Selective Activation of Alternative MYC Core Promoters by Wnt-Responsive Enhancers
Abstract
1. Introduction
2. Materials and Methods
2.1. CAGE Data Analysis
2.2. Cell Culture
2.3. Cell Growth Curves
2.4. Quantification of P1 and P2 Promoter Activities
2.5. Reporter Luciferase Assay
2.6. Real Time qPCR
2.7. Genome Editing
3. Results
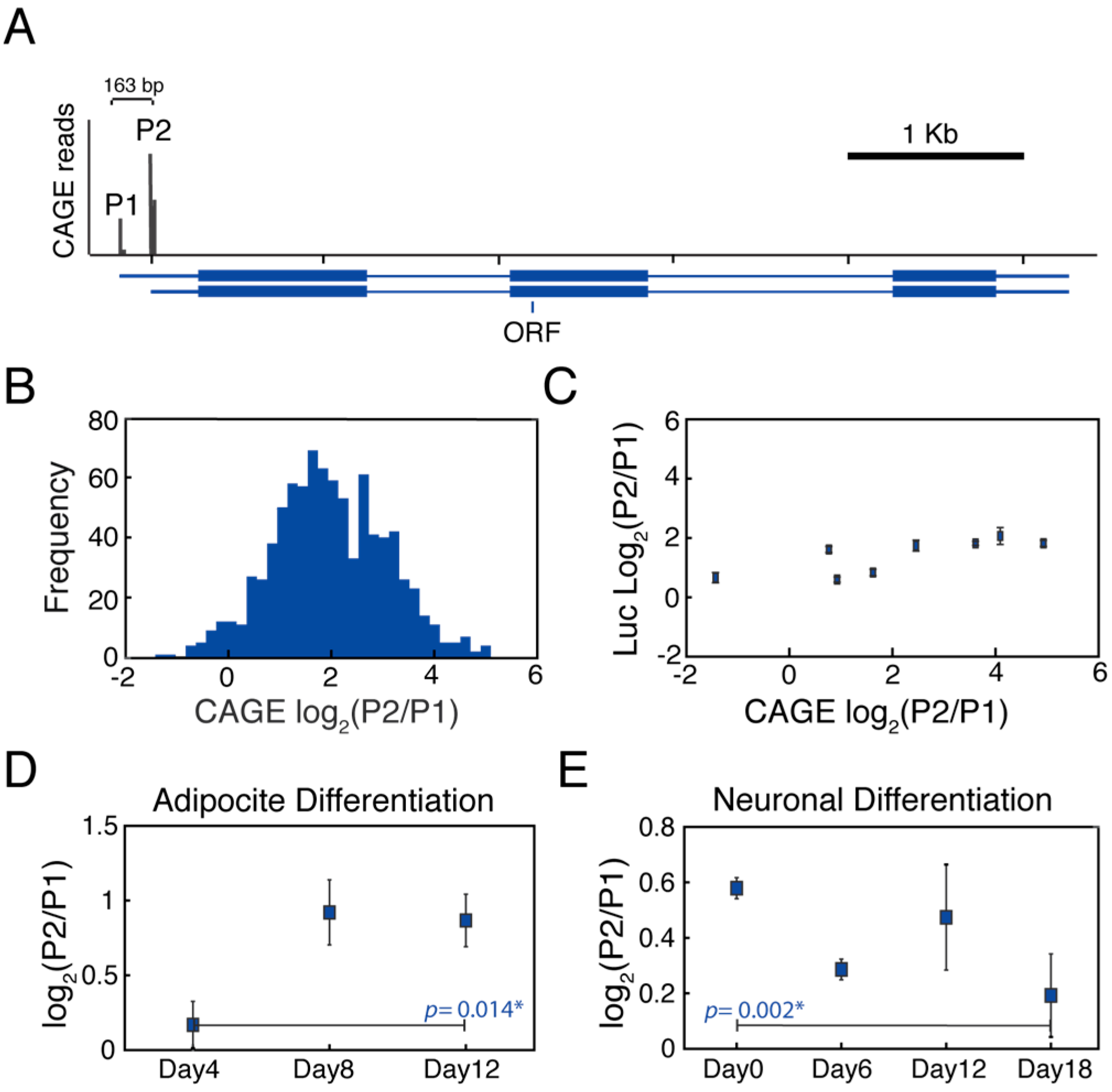
3.1. MYC Alternative Promoters Can Be Differentially Regulated
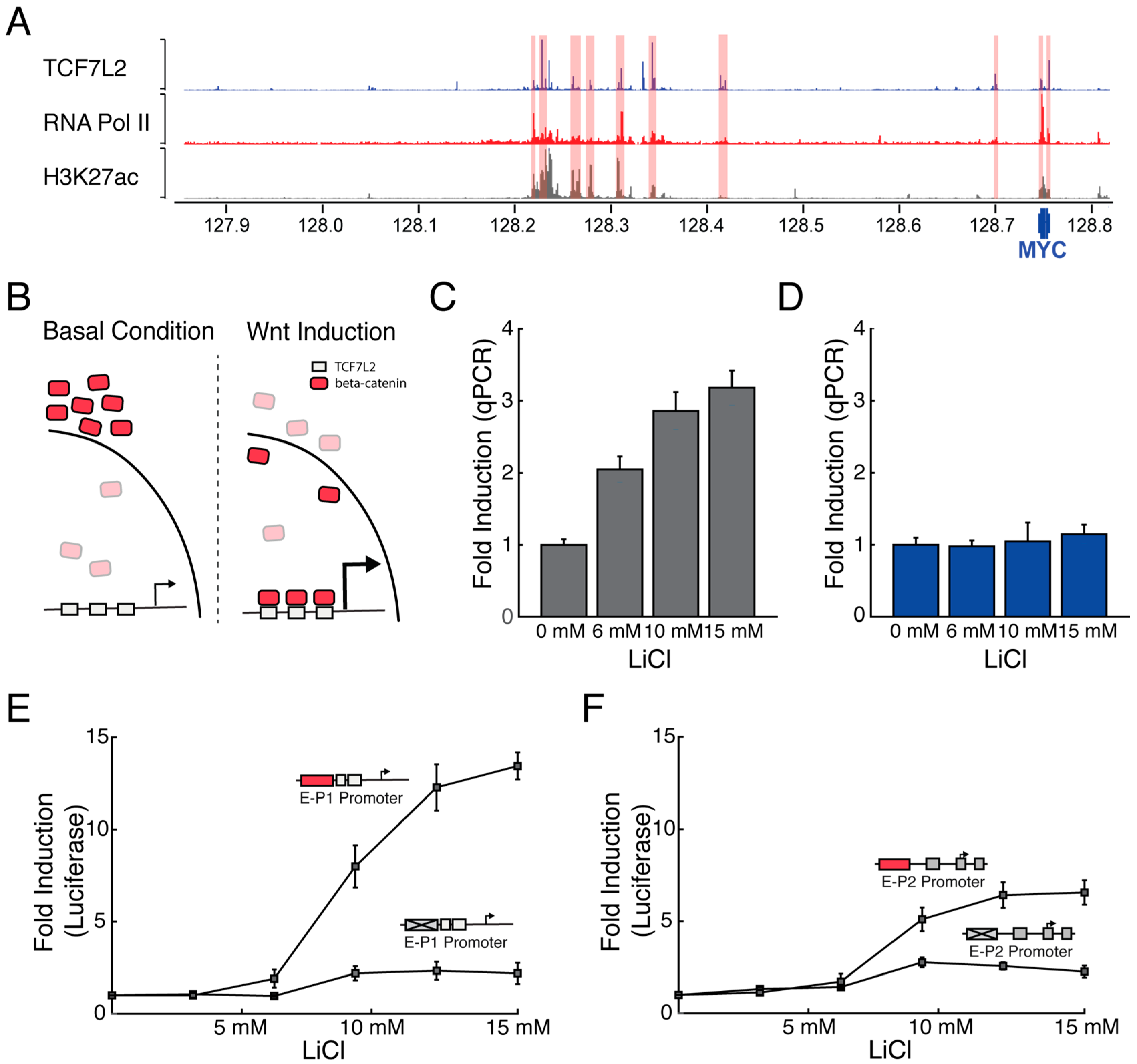
3.2. Wnt-Responsive Enhancers Preferentially Activate the P1 Promoter
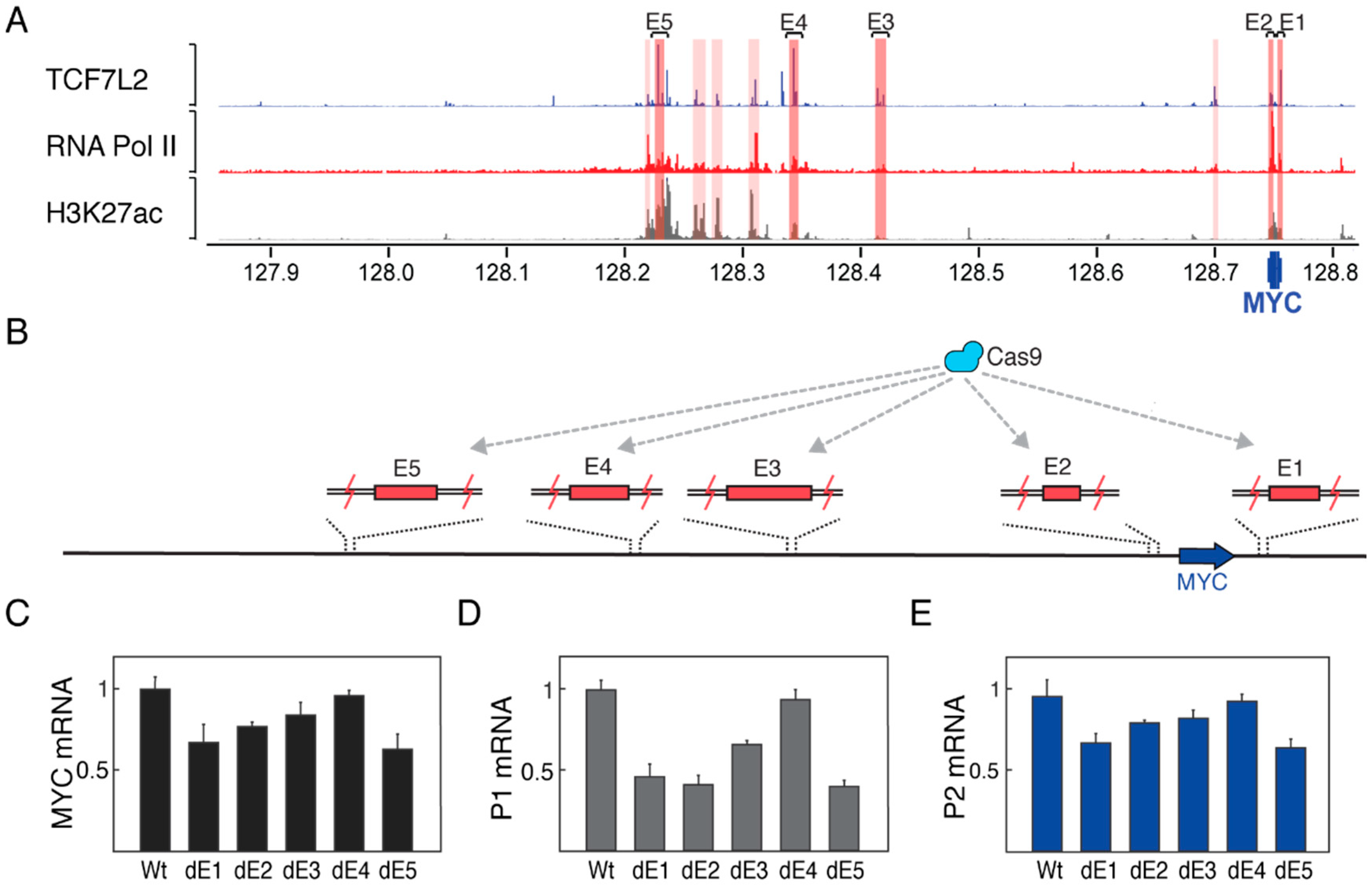
3.3. Enhancer Deletions Preferentially Downregulate Transcription from the P1 Promoter
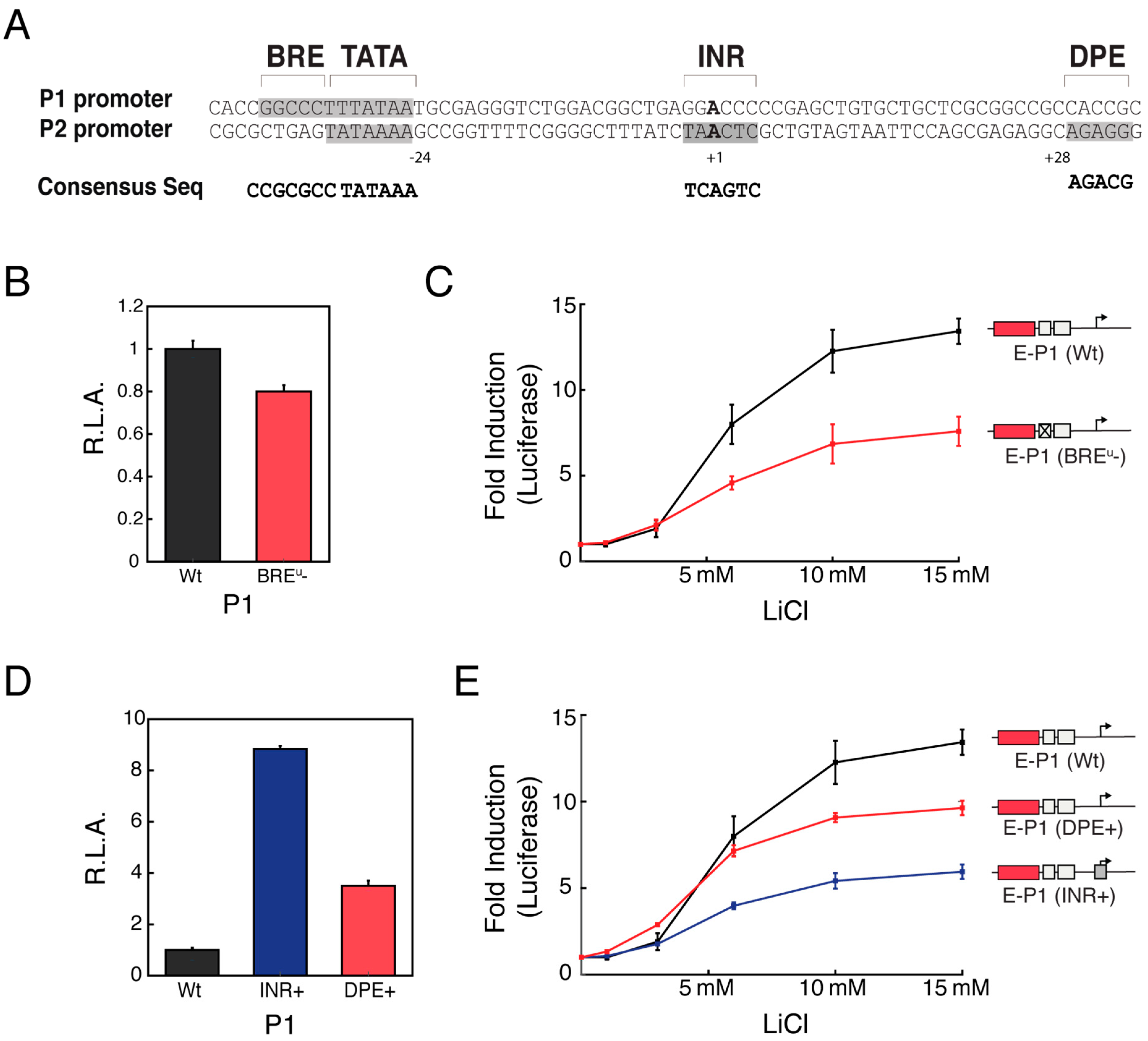
3.4. Distinct Core Promoter Architecture Influences Specific Enhancer-Promoter Communication
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, L.V.; Wang, Y.-L.; Kassavetis, G.A.; Kadonaga, J.T. The punctilious RNA polymerase II core promoter. Genes Dev. 2017, 31, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Lenhard, B.; Sandelin, A.; Carninci, P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012, 13, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Tora, L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim. Biophys. Acta 2014, 1839, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E.; Louder, R.K.; He, Y. Structural insights into the eukaryotic transcription initiation machinery. Annu. Rev. Biophys. 2017, 46, 59–83. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.F.; Kadonaga, J.T. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001, 15, 2515–2519. [Google Scholar] [CrossRef] [PubMed]
- Davis, W.; Schultz, R.M. Developmental change in TATA-box utilization during preimplantation mouse development. Dev. Biol. 2000, 218, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Hsu, J.-Y.; Kadonaga, J.T. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008, 22, 2823–2830. [Google Scholar] [CrossRef] [PubMed]
- Zabidi, M.A.; Stark, A. Regulatory enhancer–core-promoter communication via transcription factors and cofactors. Trends Genet. 2016, 32, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Davuluri, R.V.; Suzuki, Y.; Sugano, S.; Plass, C.; Huang, T.H.-M. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008, 24, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Singer, G.A.; Wu, J.; Yan, P.; Plass, C.; Huang, T.H.; Davuluri, R.V. Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genom. 2008, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wu, J.; Wickramasinghe, P.; Pal, S.; Gupta, R.; Bhattacharyya, A.; Agosto-Perez, F.J.; Showe, L.C.; Huang, T.H.; Davuluri, R.V. Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res. 2011, 39, 190–201. [Google Scholar] [CrossRef] [PubMed]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar]
- Steinthorsdottir, V.; Stefansson, H.; Ghosh, S.; Birgisdottir, B.; Bjornsdottir, S.; Fasquel, A.C.; Olafsson, O.; Stefansson, K.; Gulcher, J.R. Multiple novel transcription initiation sites for NRG1. Gene 2004, 342, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Li, N.; Hadzhiev, Y.; Plessy, C.; Previti, C.; Nepal, C.; Gehrig, J.; Dong, X.; Akalin, A.; Suzuki, A.M.; et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 2014, 507, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Broome, H.E.; Reed, J.C.; Godillot, E.P.; Hoover, R.G. Differential promoter utilization by the c-myc gene in mitogen- and interleukin-2-stimulated human lymphocytes. Mol. Cell Biol. 1987, 7, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.R.; Bulun, S.E.; Leitch, M.; Rohrich, R.; Simpson, E.R. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J. Clin. Endocrinol. Metab. 1996, 81, 3843–3849. [Google Scholar] [PubMed]
- Pedersen, I.S.; Dervan, P.; McGoldrick, A.; Harrison, M.; Ponchel, F.; Speirs, V.; Isaacs, J.D.; Gorey, T.; McCann, A. Promoter switch: A novel mechanism causing biallelic PEG1/MEST expression in invasive breast cancer. Hum. Mol. Genet. 2002, 11, 1449–1453. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Wang, Y.; Gold, B.; Chen, J.; Dean, M.; Harrison, P.J.; Weinberger, D.R.; Law, A.J. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with Schizophrenia. J. Biol. Chem. 2007, 282, 24343–24351. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, D.; Danilkovitch-Miagkova, A.; Ivanova, T.; Braga, E.; Zabarovsky, E.; Lerman, M.I. Hypermethylation of Ron proximal promoter associates with lack of full-length Ron and transcription of oncogenic short-Ron from an internal promoter. Oncogene 2007, 26, 4499–4512. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.D.; Pelascini, L.P.L.; Macedo, J.A.; Muller, C.P. Highly individual methylation patterns of alternative glucocorticoid receptor promoters suggest individualized epigenetic regulatory mechanisms. Nucleic Acids Res. 2008, 36, 7207–7218. [Google Scholar] [CrossRef] [PubMed]
- Archey, W.B.; Sweet, M.P.; Alig, G.C.; Arrick, B.A. Methylation of CpGs as a determinant of transcriptional activation at alternative promoters for transforming growth factor-β3. Cancer Res. 1999, 59, 2292–2296. [Google Scholar] [PubMed]
- Albert, T.; Wells, J.; Funk, J.-O.; Pullner, A.; Raschke, E.-E.; Stelzer, G.; Meisterernst, M.; Farnham, P.J.; Eick, D. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 2001, 276, 20482–20490. [Google Scholar] [CrossRef] [PubMed]
- Ngondo, R.P.; Carbon, P. Transcription factor abundance controlled by an auto-regulatory mechanism involving a transcription start site switch. Nucleic Acids Res. 2014, 42, 2171–2184. [Google Scholar] [CrossRef] [PubMed]
- Pozner, A.; Lotem, J.; Xiao, C.; Goldenberg, D.; Brenner, O.; Negreanu, V.; Levanon, D.; Groner, Y. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev. Biol. 2007, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Vitezic, M.; Lassmann, T.; Forrest, A.R.R.; Suzuki, M.; Tomaru, Y.; Kawai, J.; Carninci, P.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Building promoter aware transcriptional regulatory networks using siRNA perturbation and deepCAGE. Nucleic Acids Res. 2010, 38, 8141–8148. [Google Scholar] [CrossRef] [PubMed]
- Conacci-Sorrell, M.; McFerrin, L.; Eisenman, R.N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 2014, 4, a014357. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Pelengaris, S.; Khan, M.; Evan, G. c-MYC: More than just a matter of life and death. Nat. Rev. Cancer 2002, 2, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I.; Alves, J. The c-myc Promoter: Still MysterY and Challenge. Adv. Cancer Res. 2008, 99, 113–333. [Google Scholar] [PubMed]
- Fulco, C.P.; Munschauer, M.; Anyoha, R.; Munson, G.; Grossman, S.R.; Perez, E.M.; Kane, M.; Cleary, B.; Lander, E.S.; Engreitz, J.M. Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science 2016, 354, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Rennoll, S.; Yochum, G. Regulation of MYC gene expression by aberrant Wnt/β-catenin signaling in colorectal cancer. World J. Biol. Chem. 2015, 6, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Uslu, V.V.; Petretich, M.; Ruf, S.; Langenfeld, K.; Fonseca, N.A.; Marioni, J.C.; Spitz, F. Long-range enhancers regulating MYC expression are required for normal facial morphogenesis. Nat. Genet. 2014, 46, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Choi, P.S.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016, 48, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Hay, N.; Bishop, J.M.; Levens, D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987, 1, 659–671. [Google Scholar] [CrossRef] [PubMed]
- DesJardins, E.; Hay, N. Repeated CT elements bound by zinc finger proteins control the absolute and relative activities of the two principal human c-myc promoters. Mol. Cell Biol. 1993, 13, 5710–5724. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Yue, C.; Li, G.; He, B.; Cheng, W.; Wang, X.; Yan, M.; Long, Z.; Qiu, W.; Yuan, Z. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat. Commun. 2016, 7, 10180. [Google Scholar] [CrossRef] [PubMed]
- FANTOM5. Available online: http://fantom.gsc.riken.jp/ (accessed on 13 October 2017).
- ZENBU. Available online: https://bit.ly/2JmEnVR (accessed on 13 October 2017).
- Arner, E.; Daub, C.O.; Vitting-Seerup, K.; Andersson, R.; Lilje, B.; Drabløs, F.; Lennartsson, A.; Rönnerblad, M.; Hrydziuszko, O.; Vitezic, M.; et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015, 347, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Veeman, M.T.; Slusarski, D.C.; Kaykas, A.; Louie, S.H.; Moon, R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003, 13, 680–685. [Google Scholar] [CrossRef]
- Primer3 Input. Available online: http://bioinfo.ut.ee/primer3-0.4.0 (accessed on 13 December 2016).
- CRISPR Design. Available online: http://crispr.mit.edu/ (accessed on 20 May 2017).
- Yochum, G.S.; Sherrick, C.M.; MacPartlin, M.; Goodman, R.H. A β-catenin/TCF-coordinated chromatin loop at MYC integrates 5′ and 3′ Wnt responsive enhancers. Proc. Natl. Acad. Sci. USA 2010, 107, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Jäger, R.; Migliorini, G.; Henrion, M.; Kandaswamy, R.; Speedy, H.E.; Heindl, A.; Whiffin, N.; Carnicer, M.J.; Broome, L.; Dryden, N. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat. Commun. 2015, 6, 6178. [Google Scholar] [CrossRef] [PubMed]
- Yochum, G.S.; Cleland, R.; Goodman, R.H. A genome-wide screen for β-catenin binding sites identifies a downstream enhancer element that controls c-myc gene expression. Mol. Cell Biol. 2008, 28, 7368–7379. [Google Scholar] [CrossRef] [PubMed]
- Tuupanen, S.; Yan, J.; Turunen, M.; Gylfe, A.E.; Kaasinen, E.; Li, L.; Eng, C.; Culver, D.A.; Kalady, M.F.; Pennison, M.J. Characterization of the colorectal cancer–associated enhancer MYC-335 at 8q24: The role of rs67491583. Cancer Genet. 2012, 205, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Meulia, T.; Krumm, A.; Spencer, C.; Groudine, M. Sequences in the human c-myc P2 promoter affect the elongation and premature termination of transcripts initiated from the upstream P1 promoter. Mol. Cell. Biol. 1992, 12, 4590–4600. [Google Scholar] [CrossRef] [PubMed]
- Merli, C.; Bergstrom, D.E.; Cygan, J.A.; Blackman, R.K. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996, 10, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Van Arensbergen, J.; van Steensel, B.; Bussemaker, H.J. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol. 2014, 24, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Noll, M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994, 13, 400–406. [Google Scholar] [PubMed]
- Shlyueva, D.; Stelzer, C.; Gerlach, D.; Yáñez-Cuna, J.O.; Rath, M.; Boryń, Ł.M.; Arnold, C.D.; Stark, A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol. Cell 2014, 54, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Li, Y.; Dixon, J.R.; Selvaraj, S.; Ye, Z.; Lee, A.Y.; Yen, C.A.; Schmitt, A.D.; Espinoza, C.A.; Ren, B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013, 503, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Visel, A.; Blow, M.J.; Li, Z.; Zhang, T.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; Chen, F. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009, 457, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Davis, C.; Ewing, B.; Gordon, D.; Green, P. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007, 17, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, J.; Esposito, D.; Duhagon, M.A.; Banfield, K.; Mehalko, J.; Liao, H.; Stephens, R.M.; Harris, T.J.; Munroe, D.J.; Wu, X. Long-range enhancers on 8q24 regulate c-Myc. Proc. Natl. Acad. Sci. USA 2010, 107, 3001–3005. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bardales, J.A.; Wieser, E.; Kawaji, H.; Murakawa, Y.; Darzacq, X. Selective Activation of Alternative MYC Core Promoters by Wnt-Responsive Enhancers. Genes 2018, 9, 270. https://doi.org/10.3390/genes9060270
Bardales JA, Wieser E, Kawaji H, Murakawa Y, Darzacq X. Selective Activation of Alternative MYC Core Promoters by Wnt-Responsive Enhancers. Genes. 2018; 9(6):270. https://doi.org/10.3390/genes9060270
Chicago/Turabian StyleBardales, Jorge A., Evin Wieser, Hideya Kawaji, Yasuhiro Murakawa, and Xavier Darzacq. 2018. "Selective Activation of Alternative MYC Core Promoters by Wnt-Responsive Enhancers" Genes 9, no. 6: 270. https://doi.org/10.3390/genes9060270
APA StyleBardales, J. A., Wieser, E., Kawaji, H., Murakawa, Y., & Darzacq, X. (2018). Selective Activation of Alternative MYC Core Promoters by Wnt-Responsive Enhancers. Genes, 9(6), 270. https://doi.org/10.3390/genes9060270




