Google Scholar] [CrossRef] [PubMed]
- van der Weerden, N.L.; Hancock, R.E.; Anderson, M.A. Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 2010, 285, 37513–37520. [Google Scholar] [CrossRef] [PubMed]
- Koike, M.; Okamoto, T.; Tsuda, S.; Imai, R. A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem. Biophys. Res. Commun. 2002, 298, 46–53. [Google Scholar] [CrossRef]
- Wijaya, R.; Neumann, G.M.; Condron, R.; Hughes, A.B.; Polya, G.M. Defense proteins from seed of Cassia fistula include a lipid transfer protein homologue and a protease inhibitory plant defensin. Plant Sci. 2000, 159, 243–255. [Google Scholar] [CrossRef]
- Aerts, A.M.; François, I.E.; Meert, E.M.; Li, Q.-T.; Cammue, B.P.; Thevissen, K. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J. Mol. Microbiol. Biotechnol. 2007, 13, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Spelbrink, R.G.; Dilmac, N.; Allen, A.; Smith, T.J.; Shah, D.M.; Hockerman, G.H. Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol. 2004, 135, 2055–2067. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Thomson, J.; Wang, Y. Plant defensins: Defense, development and application. Plant Signal. Behav. 2009, 4, 1010–1012. [Google Scholar] [CrossRef] [PubMed]
- García-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodríguez-Palenzuéla, P. Plant defense peptides. Pept. Sci. 1998, 47, 479–491. [Google Scholar] [CrossRef]
- De Paula, V.S.; Razzera, G.; Barreto-Bergter, E.; Almeida, F.C.; Valente, A.P. Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR: Multiple motions associated with membrane interaction. Structure 2011, 19, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.F.; Vasconcelos, É.A.; Pelegrini, P.B.; de Sa, M.F.G. Antifungal defensins and their role in plant defense. Front. Microbiol. 2014, 5, 116. [Google Scholar] [CrossRef] [PubMed]
- Vriens, K.; Peigneur, S.; De Coninck, B.; Tytgat, J.; Cammue, B.P.; Thevissen, K. The antifungal plant defensin AtPDF2.3 from Arabidopsis thaliana blocks potassium channels. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.S.; Cabral, K.M.; Kurtenbach, E.; Almeida, F.C.; Valente, A.P. Solution structure of Pisum sativum defensin 1 by high resolution NMR: Plant defensins, identical backbone with different mechanisms of action. J. Mol. Biol. 2002, 315, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.; Graham, M.A.; Paape, T.D.; VandenBosch, K.A. Genome organization of more than 300 defensin-like genes in arabidopsis. Plant Physiol. 2005, 138, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.A.; Silverstein, K.A.; Cannon, S.B.; VandenBosch, K.A. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004, 135, 1179–1197. [Google Scholar] [CrossRef] [PubMed]
- Van der Weerden, N.L.; Anderson, M.A. Plant defensins: Common fold, multiple functions. Fungal Biol. Rev. 2013, 26, 121–131. [Google Scholar] [CrossRef]
- Wang, S.; Rao, P.; Ye, X. Isolation and biochemical characterization of a novel leguminous defense peptide with antifungal and antiproliferative potency. Appl. Microbiol. Biotechnol. 2009, 82, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Ming, D.; Hellekant, G. Brazzein, a new high-potency thermostable sweet protein from Pentadiplandra brazzeana B. FEBS Lett. 1994, 355, 106–108. [Google Scholar] [CrossRef]
- Wel, H.; Loeve, K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii benth. FEBS J. 1972, 31, 221–225. [Google Scholar] [CrossRef]
- Osmond, R.I.; Hrmova, M.; Fontaine, F.; Imberty, A.; Fincher, G.B. Binding interactions between barley thaumatin-like proteins and (1,3)-β-d-glucans. FEBS J. 2001, 268, 4190–4199. [Google Scholar] [CrossRef]
- Bryngelsson, T.; Green, B. Characterization of a pathogenesis-related, thaumatin-like protein isolated from barley challenged with an incompatible race of mildew. Physiol. Mol. Plant Pathol. 1989, 35, 45–52. [Google Scholar] [CrossRef]
- Trümper, C.; Paffenholz, K.; Smit, I.; Kössler, P.; Karlovsky, P.; Braun, H.-P.; Pawelzik, E. Identification of regulated proteins in naked barley grains (Hordeum vulgare nudum) after Fusarium graminearum infection at different grain ripening stages. J. Proteom. 2016, 133, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Hejgaard, J.; Jacobsen, S.; Svendsen, I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991, 291, 127–131. [Google Scholar] [CrossRef] [Green Version]
- Vigers, A.J.; Roberts, W.K.; Selitrennikoff, C.P. A new family of plant antifungal proteins. Mol. Plant Microbe Interact. 1991, 4, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.K.; Selitrennikoff, C.P. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. Microbiology 1990, 136, 1771–1778. [Google Scholar] [CrossRef] [Green Version]
- Abad, L.R.; D’Urzo, M.P.; Liu, D.; Narasimhan, M.L.; Reuveni, M.; Zhu, J.K.; Niu, X.; Singh, N.K.; Hasegawa, P.M.; Bressan, R.A. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996, 118, 11–23. [Google Scholar] [CrossRef]
- Skadsen, R.; Sathish, P.; Kaeppler, H. Expression of thaumatin-like permatin PR-5 genes switches from the ovary wall to the aleurone in developing barley and oat seeds. Plant Sci. 2000, 156, 11–22. [Google Scholar] [CrossRef]
- Hahn, M.; Jüngling, S.; Knogge, W. Cultivar-specific elicitation of barley defense reactions by the phytotoxic peptide NIP1 from Rhynchosporium secalis. Mol. Plant Microbe Interact. 1993, 6, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Rebmann, G.; Mauch, F.; Dudler, R. Sequence of a wheat cDNA encoding a pathogen-induced thaumatin-like protein. Plant Mol. Biol. 1991, 17, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-C.; Bushnell, W.R.; Szabo, L.J.; Smith, A.G. Isolation and expression of a host response gene family encoding thaumatin-like proteins in incompatible oat-stem rust fungus interactions. Mol. Plant Microbe Interact. 1996, 9, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Tripathi, R.K.; Lemaux, P.G.; Buchanan, B.B.; Singh, J. Redox-dependent interaction between thaumatin-like protein and β-glucan influences malting quality of barley. Proc. Natl. Acad. Sci. USA 2017, 201701824. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.; Van Strien, E. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Ebrahim, S.; Usha, K.; Singh, B. Pathogenesis related (PR) proteins in plant defense mechanism. Sci. Against Microb Pathog. 2011, 2, 1043–1054. [Google Scholar]
- Sinha, M.; Singh, R.P.; Kushwaha, G.S.; Iqbal, N.; Singh, A.; Kaushik, S.; Kaur, P.; Sharma, S.; Singh, T.P. Current overview of allergens of plant pathogenesis related protein families. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Kettles, G.J.; Drurey, C.; Schoonbeek, H.J.; Maule, A.J.; Hogenhout, S.A. Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAas. New Phytol. 2013, 198, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I.E. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Hossain, S.; Snitynsky, R.B. Detoxification of cruciferous phytoalexins in Botrytis cinerea: Spontaneous dimerization of a camalexin metabolite. Phytochemistry 2011, 72, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Weibull, J.; Niemeyer, H. Changes in dihydroxymethoxybenzoxazinone glycoside content in wheat plants infected by three plant pathogenic fungi. Physiol. Mol. Plant Pathol. 1995, 47, 201–212. [Google Scholar] [CrossRef]
- Wouters, F.C.; Blanchette, B.; Gershenzon, J.; Vassão, D.G. Plant defense and herbivore counter-defense: Benzoxazinoids and insect herbivores. Phytochem. Rev. 2016, 15, 1127–1151. [Google Scholar] [CrossRef] [PubMed]
- Givovich, A.; Niemeyer, H. Hydroxamic acids affecting barley yellow dwarf virus transmission by the aphid Rhopalosiphum padi. Entomol. Exp. Appl. 1991, 59, 79–85. [Google Scholar] [CrossRef]
- Friebe, A.; Vilich, V.; Hennig, L.; Kluge, M.; Sicker, D. Detoxification of benzoxazolinone allelochemicals from wheat by Gaeumannomyces graminis var. tritici, G. graminis var. graminis, G. graminis var. avenae, and Fusarium culmorum. Appl. Environ. Microbiol. 1998, 64, 2386–2391. [Google Scholar] [PubMed]
- Niemeyer, H.M.; Jerez, J.M. Chromosomal location of genes for hydroxamic acid accumulation in Triticum aestivum L.(wheat) using wheat aneuploids and wheat substitution lines. Heredity 1997, 79. [Google Scholar] [CrossRef]
- Osbourn, A.; Clarke, B.; Lunness, P.; Scott, P.; Daniels, M. An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiol. Mol. Plant Pathol. 1994, 45, 457–467. [Google Scholar] [CrossRef]
- Morrissey, J.P.; Osbourn, A.E. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 1999, 63, 708–724. [Google Scholar] [PubMed]
- González-Lamothe, R.; Mitchell, G.; Gattuso, M.; Diarra, M.S.; Malouin, F.; Bouarab, K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009, 10, 3400–3419. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.; Clarke, B.; Dow, J.; Daniels, M. Partial characterization of avenacinase from Gaeumannomyces graminis var. avenae. Physiol. Mol. Plant Pathol. 1991, 38, 301–312. [Google Scholar] [CrossRef]
- Wu, H.; Pratley, J.; Lemerle, D.; Haig, T. Allelopathy in wheat (Triticum aestivum). Ann. Appl. Biol. 2001, 139, 1–9. [Google Scholar] [CrossRef]
- Adhikari, K.B.; Tanwir, F.; Gregersen, P.L.; Steffensen, S.K.; Jensen, B.M.; Poulsen, L.K.; Nielsen, C.H.; Høyer, S.; Borre, M.; Fomsgaard, I.S. Benzoxazinoids: Cereal phytochemicals with putative therapeutic and health-protecting properties. Mol. Nutr. Food Res. 2015, 59, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.A.; Loening, U.E.; Graham, M.C. Impacts of glyphosate-based herbicides on disease resistance and health of crops: A review. Environ. Sci. Eur. 2018, 30, 2. [Google Scholar] [CrossRef] [PubMed]
- Turlings, T.C.; Tumlinson, J.H.; Lewis, W.J. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 1990, 250, 1251. [Google Scholar] [CrossRef] [PubMed]
- Turlings, T.C.; Tumlinson, J.H.; Heath, R.R.; Proveaux, A.T.; Doolittle, R.E. Isolation and identification of allelochemicals that attract the larval parasitoid, Cotesia marginiventris (cresson), to the microhabitat of one of its hosts. J. Chem. Ecol. 1991, 17, 2235–2251. [Google Scholar] [CrossRef] [PubMed]
- Turlings, T.C.; McCall, P.J.; Alborn, H.T.; Tumlinson, J.H. An elicitor in caterpillar oral secretions that induces corn seedlings to emit chemical signals attractive to parasitic wasps. J. Chem. Ecol. 1993, 19, 411–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKay, S.A.B.; Hunter, W.L.; Godard, K.-A.; Wang, S.X.; Martin, D.M.; Bohlmann, J.; Plant, A.L. Insect attack and wounding induce traumatic resin duct development and gene expression of (−)-pinene synthase in Sitka spruce. Plant Physiol. 2003, 133, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.A.; Konno, K. Latex: A model for understanding mechanisms, ecology, and evolution of plant defense against herbivory. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 311–331. [Google Scholar] [CrossRef]
- Becerra, J.X. Squirt-gun defense in bursera and the chrysomelid counterploy. Ecology 1994, 75, 1991–1996. [Google Scholar] [CrossRef]
- Thomson, V.P.; Cunningham, S.A.; Ball, M.C.; Nicotra, A.B. Compensation for herbivory by Cucumis sativus through increased photosynthetic capacity and efficiency. Oecologia 2003, 134, 167–175. [Google Scholar] [CrossRef] [PubMed]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, R.J.; Thomashow, L.S.; Weller, D.M.; Fujimoto, D.; Mazzola, M.; Bangera, G.; Kim, D.-S. Molecular mechanisms of defense by rhizobacteria against root disease. Proc. Natl. Acad. Sci. USA 1995, 92, 4197–4201. [Google Scholar] [CrossRef] [PubMed]
- Campos-Soriano, L.; García-Martínez, J.; Segundo, B.S. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol. 2012, 13, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Y.; Cao, M.; Xie, L.J.; Liang, X.T.; Zeng, R.S.; Su, Y.J.; Huang, J.H.; Wang, R.L.; Luo, S.M. Induction of dimboa accumulation and systemic defense responses as a mechanism of enhanced resistance of mycorrhizal corn (Zea mays L.) to sheath blight. Mycorrhiza 2011, 21, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssières, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, L.; Martínez, A.; Sánchez, F.; Quinto, C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 2008, 56, 802–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, R.M.; Long, S.R. Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 2004, 134, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Thrall, P.H.; Papaïx, J.; Xie, L.; Burdon, J.J. Playing on a pathogen’s weakness: Using evolution to guide sustainable plant disease control strategies. Ann. Rev. Phytopathol. 2015, 53, 19–43. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Golstein, C.; Ade, J.; Stoutemyer, M.; Dixon, J.E.; Innes, R.W. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 2003, 301, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; Dubiella, U.; Kim, S.H.; Sloss, D.I.; Dowen, R.H.; Dixon, J.E.; Innes, R.W. Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein resistance to PSEUDOMONAS SYRINGIAE5 is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 2014, 164, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Sarma, B.K.; Singh, H.B.; Fernando, D.; Silva, R.N.; Gupta, V.K. Enhancing plant disease resistance without R genes. Trends Biotechnol. 2016, 34, 523–525. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 1994, 367, 384–386. [Google Scholar]
- Zhou, H.; Steffenson, B. Genome-wide association mapping reveals genetic architecture of durable spot blotch resistance in US barley breeding germplasm. Mol. Breed. 2013, 32, 139–154. [Google Scholar] [CrossRef]
- Wilcoxson, R.; Rasmusson, D.; Miles, M. Development of barley resistant to spot blotch and genetics of resistance. Plant Dis. 1990, 74, 207–210. [Google Scholar] [CrossRef]
- Haas, M.; Menke, J.; Chao, S.; Steffenson, B.J. Mapping quantitative trait loci conferring resistance to a widely virulent isolate of Cochliobolus sativus in wild barley accession PI 466423. Theor. Appl. Genet. 2016, 129, 1831–1842. [Google Scholar] [CrossRef]
- Valjavec-Gratian, M.; Steffenson, B. Pathotypes of Cochliobolus sativus on barley in North Dakota. Plant Dis. 1997, 81, 1275–1278. [Google Scholar] [CrossRef]
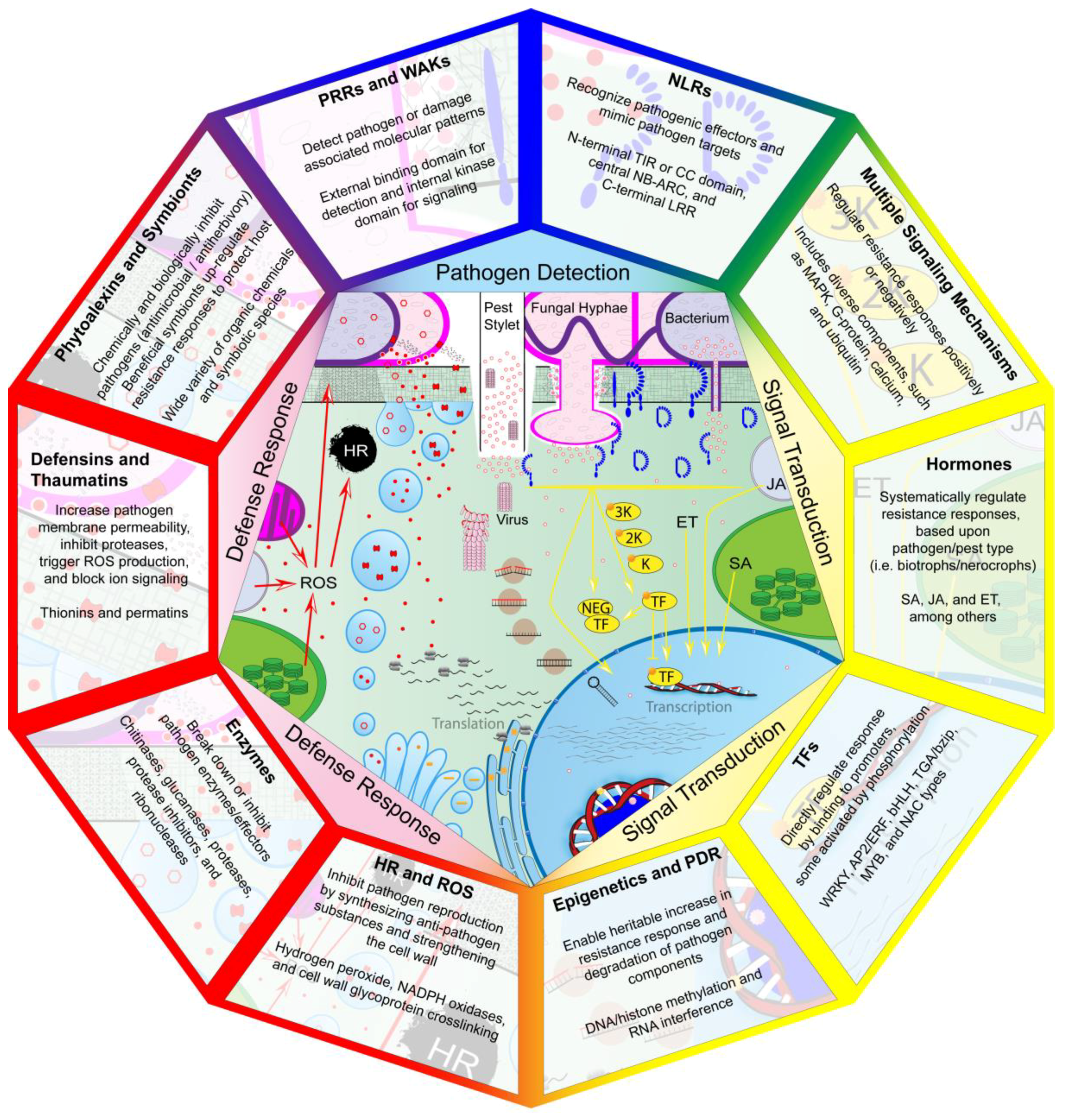
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease Resistance Mechanisms in Plants. Genes 2018, 9, 339. https://doi.org/10.3390/genes9070339
Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP. Disease Resistance Mechanisms in Plants. Genes. 2018; 9(7):339. https://doi.org/10.3390/genes9070339
Chicago/Turabian StyleAndersen, Ethan J., Shaukat Ali, Emmanuel Byamukama, Yang Yen, and Madhav P. Nepal. 2018. "Disease Resistance Mechanisms in Plants" Genes 9, no. 7: 339. https://doi.org/10.3390/genes9070339
APA StyleAndersen, E. J., Ali, S., Byamukama, E., Yen, Y., & Nepal, M. P. (2018). Disease Resistance Mechanisms in Plants. Genes, 9(7), 339. https://doi.org/10.3390/genes9070339





