Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Tissue Culture
2.2. Tumor Tissue
2.3. Somatostatin Analogues
2.4. Transfection Studies and Cell Growth Analyses
2.5. RNA Extraction
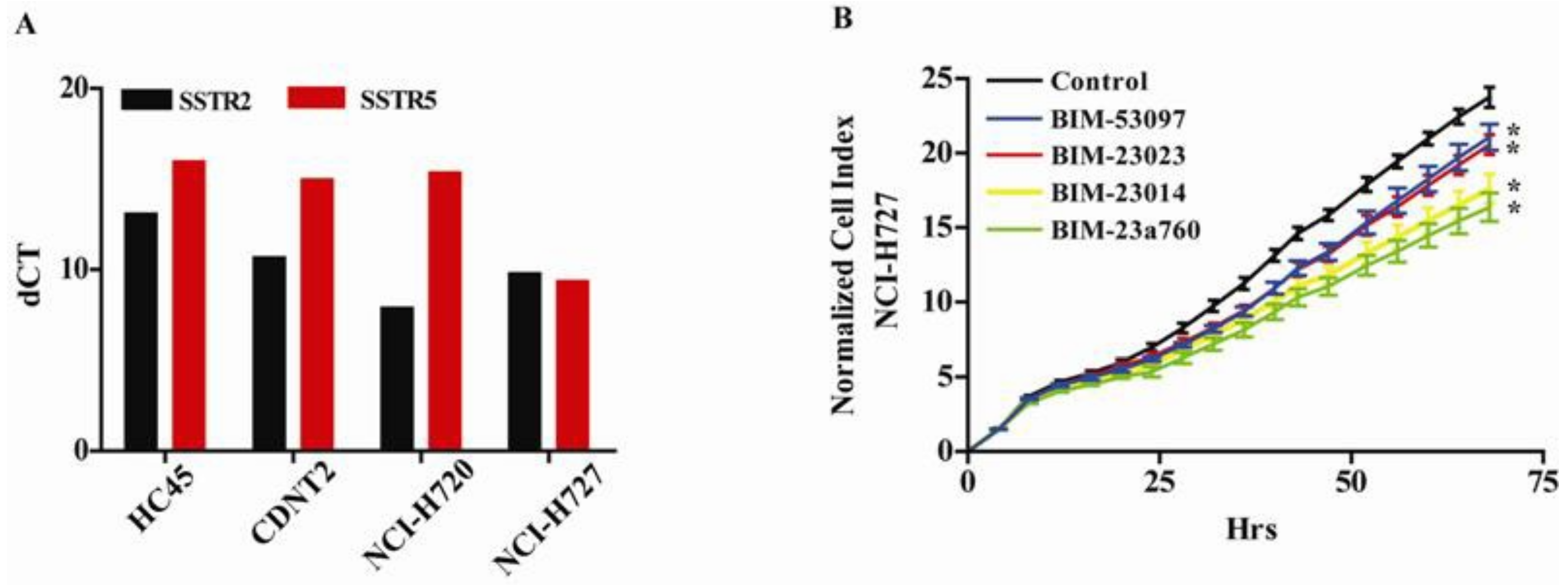
2.6. Somatostatin Receptors mRNA Quantification
2.7. Laser Capture Microdissection
2.8. qPCR of microRNA Expression
2.9. In Situ Hybridization
2.10. MicroRNA Microarray
2.11. RNA Profiling
2.12. Data Analysis
2.13. Statistical Analyses
3. Results
3.1. Somatostatin and Dopamine Analogues Inhibit the Growth of a Carcinoid Cell Line NCI-H727
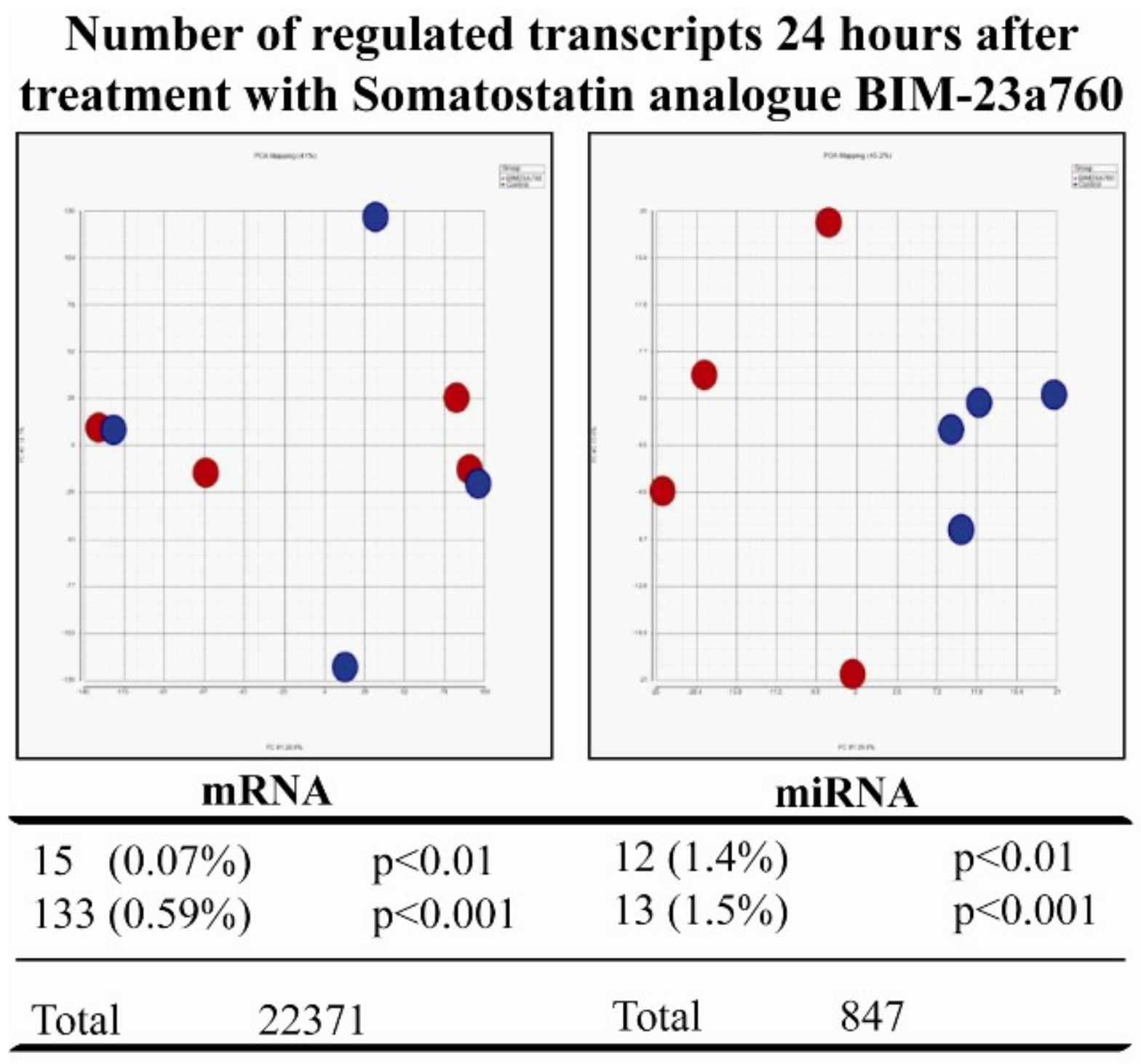
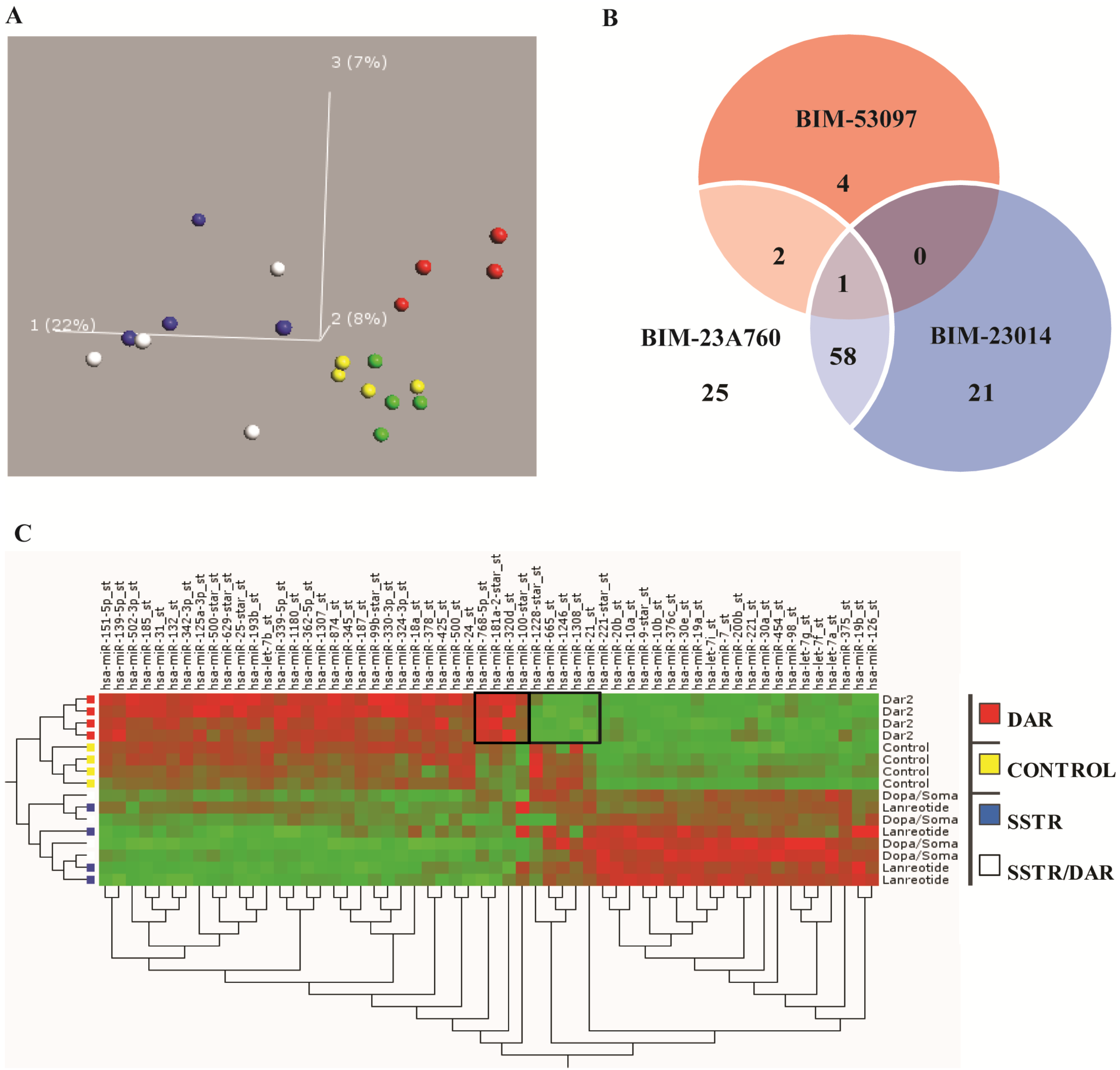
3.2. Somatostatin Receptor Activation Primarily Induces microRNA Expression Changes
3.3. Somatostatin Induces Distinct Receptor Based/Activated microRNA Expression Profiles and Particularly Up-Regulates miR-7 and miR-148a
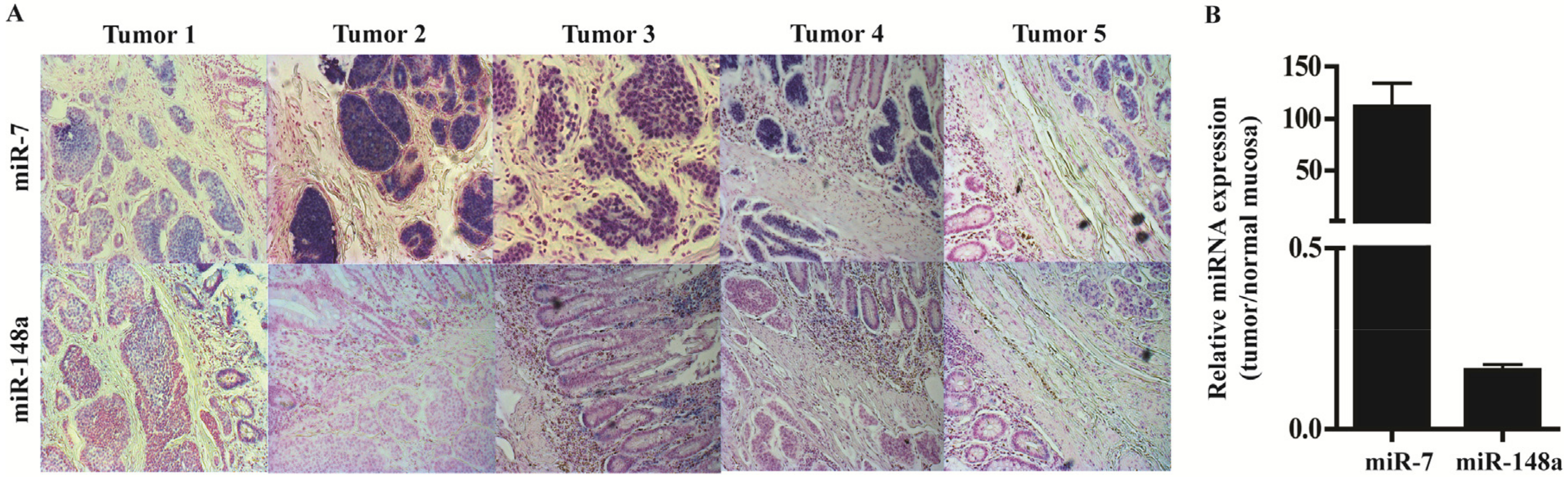
3.4. In Situ Hybridization and qPCR on Neuroendocrine Tumors and Neuroendocrine Tumor Laser Capture Microdissected Tissue Confirms the Presence of miR-7 and miR-148a
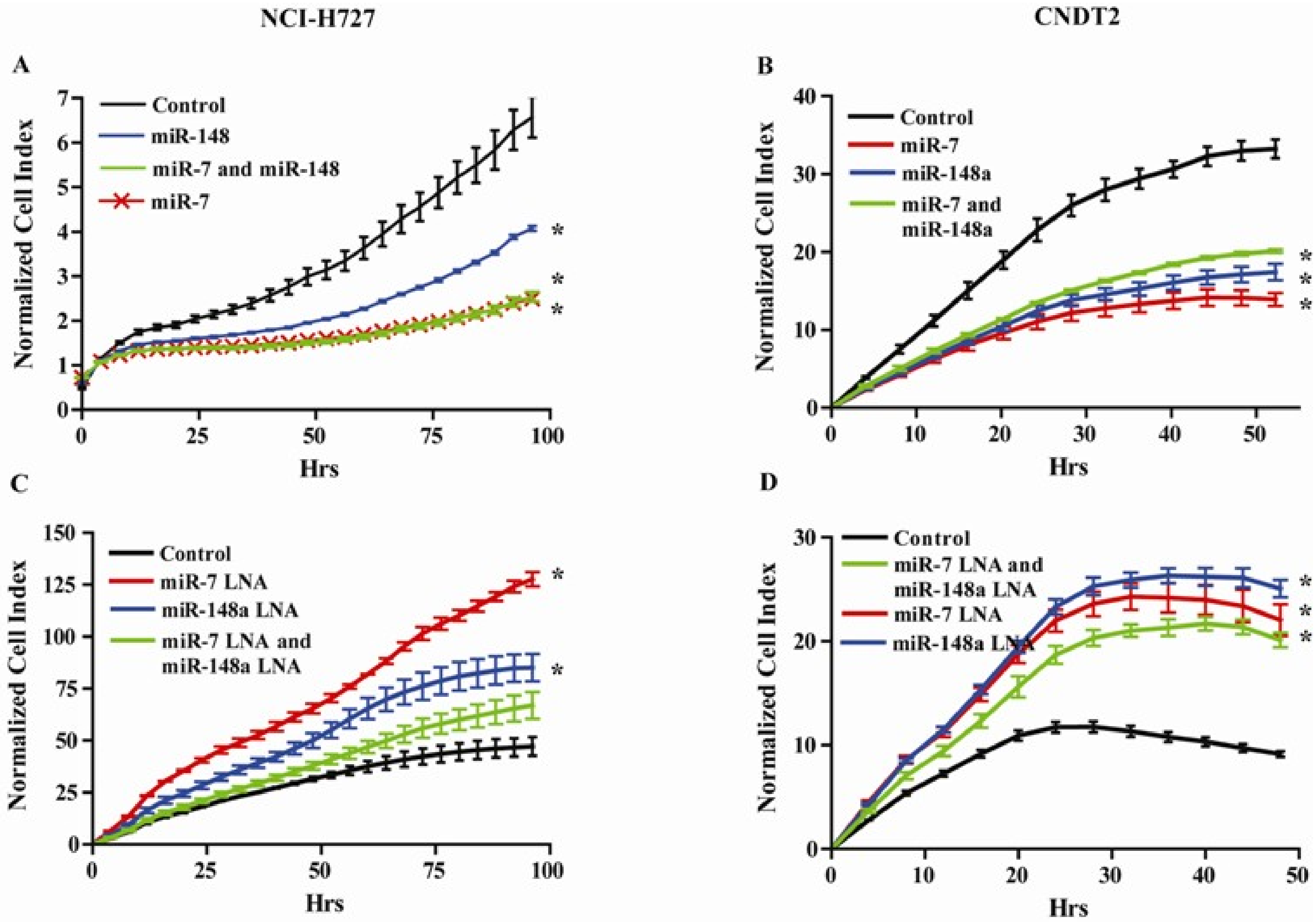
3.5. miR-7 and miR-148a Modulate the Growth of NCI-H727 and CNDT2 Carcinoid Cell Lines
3.6. Somatostatin Modulates the Expression of the Let-7 Family
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Modlin, I.M.; Oberg, K.; Chung, D.C.; Jensen, R.T.; de Herder, W.W.; Thakker, R.V.; Caplin, M.; Delle Fave, G.; Kaltsas, G.A.; Krenning, E.P.; et al. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol. 2008, 9, 61–72. [Google Scholar] [CrossRef]
- Kloppel, G. Classification and pathology of gastroenteropancreatic neuroendocrine neoplasms. Endocr. Relat. Cancer 2011, 18 (Suppl. 1), S1–S16. [Google Scholar] [CrossRef] [PubMed]
- Janson, E.T.; Sorbye, H.; Welin, S.; Federspiel, B.; Gronbaek, H.; Hellman, P.; Ladekarl, M.; Langer, S.W.; Mortensen, J.; Schalin-Jantti, C.; et al. Nordic guidelines 2014 for diagnosis and treatment of gastroenteropancreatic neuroendocrine neoplasms. Acta Oncol. 2014, 53, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D. The 2015 World Health Organization classification of lung tumors. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.A.; Beyer, D.T.; Diebold, A.E.; Voros, B.A.; Chester, M.M.; Wang, Y.Z.; Boudreaux, J.P.; Woltering, E.A.; Uhlhorn, A.P.; Ryan, P.; et al. Prognostic factors in typical and atypical pulmonary carcinoids. Ochsner. J. 2017, 17, 335–340. [Google Scholar] [PubMed]
- Lepage, C.; Rachet, B.; Coleman, M.P. Survival from malignant digestive endocrine tumors in England and Wales: A population-based study. Gastroenterology 2007, 132, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Niederle, M.B.; Hackl, M.; Kaserer, K.; Niederle, B. Gastroenteropancreatic neuroendocrine tumours: The current incidence and staging based on the Who and European neuroendocrine tumour society classification: An analysis based on prospectively collected parameters. Endocr. Relat. Cancer 2010, 17, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Caplin, M.E.; Baudin, E.; Ferolla, P.; Filosso, P.; Garcia-Yuste, M.; Lim, E.; Oberg, K.; Pelosi, G.; Perren, A.; Rossi, R.E.; et al. Pulmonary neuroendocrine (carcinoid) tumors: European neuroendocrine tumor society expert consensus and recommendations for best practice for typical and atypical pulmonary carcinoids. Ann. Oncol. 2015, 26, 1604–1620. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, H.; Vanberkel, V.; Bade, B.C.; Kloecker, G. Clinical presentation, diagnosis, and management of typical and atypical bronchopulmonary carcinoid. J. Community Support 2017, 15, E303–E308. [Google Scholar] [CrossRef]
- Eriksson, B.; Arnberg, H.; Lindgren, P.G.; Lorelius, L.E.; Magnusson, A.; Lundqvist, G.; Skogseid, B.; Wide, L.; Wilander, E.; Oberg, K. Neuroendocrine pancreatic tumours: Clinical presentation, biochemical and histopathological findings in 84 patients. J. Intern. Med. 1990, 228, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Hendifar, A.E.; Marchevsky, A.M.; Tuli, R. Neuroendocrine tumors of the lung: Current challenges and advances in the diagnosis and management of well-differentiated disease. J. Thorac. Oncol. 2017, 12, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Ramage, J.K.; Davies, A.H.; Ardill, J.; Bax, N.; Caplin, M.; Grossman, A.; Hawkins, R.; McNicol, A.M.; Reed, N.; Sutton, R.; et al. Guidelines for the management of gastroenteropancreatic neuroendocrine (including carcinoid) tumours. Gut 2005, 54 (Suppl. 4), iv1–iv16. [Google Scholar] [CrossRef] [PubMed]
- Plockinger, U.; Wiedenmann, B. Treatment of gastroenteropancreatic neuroendocrine tumors. Virchows Arch. 2007, 451 (Suppl. 1), S71–S80. [Google Scholar] [CrossRef] [PubMed]
- Cameselle-Teijeiro, J.M.; Mato Mato, J.A.; Fernandez Calvo, O.; Garcia Mata, J. Neuroendocrine pulmonary tumors of low, intermediate and high grade: Anatomopathological diagnosis-prognostic and predictive factors. Mol. Diagn. Ther. 2018, 22, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Reichlin, S. Secretion of somatostatin and its physiologic function. J. Lab. Clin. Med. 1987, 109, 320–326. [Google Scholar] [PubMed]
- Larsson, L.I.; Goltermann, N.; de Magistris, L.; Rehfeld, J.F.; Schwartz, T.W. Somatostatin cell processes as pathways for paracrine secretion. Science 1979, 205, 1393–1395. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Post, S.R.; Wang, K.; Tager, H.S.; Bell, G.I.; Seino, S. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc. Natl. Acad. Sci. USA 1992, 89, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Papotti, M.; Bongiovanni, M.; Volante, M.; Allia, E.; Landolfi, S.; Helboe, L.; Schindler, M.; Cole, S.L.; Bussolati, G. Expression of somatostatin receptor types 1–5 in 81 cases of gastrointestinal and pancreatic endocrine tumors. A correlative immunohistochemical and reverse-transcriptase polymerase chain reaction analysis. Virchows Arch. 2002, 440, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Modlin, I.M.; Pavel, M.; Kidd, M.; Gustafsson, B.I. Review article: Somatostatin analogues in the treatment of gastroenteropancreatic neuroendocrine (carcinoid) tumours. Aliment. Pharmacol. Ther. 2010, 31, 169–188. [Google Scholar] [PubMed]
- Binderup, T.; Knigge, U.; Loft, A.; Mortensen, J.; Pfeifer, A.; Federspiel, B.; Hansen, C.P.; Hojgaard, L.; Kjaer, A. Functional imaging of neuroendocrine tumors: A head-to-head comparison of somatostatin receptor scintigraphy, 123i-MIBG scintigraphy, and 18F-FDG PET. J. Nucl. Med. 2010, 51, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Buscail, L.; Saint-Laurent, N.; Chastre, E.; Vaillant, J.C.; Gespach, C.; Capella, G.; Kalthoff, H.; Lluis, F.; Vaysse, N.; Susini, C. Loss of sst2 somatostatin receptor gene expression in human pancreatic and colorectal cancer. Cancer Res. 1996, 56, 1823–1827. [Google Scholar] [PubMed]
- Sheppard, M.; Shapiro, B.; Pimstone, B.; Kronheim, S.; Berelowitz, M.; Gregory, M. Metabolic clearance and plasma half-disappearance time of exogenous somatostatin in man. J. Clin. Endocrinol. Metab. 1979, 48, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Appetecchia, M.; Baldelli, R. Somatostatin analogues in the treatment of gastroenteropancreatic neuroendocrine tumours, current aspects and new perspectives. J. Exp. Clin. Cancer Res. 2010, 29, 19. [Google Scholar] [CrossRef] [PubMed]
- Cives, M.; Kunz, P.L.; Morse, B.; Coppola, D.; Schell, M.J.; Campos, T.; Nguyen, P.T.; Nandoskar, P.; Khandelwal, V.; Strosberg, J.R. Phase II clinical trial of pasireotide long-acting repeatable in patients with metastatic neuroendocrine tumors. Endocr. Relat. Cancer 2015, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alexandraki, K.I.; Karapanagioti, A.; Karoumpalis, I.; Boutzios, G.; Kaltsas, G.A. Advances and current concepts in the medical management of gastroenteropancreatic neuroendocrine neoplasms. Biomed Res. Int. 2017, 2017, 9856140. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.Y. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune Netw. 2011, 11, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Van Buren, G., 2nd; Rashid, A.; Yang, A.D.; Abdalla, E.K.; Gray, M.J.; Liu, W.; Somcio, R.; Fan, F.; Camp, E.R.; Yao, J.C.; et al. The development and characterization of a human midgut carcinoid cell line. Clin. Cancer Res. 2007, 13, 4704–4712. [Google Scholar] [CrossRef] [PubMed]
- Stilling, G.A.; Zhang, H.; Ruebel, K.H.; Leontovich, A.A.; Jin, L.; Tanizaki, Y.; Zhang, S.; Erickson, L.A.; Hobday, T.; Lloyd, R.V. Characterization of the functional and growth properties of cell lines established from ileal and rectal carcinoid tumors. Endocr. Pathol. 2007, 18, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Kidd, M.; Drozdov, I.; Joseph, R.; Pfragner, R.; Culler, M.; Modlin, I. Differential cytotoxicity of novel somatostatin and dopamine chimeric compounds on bronchopulmonary and small intestinal neuroendocrine tumor cell lines. Cancer 2008, 113, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Arvigo, M.; Gatto, F.; Ruscica, M.; Ameri, P.; Dozio, E.; Albertelli, M.; Culler, M.D.; Motta, M.; Minuto, F.; Magni, P.; et al. Somatostatin and dopamine receptor interaction in prostate and lung cancer cell lines. J. Endocrinol. 2010, 207, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Kustermann, S.; Boess, F.; Buness, A.; Schmitz, M.; Watzele, M.; Weiser, T.; Singer, T.; Suter, L.; Roth, A. A label-free, impedance-based real time assay to identify drug-induced toxicities and differentiate cytostatic from cytotoxic effects. Toxicol. In Vitro 2013, 27, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Ke, N.; Wang, X.; Xu, X.; Abassi, Y.A. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 2011, 740, 33–43. [Google Scholar] [PubMed]
- Binderup, T.; Knigge, U.; Mellon Mogensen, A.; Palnaes Hansen, C.; Kjaer, A. Quantitative gene expression of somatostatin receptors and noradrenaline transporter underlying scintigraphic results in patients with neuroendocrine tumors. Neuroendocrinology 2008, 87, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.; Baker, A.; Moller, S.; Nielsen, B.S. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 2010, 52, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Dossing, K.B.; Binderup, T.; Kaczkowski, B.; Jacobsen, A.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjaer, A.; Friis-Hansen, L. Down-regulation of miR-129-5p and the let-7 family in neuroendocrine tumors and metastases leads to up-regulation of their targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2014, 6, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Strosberg, J.R.; Fisher, G.A.; Benson, A.B.; Malin, J.L.; Panel, G.T.C.; Cherepanov, D.; Broder, M.S.; Anthony, L.B.; Arslan, B.; Fisher, G.A.; et al. Systemic treatment in unresectable metastatic well-differentiated carcinoid tumors: Consensus results from a modified delphi process. Pancreas 2013, 42, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Rinke, A.; Muller, H.H.; Schade-Brittinger, C.; Klose, K.J.; Barth, P.; Wied, M.; Mayer, C.; Aminossadati, B.; Pape, U.F.; Blaker, M.; et al. Placebo-controlled, double-blind, prospective, randomized study on the effect of octreotide lar in the control of tumor growth in patients with metastatic neuroendocrine midgut tumors: A report from the PROMID study group. J. Clin. Oncol. 2009, 27, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Caplin, M.E.; Pavel, M.; Ruszniewski, P. Lanreotide in metastatic enteropancreatic neuroendocrine tumors. N. Engl. J. Med. 2014, 371, 1556–1557. [Google Scholar] [CrossRef] [PubMed]
- Kidd, M.; Schally, A.V.; Pfragner, R.; Malfertheiner, M.V.; Modlin, I.M. Inhibition of proliferation of small intestinal and bronchopulmonary neuroendocrine cell lines by using peptide analogs targeting receptors. Cancer 2008, 112, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Martijn, C.; Cui, T.; Essaghir, A.; Luque, R.M.; Demoulin, J.B.; Castano, J.P.; Oberg, K.; Giandomenico, V. The somatostatin analogue octreotide inhibits growth of small intestine neuroendocrine tumour cells. PLoS ONE 2012, 7, e48411. [Google Scholar] [CrossRef] [PubMed]
- Kharmate, G.; Rajput, P.S.; Lin, Y.C.; Kumar, U. Inhibition of tumor promoting signals by activation of SSTR2 and opioid receptors in human breast cancer cells. Cancer Cell Int. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, I.; Pacheco, T.R.; Costa, A.; Santos, A.C.; Fernandes, A.R.; Santos, M.; Oliveira, A.G.; Casimiro, S.; Quintela, A.; Fernandes, A.; et al. Prognostic significance of AKT/mTOR signaling in advanced neuroendocrine tumors treated with somatostatin analogs. Onco Targets Ther. 2012, 5, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/AKT/mTOR signaling in cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Johnbeck, C.B.; Munk Jensen, M.; Haagen Nielsen, C.; Fisker Hag, A.M.; Knigge, U.; Kjaer, A. 18F-FDG and 18F-FLT-pet imaging for monitoring everolimus effect on tumor-growth in neuroendocrine tumors: Studies in human tumor xenografts in mice. PLoS ONE 2014, 9, e91387. [Google Scholar]
- Raymond, E.; Dahan, L.; Raoul, J.L.; Bang, Y.J.; Borbath, I.; Lombard-Bohas, C.; Valle, J.; Metrakos, P.; Smith, D.; Vinik, A.; et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011, 364, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.C.; Shah, M.H.; Ito, T.; Bohas, C.L.; Wolin, E.M.; Van Cutsem, E.; Hobday, T.J.; Okusaka, T.; Capdevila, J.; de Vries, E.G.; et al. Everolimus for advanced pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011, 364, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Pavel, M.E.; Hainsworth, J.D.; Baudin, E.; Peeters, M.; Horsch, D.; Winkler, R.E.; Klimovsky, J.; Lebwohl, D.; Jehl, V.; Wolin, E.M.; et al. Everolimus plus octreotide long-acting repeatable for the treatment of advanced neuroendocrine tumours associated with carcinoid syndrome (RADIANT-2): A randomised, placebo-controlled, phase 3 study. Lancet 2011, 378, 2005–2012. [Google Scholar] [CrossRef]
- Bousquet, C.; Guillermet-Guibert, J.; Saint-Laurent, N.; Archer-Lahlou, E.; Lopez, F.; Fanjul, M.; Ferrand, A.; Fourmy, D.; Pichereaux, C.; Monsarrat, B.; et al. Direct binding of p85 to sst2 somatostatin receptor reveals a novel mechanism for inhibiting PI3K pathway. EMBO J. 2006, 25, 3943–3954. [Google Scholar] [CrossRef] [PubMed]
- Briest, F.; Grabowski, P. PI3K-AKT-mTOR-signaling and beyond: The complex network in gastroenteropancreatic neuroendocrine neoplasms. Theranostics 2014, 4, 336–365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, X.; Wu, C.W.; Dong, Y.; Cai, M.; Mok, M.T.; Wang, H.; Chen, J.; Ng, S.S.; Chen, M.; et al. MicroRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 2013, 32, 5078–5088. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Dou, W.; He, L.; Liang, S.; Tie, J.; Liu, C.; Li, T.; Lu, Y.; Mo, P.; Shi, Y.; et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 2013, 32, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Kredo-Russo, S.; Mandelbaum, A.D.; Ness, A.; Alon, I.; Lennox, K.A.; Behlke, M.A.; Hornstein, E. Pancreas-enriched miRNA refines endocrine cell differentiation. Development 2012, 139, 3021–3031. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.N.; Huang, Q.; Tian, L. The molecular mechanisms and therapeutic potential of microRNA-7 in cancer. Expert Opin. Ther. Targets 2015, 19, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wen, Z.; Zhou, Y.; Liu, Z.; Li, Q.; Fei, G.; Luo, J.; Ren, T. MicroRNA-7-regulated TLR9 signaling-enhanced growth and metastatic potential of human lung cancer cells by altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt pathway. Mol. Biol. Cell 2013, 24, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xue, J.L.; Shen, Q.; Chen, J.; Tian, L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012, 55, 1852–1862. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Xiong, Y.; Xu, W.; Wang, Y.; Chen, F.; Wang, Z.; Yan, Z. A two-microRNA signature as a potential biomarker for early gastric cancer. Oncol. Lett. 2014, 7, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Hanoun, N.; Delpu, Y.; Suriawinata, A.A.; Bournet, B.; Bureau, C.; Selves, J.; Tsongalis, G.J.; Dufresne, M.; Buscail, L.; Cordelier, P.; et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin. Chem. 2010, 56, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Shivapurkar, N.; Weiner, L.M.; Marshall, J.L.; Madhavan, S.; Deslattes Mays, A.; Juhl, H.; Wellstein, A. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS ONE 2014, 9, e84686. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, N.; Naito, Y.; Oue, N.; Sentani, K.; Uraoka, N.; Zarni Oo, H.; Yanagihara, K.; Aoyagi, K.; Sasaki, H.; Yasui, W. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci. 2014, 105, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Song, Y.; Wang, Z.; Wang, G.; Gao, P.; Chen, X.; Gao, Z.; Xu, H. Clinical significance of promoter region hypermethylation of microRNA-148a in gastrointestinal cancers. Onco Targets Ther. 2014, 7, 853–863. [Google Scholar] [PubMed]
- Murray, R.D.; Kim, K.; Ren, S.G.; Chelly, M.; Umehara, Y.; Melmed, S. Central and peripheral actions of somatostatin on the growth hormone-IGF-I axis. J. Clin. Investig. 2004, 114, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Libutti, S.K. Therapy: Blockade of IGF-1R-not effective in neuroendocrine tumours. Nat. Rev. Endocrinol. 2013, 9, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Gregory, R.I. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012, 22, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Wu, R.; Liu, B.; Dong, W.; Tu, Z.; Yang, J.; Xu, Z.; Pan, T. Upstream and downstream mechanisms for the promoting effects of IGF-1 on differentiation of spermatogonia to primary spermatocytes. Life Sci. 2014, 101, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Alajez, N.M.; Shi, W.; Wong, D.; Lenarduzzi, M.; Waldron, J.; Weinreb, I.; Liu, F.F. Lin28b promotes head and neck cancer progression via modulation of the insulin-like growth factor survival pathway. Oncotarget 2012, 3, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
| Patient | Age/Sex | Location | Ki67 Index | Treatment |
|---|---|---|---|---|
| 1 | 65/F | Colon | 1% | Surgery |
| 2 | 69/F | Small intestine | 3% | Surgery/SSA |
| 3 | 62/M | Ileum | 2% | Surgery |
| 4 | 55/F | Small intestine | 7–8% | Surgery |
| 5 | 64/F | Ileocecal | 1% | Surgery/SSA |
| Compound | Somatostatin Receptor Subtype | Dopamine | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| Somatostatin 14 | 1.95 | 0.25 | 1.2 | 1.77 | 1.4 | ||
| Somatostatin 28 | 1.86 | 0.31 | 1.3 | 5.4 | 0.4 | ||
| BIM-23014 (Lanreotide) | >1000 | 0.75 | 98 | >1000 | 12.7 | ||
| BIM-23023 | >1000 | 0.42 | 87 | 2.7 | 4.2 | ||
| BIM-53097 | 22.1 | ||||||
| BIM-23A760 | 622 | 0.03 | 160 | >1000 | 42.0 | 15 | |
| MiRNA | Mature Sequence | Product ID |
|---|---|---|
| miR-7 | UGGAAGACUAGUGAUUUUGUUGU | 000268 |
| miR-148a | UCAGUGCACUACAGAACUUUGU | 000470 |
| MiR-191 | CAACGGAAUCCCAAAAGCAGCUG | 002299 |
| RNU-44 | CCTGGATGATGATAGCAAATGCTG-ACTGAACATGAAGGTCTTAATTAGCTCTAACTGACT | 001094 |
| miRNA | Change | BIM-23014 | BIM-23A760 | BIM-53097 | |||
|---|---|---|---|---|---|---|---|
| FC | p-Value | FC | p-Value | FC | p-Value | ||
| miR-769-3p | −2.8 | 0.001 | −2.6 | 0.002 | −1.1 | NS | |
| miR-663b | −2.0 | 0.009 | −2.5 | 0.001 | −2.9 | 0.00 | |
| miR-663 |  | −2.0 | 0.007 | −2.3 | 0.002 | −3.3 | 0.00 |
| miR-30b-star | −2.0 | 0.001 | −2.2 | 0.000 | −1.3 | NS | |
| miR-297 | −1.7 | 0.008 | −2.0 | 0.001 | −1.7 | 0.01 | |
| miR-483-5p | −1.9 | 0.003 | −2.0 | 0.002 | −1.0 | NS | |
| miR-376c | 2.1 | 0.000 | 2.0 | 0.000 | −1.2 | NS | |
| Let-7f | 1.8 | 0.001 | 2.1 | 0.000 | −1.2 | NS | |
| miR-10a-star |  | 1.9 | 0.001 | 2.1 | 0.000 | 1.1 | NS |
| miR-495 | 2.0 | 0.001 | 2.2 | 0.000 | 1.6 | 0.01 | |
| miR-7 | 2.3 | 0.000 | 2.2 | 0.000 | −1.1 | NS | |
| miR-9-star | 2.3 | 0.000 | 2.3 | 0.000 | −1.4 | NS | |
| miR-454 | 1.9 | 0.001 | 2.3 | 0.000 | −1.0 | NS | |
| miR-26b | 2.2 | 0.010 | 2.6 | 0.003 | 1.0 | NS | |
| miR-429 | 2.9 | 0.004 | 2.8 | 0.004 | −1.4 | NS | |
| miR-9 | 1.5 | NS | 2.9 | 0.002 | −1.5 | NS | |
| miR-148a | 2.5 | 0.006 | 2.9 | 0.002 | 2.0 | 0.03 | |
| miR-30e-star | 2.8 | 0.000 | 3.1 | 0.000 | 1.5 | NS | |
| miRNA | Change | BIM-23014 | BIM-23A760 | BIM-53097 | |||
|---|---|---|---|---|---|---|---|
| FC | p-Value | FC | p-Value | FC | p-Value | ||
| let-7a | → | 1.2 | 0.000 | 1.2 | 0.000 | −1.0 | NS |
| let-7b | ↓ | −1.4 | 0.000 | −1.4 | 0.001 | −1.0 | NS |
| let-7c | → | −1.0 | NS | 1.0 | NS | −1.1 | NS |
| let-7d | → | −1.0 | NS | 1.0 | NS | 1.1 | NS |
| let-7e | → | −1.2 | NS | −1.1 | NS | 1.1 | NS |
| let-7f | ↑ | 1.8 | 0.002 | 2.1 | 0.000 | −1.2 | NS |
| let-7g | ↑ | 1.7 | 0.001 | 1.9 | 0.000 | −1.1 | NS |
| let-7i | ↑ | 1.3 | 0.000 | 1.4 | 0.000 | −1.1 | NS |
| miR-98 | ↑ | 1.8 | 0.004 | 2.2 | 0.000 | −1.1 | NS |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Døssing, K.B.V.; Kjær, C.; Vikeså, J.; Binderup, T.; Knigge, U.; Culler, M.D.; Kjær, A.; Federspiel, B.; Friis-Hansen, L. Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells. Genes 2018, 9, 337. https://doi.org/10.3390/genes9070337
Døssing KBV, Kjær C, Vikeså J, Binderup T, Knigge U, Culler MD, Kjær A, Federspiel B, Friis-Hansen L. Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells. Genes. 2018; 9(7):337. https://doi.org/10.3390/genes9070337
Chicago/Turabian StyleDøssing, Kristina B. V., Christina Kjær, Jonas Vikeså, Tina Binderup, Ulrich Knigge, Michael D. Culler, Andreas Kjær, Birgitte Federspiel, and Lennart Friis-Hansen. 2018. "Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells" Genes 9, no. 7: 337. https://doi.org/10.3390/genes9070337
APA StyleDøssing, K. B. V., Kjær, C., Vikeså, J., Binderup, T., Knigge, U., Culler, M. D., Kjær, A., Federspiel, B., & Friis-Hansen, L. (2018). Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells. Genes, 9(7), 337. https://doi.org/10.3390/genes9070337




