Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.)
Abstract
1. Introduction
2. Material and Methods
2.1. The Identification of MADS-Box Genes in Carnation’s Genome
2.2. Conserved Sequence and Structure Model Analysis
2.3. Phylogenetic Analysis of MADS-Box Proteins
2.4. RNA Sample Preparation and Quantitative PCR Expression Analysis
2.5. Subcellular Localization
3. Results
3.1. The Identification and Annotation Information of Carnation MADS-Box Domain Genes
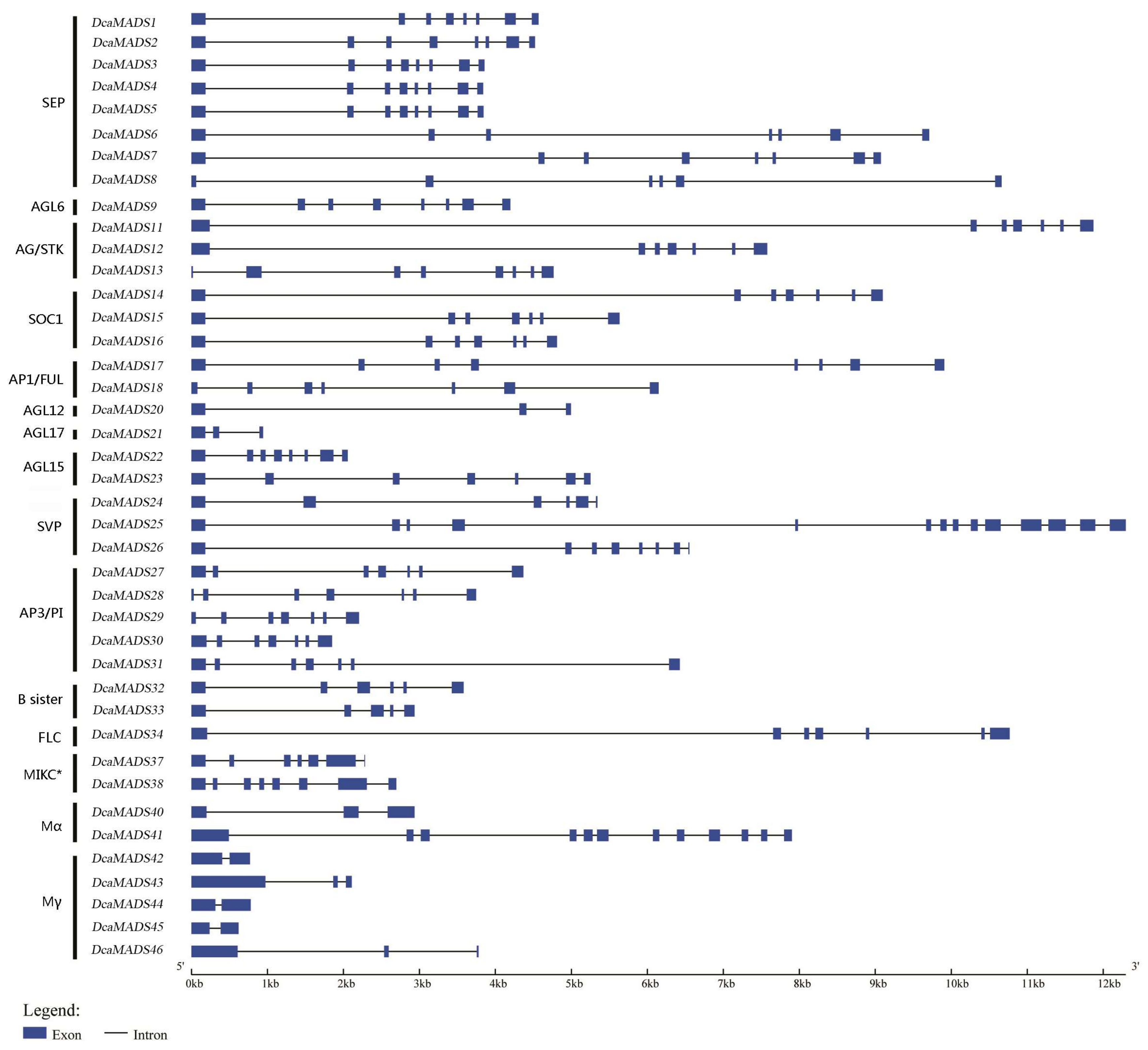
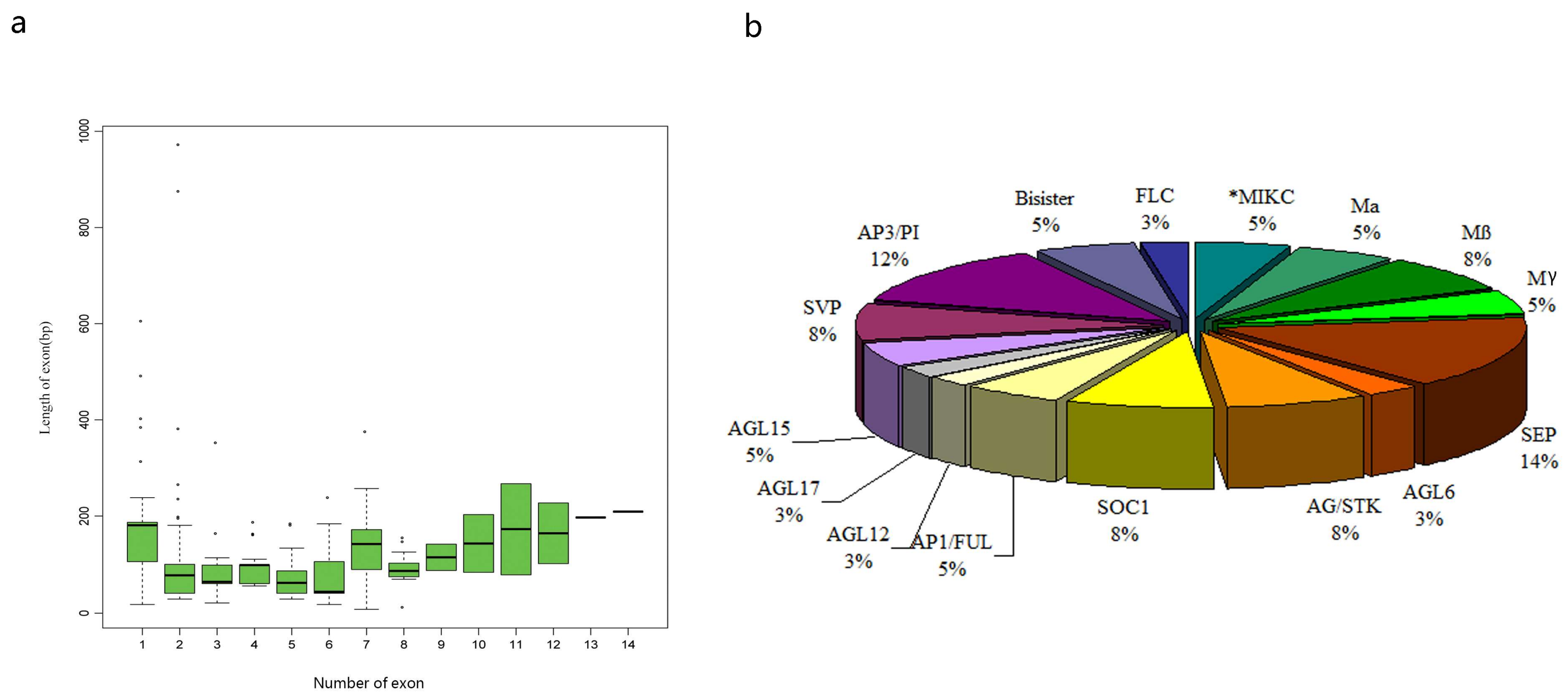
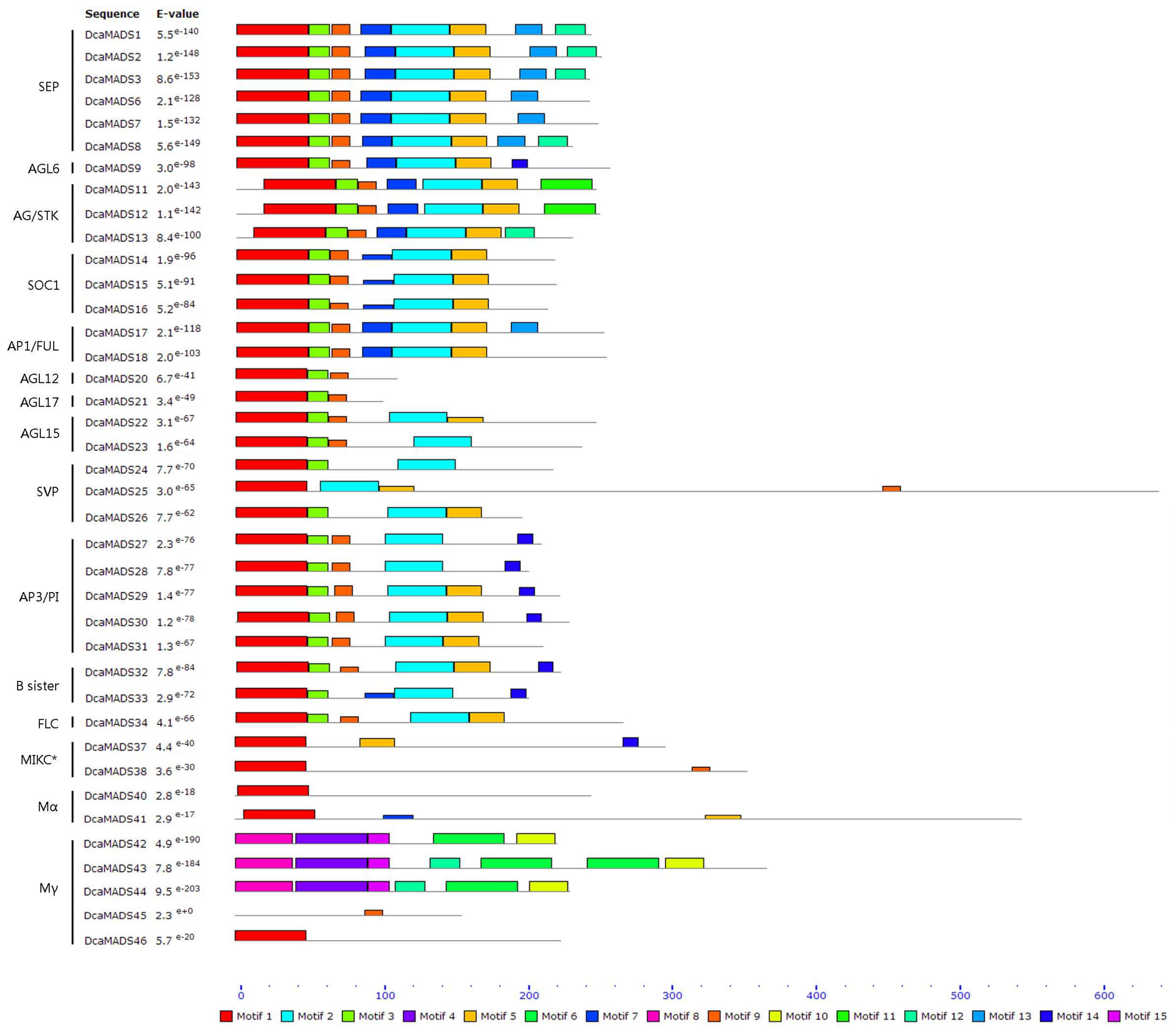
3.2. Analysis of the Gene Structure and Conserved Sequence
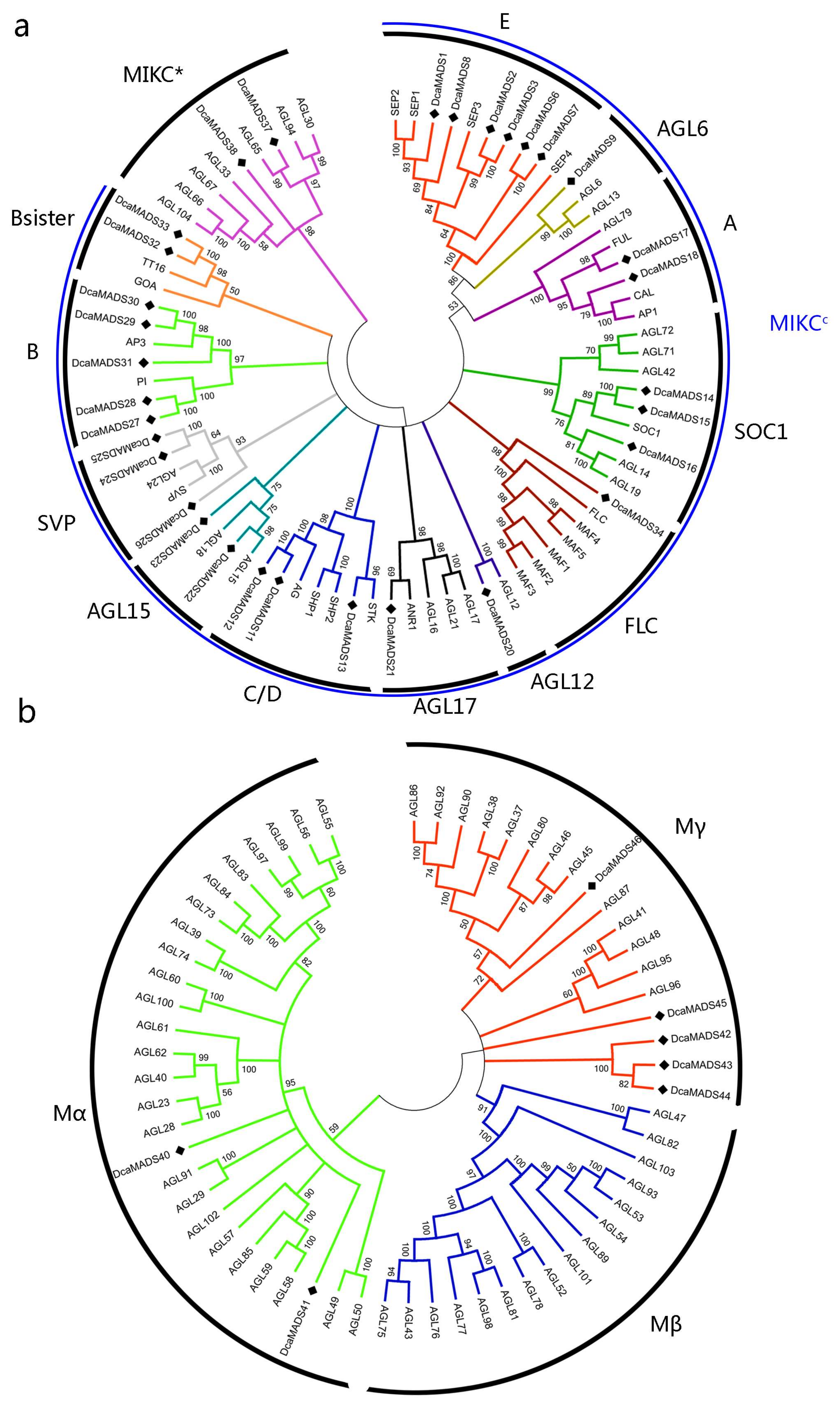
3.3. Phylogenetic Analysis of Carnation MADS-Box Domain
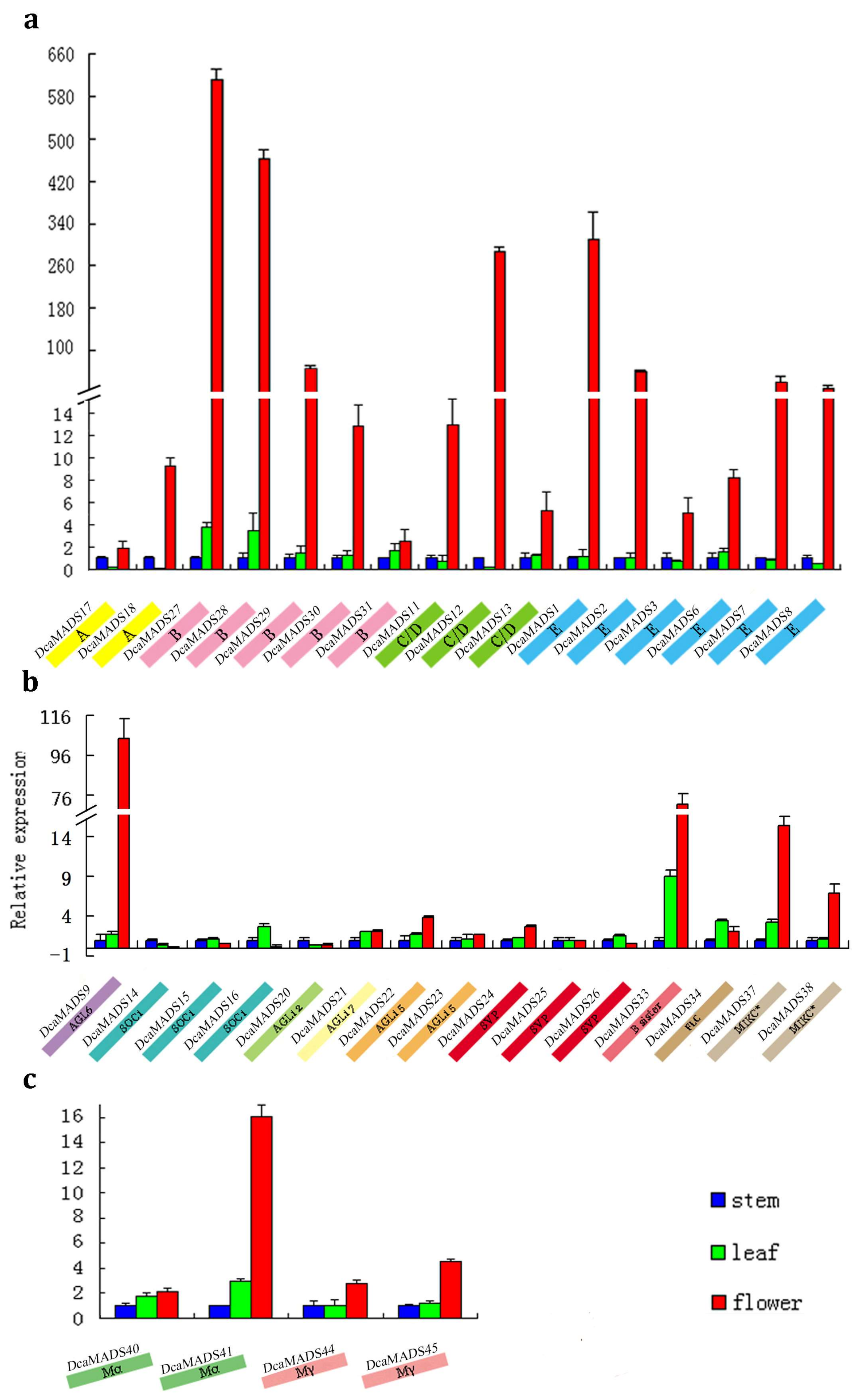
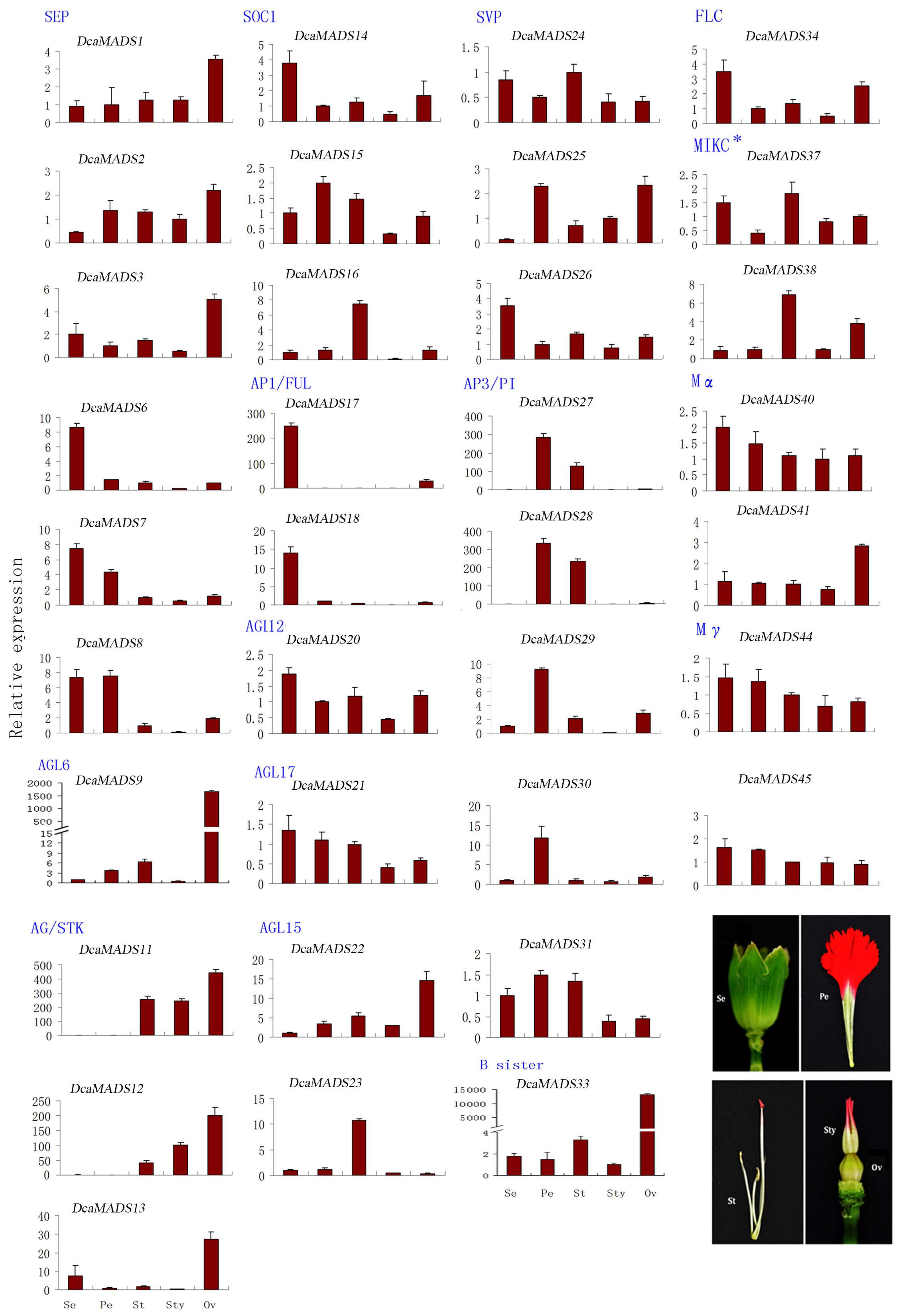
3.4. Expression of DcaMADS Genes in Floral Organs
3.4.1. SEPALLATA
3.4.2. AGAMOUS-LIKE 6
3.4.3. AGAMOUS/SEEDSTICK
3.4.4. SUPPRESSOR OF OVEREXPRESSION OF CO1
3.4.5. APETALA 1/FRUITFUL
3.4.6. AGAMOUS-LIKE 12
3.4.7. AGAMOUS-LIKE17
3.4.8. AGAMOUS-LIKE 15
3.4.9. SHORT VEGETATIVE PHASE/AGAMOUS-LIKE 24
3.4.10. APETALA 3/PISTILLATA
3.4.11. B sister
3.4.12. FLOWERING LOCUS
3.4.13. MIKC*
3.4.14. Mα
3.4.15. Mγ
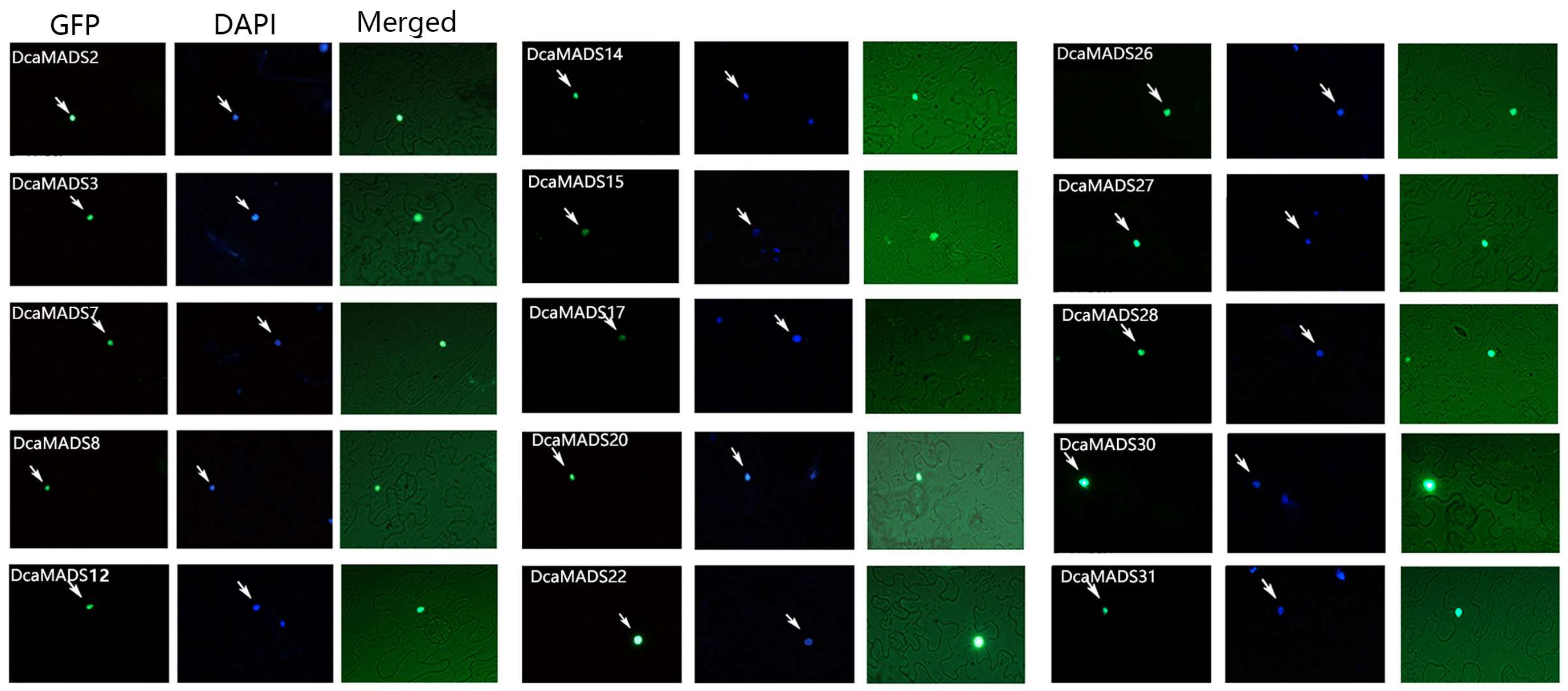
3.5. Subcellular Localization of DcaMADS
4. Discussion
4.1. AP3/PI and SEP Subfamily with Duplication Influences Evolution and Divergence
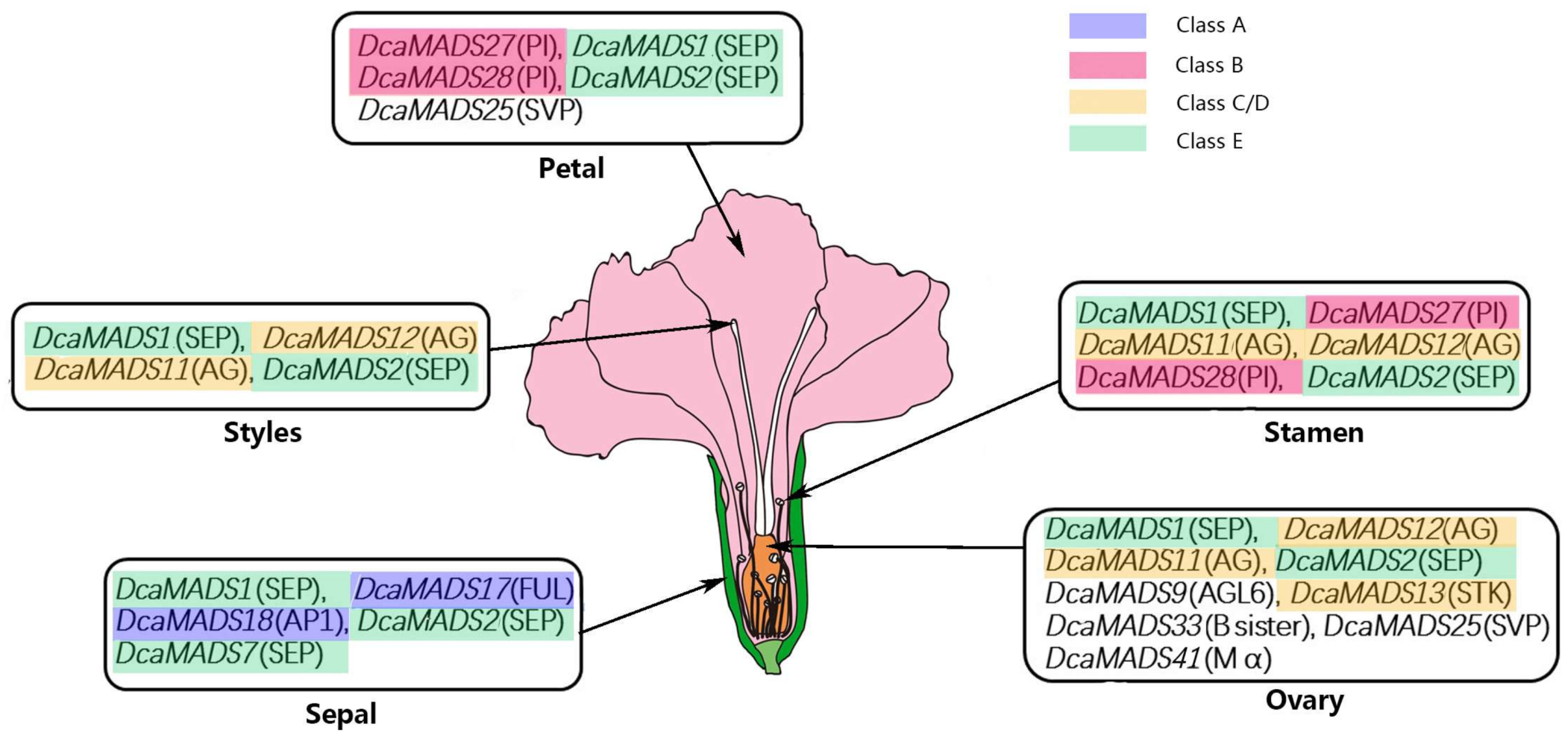
4.2. A Model in Flower Organ Identity of Carnation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Pellegrini, L.; Tan, S.; Richmond, T.J. Structure of serum response factor core bound to DNA. Nature 1995, 376, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. FEBS J. 1995, 229, 1–13. [Google Scholar]
- Sasaki, K.; Aida, R.; Yamaguchi, H.; Shikata, M.; Niki, T.; Nishijima, T.; Ohtsubo, N. Functional divergence within class B MADS-box genes TfGLO and TfDEF in Torenia fournieri Lind. Mol. Genet. Genom. 2010, 284, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Alvarezbuylla, E.R.; Liljegren, S.J.; Pelaz, S.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Vergarasilva, F.; Yanofsky, M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. Cell Mol. Biol. 2000, 24, 457–466. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, S.; Wu, F.; Yan, S.; Lin, X.; Du, X.; Chong, K.; Schilling, S.; Theißen, G.; Meng, Z. Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 2013, 25, 1288–1303. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; De, F.S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B. Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis: New Openings to the MADS World. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theißen, G. Phylogenomics of MADS-Box genes in plants—Two opposing life styles in one gene family. Biology 2013, 2, 1150–1164. [Google Scholar] [CrossRef] [PubMed]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theissen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Bodt, S.D.; Raes, J.; Florquin, K.; Rombauts, S.; Rouzé, P.; Theißen, G.; Peer, Y.V.D. Genomewide structural annotation and evolutionary analysis of the type I MADS-Box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.E.; Vendramin, E.; Jimenez, T.S.; Verde, I.; Bielenberg, D.G. A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.Y.; Meaux, J.D. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 2014, 26, 2024–2037. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Tapialópez, R.; Garcíaponce, B.; Dubrovsky, J.G.; Garayarroyo, A.; Pérezruíz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarezbuylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Kumimoto, R.W.; Wong, B.J.; Riechmann, J.L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 2003, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Heuer, S.; Lörz, H.; Dresselhaus, T. The MADS box gene ZmMADS2 is specifically expressed in maize pollen and during maize pollen tube growth. Plant Reprod. 2000, 13, 21–27. [Google Scholar] [CrossRef]
- Zobell, O.; Faigl, W.; Saedler, H.; Münster, T. MIKC* MADS-box proteins: Conserved regulators of the gametophytic generation of land plants. Mol. Biol. Evol. 2010, 27, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Barker, E.I.; Ashton, N.W. A parsimonious model of lineage-specific expansion of MADS-box genes in Physcomitrella patens. Plant Cell Rep. 2013, 32, 1161–1177. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M. Recent progress in genomic analysis of ornamental plants, with a focus on Carnation. Hortic. J. 2015, 84, 3–13. [Google Scholar] [CrossRef]
- Hileman, L.C.; Sundstrom, J.F.; Litt, A.; Chen, M.; Shumba, T.; Irish, V.F. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 2006, 23, 2245–2258. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, S12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, X.; Chen, W.; Peng, X.; Cheng, X.; Zhu, S.; Cheng, B. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tissue Organ Cult. 2011, 105, 159–173. [Google Scholar] [CrossRef]
- Gan, D.F. Genome-wide sequence characterization analysis of MADS-Box transcription factor gene family in cucumber (Cucumis sativus L.). J. Nucl. Agric. Sci. 2012, 26, 1249–1256. [Google Scholar]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, L.; Yu, J.; Zhang, Y.; Li, D.; Zhang, X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 2015, 569, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-wide characterization of the MADS-Box gene family in Radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Kosugi, S.; Hirakawa, H.; Ohmiya, A.; Tanase, K.; Harada, T.; Kishimoto, K.; Nakayama, M.; Ichimura, K.; Onozaki, T. Sequence Analysis of the genome of Carnation (Dianthus caryophyllus L.). DNA Res. 2014, 21, 2319–2321. [Google Scholar] [CrossRef] [PubMed]
- TAIR Website. Available online: http://www.arabidopsis.org/ (accessed on 8 March 2017).
- The Rice Genome Annotation Project. Available online: http://rice.plantbiology.msu.edu/ (accessed on 9 March 2017).
- The National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 2 April 2017).
- SMART. Available online: http://smart.embl.de/ (accessed on 5 April 2017).
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acid Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Carnation genome. Available online: http://carnation.kazusa.or.jp/ (accessed on 23 February 2017).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Comput. Appl. Biosci. CABIOS 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.L.; Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 2001, 13, 2175–2190. [Google Scholar]
- Baudinette, S.C.; Stevenson, T.W.; Savin, K.W. Isolation and characterisation of the carnation floral-specific MADS box gene, CMB2. Plant Sci. 2000, 155, 123–131. [Google Scholar] [CrossRef]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Johansen, B.; Pedersen, L.B.; Skipper, M.; Frederiksen, S. MADS-box gene evolution-structure and transcription patterns. Mol. Phylogenetics Evol. 2002, 23, 458–480. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Ferrario, S.; Immink, R.G.; Shchennikova, A.; Busscherlange, J.; Angenent, G.C. The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 2003, 15, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Malcomber, S.T.; Kellogg, E.A. SEPALLATA gene diversification: Brave new whorls. Trends Plant Sci 2005, 10, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Romera-Branchat, M.; Pelaz, S. A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 2005, 43, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Kimizu, M.; Sugita, M.; Miyao, A.; Hirochika, H.; Uchida, E.; Nagato, Y.; Yoshida, H. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 2009, 21, 3008–3025. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Fu, Y.; Zhao, W.; Liu, L.; Bade, R.; Hasi, A.; Hao, J. Genome-wide identification and analysis of the MADS-box gene family in Melon. J. Am. Soc. Horticult. Sci. 2016, 141, 507–519. [Google Scholar] [CrossRef]
- Pnueli, L.; Hareven, D.; Rounsley, S.D.; Yanofsky, M.F.; Lifschitz, E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 1994, 6, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Robinson-Beers, K.; Ray, S.; Baker, S.C.; Lang, J.D.; Preuss, D.; Milligan, S.B.; Gasser, C.S. Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. USA 1994, 91, 5761–5765. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.; Han, J.J.; Han, M.J.; An, G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.S.; Kim, Y.S.; Han, J.Y.; Yoon, E.S.; Yong, E.C. Panax ginseng PgMADS1, an AP1/FUL-like MADS-box gene, is activated by hormones and is involved in inflorescence growth. Plant Cell Tissue Organ Cult. 2015, 122, 161–173. [Google Scholar] [CrossRef]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Litt, A. An evaluation of A-function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 2007, 168, 73–91. [Google Scholar] [CrossRef]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [PubMed]
- Burgeff, C.; Liljegren, S.J.; Tapia-López, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Zachgo, S.; Saedler, H.; Schwarz-Sommer, Z. Pollen-specific expression of DEFH125, a MADS-box transcription factor in Antirrhinum with unusual features. Plant J. Cell Mol. Biol. 1997, 11, 1043–1050. [Google Scholar] [CrossRef]
- Mouradov, A.; Hamdorf, B.; Teasdale, R.D.; Kim, J.T.; Winter, K.U.; Theißen, G. A DEF/GLO-like MADS-box gene from a gymnosperm: Pinus radiata contains an ortholog of angiosperm B class floral homeotic genes. Genesis 1999, 25, 245–252. [Google Scholar]
- Whipple, C.J.; Ciceri, P.; Padilla, C.M.; Ambrose, B.A.; Bandong, S.L.; Schmidt, R.J. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 2004, 131, 6083–6091. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Kaufmann, K.; Freialdenhoven, A.; Vincent, C.; Li, M.A.; Saedler, H.; Theissen, G. A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genom. 2002, 266, 942–950. [Google Scholar]
- De Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G.H. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Zhang, X.; Tobón, E.; Ambrose, B.A. The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J. 2010, 62, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.; Gramzow, L.; Melzer, R.; Theissen, G.; Becker, A. GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana Bsister MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J. 2010, 63, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001, 13, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; He, Y.; Scortecci, K.C.; Amasino, R.M. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 10102–10107. [Google Scholar] [CrossRef] [PubMed]
- Leseberg, C.H.; Li, A.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Fernandez, D.E. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009, 149, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Verelst, W.; Saedler, H.; Münster, T. MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 2007, 143, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.C.; Chou, M.L.; Yue, J.J.; Hsu, C.T.; Chang, W.J.; Ko, S.S.; Liao, D.C.; Huang, Y.T.; Chen, J.J.; Yuan, J.L. BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Díazriquelme, J.; Lijavetzky, D.; Martínezzapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKC-type MADS box genes in grapevine. Plant Physiol. 2008, 149, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, Q.; Sun, L.; Du, D.; Cheng, T.; Pan, H.; Yang, W.; Wang, J. Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Mol. Genet. Genom. 2014, 289, 903–920. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Hsu, C.T.; Liao, C.; Chang, W.J.; Chou, M.L.; Huang, Y.T.; Chen, J.J.; Ko, S.S.; Chan, M.T.; Shih, M.C. Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 2015, 14, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Zahn, L.M. To B or Not to B a Flower: The Role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered. 2005, 96, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.; Uehara, K.; Imafuku, M.; Hasebe, M.; Ito, M. Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. J. Plant Res. 2004, 117, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Dorit, R.L.; Irish, V.F. Molecular Evolution of Genes Controlling Petal and Stamen Development: Duplication and Divergence within the APETALA3 and PISTILLATA MADS-Box Gene Lineages. Genetics 1998, 149, 765–783. [Google Scholar] [PubMed]
- Kramer, E.M.; Su, H.J.; Wu, C.C.; Hu, J.M. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol. Biol. 2006, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.A.; Kramer, E.M. Molecular evolution of the petal and stamen identity genes, APETALA3 and PISTILLATA, after petal loss in the Piperales. Mol. Phylogenetics Evol. 2007, 44, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Biewers, S.M. Sepallata Genes and Their Role during Floral Organ Formation. Ph.D. Thesis, University of Leeds, Leeds, UK, 2014. [Google Scholar]
- Angenent, G.C.; Franken, J.; Busscher, M.; Weiss, D.; van Tunen, A.J. Co-suppression of the petunia homeotic gene FBP2 affects the identity of the generative meristem. Plant J. 1994, 5, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Hareven, D.; Broday, L.; Hurwitz, C.; Lifschitz, E. The TM5 MADS Box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 1994, 6, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-Box Gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Heijmans, K.; Airoldi, C.; Davies, B.; Angenent, G.C. An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 2010, 154, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Rijpkema, A.S.; Royaert, S.; Zethof, J.; Van, d.W.G.; Gerats, T.; Vandenbussche, M. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 2006, 18, 1819–1832. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Name | Accession Number | ORF (bp) | Group | Clades |
|---|---|---|---|---|
| DcaMADS1 | Dca58524.1 | 738 | MIKCc | SEP |
| DcaMADS2 | Dca4751.1 | 759 | MIKCc | SEP |
| DcaMADS3 | Dca45289.1 | 735 | MIKCc | SEP |
| DcaMADS4 | Dca42307.1 | 735 | MIKCc | SEP |
| DcaMADS5 | Dca47184.1 | 735 | MIKCc | SEP |
| DcaMADS6 | Dca61854.1 | 630 | MIKCc | SEP |
| DcaMADS7 | Dca42755.1 | 753 | MIKCc | SEP |
| DcaMADS8 | Dca22562.1 | 438 | MIKCc | SEP |
| DcaMADS9 | Dca72.1 | 777 | MIKCc | AGL6 |
| DcaMADS10 | Dca19158.1 | 540 | MIKCc | |
| DcaMADS11 | Dca50159.1 | 750 | MIKCc | AG/STK |
| DcaMADS12 | Dca35398.1 | 756 | MIKCc | AG/STK |
| DcaMADS13 | Dca13518.1 | 699 | MIKCc | AG/STK |
| DcaMADS14 | Dca73.1 | 663 | MIKCc | SOC1 |
| DcaMADS15 | Dca19159.1 | 666 | MIKCc | SOC1 |
| DcaMADS16 | Dca23368.1 | 648 | MIKCc | SOC1 |
| DcaMADS17 | Dca61853.1 | 765 | MIKCc | AP1/FUL |
| DcaMADS18 | Dca59408.1 | 585 | MIKCc | AP1/FUL |
| DcaMADS19 | Dca21554.1 | 249 | MIKCc | |
| DcaMADS20 | Dca51118.1 | 339 | MIKCc | AGL12 |
| DcaMADS21 | Dca27570.1 | 309 | MIKCc | AGL17 |
| DcaMADS22 | Dca58547.1 | 756 | MIKCc | AGL15 |
| DcaMADS23 | Dca57893.1 | 726 | MIKCc | AGL15 |
| DcaMADS24 | Dca7325.1 | 666 | MIKCc | SVP |
| DcaMADS25 | Dca5233.1 | 1935 | MIKCc | SVP |
| DcaMADS26 | Dca56094.1 | 600 | MIKCc | SVP |
| DcaMADS27 | Dca17660.1 | 642 | MIKCc | AP3/PI |
| DcaMADS28 | Dca7718.1 | 456 | MIKCc | AP3/PI |
| DcaMADS29 | Dca35875.1 | 543 | MIKCc | AP3/PI |
| DcaMADS30 | Dca24326.1 | 699 | MIKCc | AP3/PI |
| DcaMADS31 | Dca28764.1 | 645 | MIKCc | AP3/PI |
| DcaMADS32 | Dca52384.1 | 672 | MIKCc | B sister |
| DcaMADS33 | Dca35154.1 | 615 | MIKCc | B sister |
| DcaMADS34 | Dca45290.1 | 813 | MIKCc | FLC |
| DcaMADS35 | Dca42306.1 | 747 | MIKCc | |
| DcaMADS36 | Dca62484.1 | 792 | ||
| DcaMADS37 | Dca41798.1 | 903 | MIKC* | MIKC* |
| DcaMADS38 | Dca21633.1 | 1074 | MIKC* | MIKC* |
| DcaMADS39 | Dca37955.1 | 1050 | ||
| DcaMADS40 | Dca20695.1 | 747 | Mα | Mα |
| DcaMADS41 | Dca37633.1 | 1641 | Mα | Mα |
| DcaMADS42 | Dca46738.1 | 669 | Mγ | Mγ |
| DcaMADS43 | Dca21085.1 | 1107 | Mγ | Mγ |
| DcaMADS44 | Dca21084.1 | 696 | Mγ | Mγ |
| DcaMADS45 | Dca50134.1 | 474 | Mγ | Mγ |
| DcaMADS46 | Dca38557.1 | 687 | Mγ | Mγ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, Q.; Yang, S.; Lin, S.; Bao, M.; Bendahmane, M.; Wu, Q.; Wang, C.; Fu, X. Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.). Genes 2018, 9, 193. https://doi.org/10.3390/genes9040193
Zhang X, Wang Q, Yang S, Lin S, Bao M, Bendahmane M, Wu Q, Wang C, Fu X. Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.). Genes. 2018; 9(4):193. https://doi.org/10.3390/genes9040193
Chicago/Turabian StyleZhang, Xiaoni, Qijian Wang, Shaozong Yang, Shengnan Lin, Manzhu Bao, Mohammed Bendahmane, Quanshu Wu, Caiyun Wang, and Xiaopeng Fu. 2018. "Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.)" Genes 9, no. 4: 193. https://doi.org/10.3390/genes9040193
APA StyleZhang, X., Wang, Q., Yang, S., Lin, S., Bao, M., Bendahmane, M., Wu, Q., Wang, C., & Fu, X. (2018). Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.). Genes, 9(4), 193. https://doi.org/10.3390/genes9040193





