Transcriptome Analysis Reveals Candidate Genes for Cold Tolerance in Drosophila ananassae
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Strains
2.2. Chill Coma Recovery Assays
2.3. RNA Extraction and RNA Sequencing
2.4. Read Mapping and Differential Gene Expression Analysis
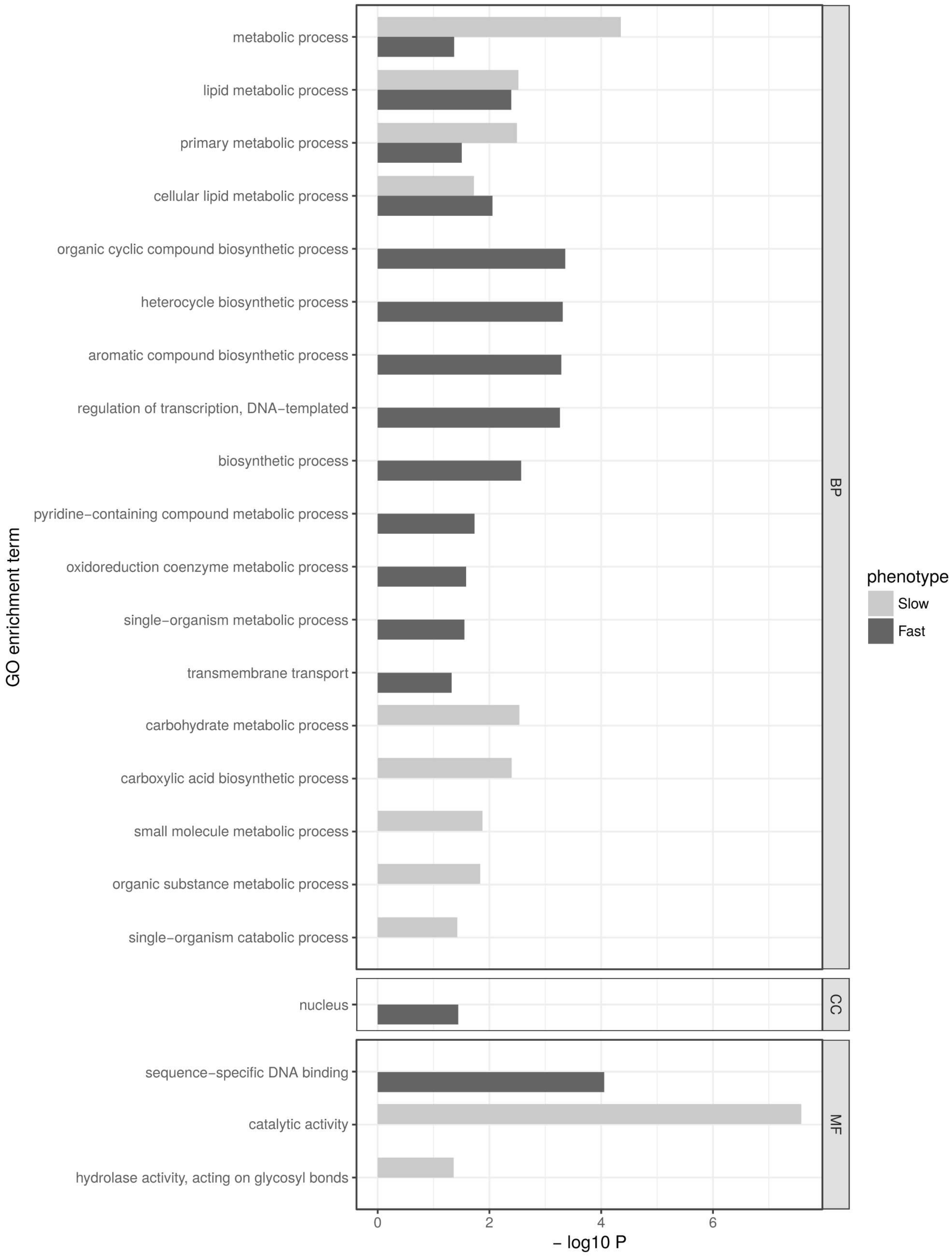
2.5. Gene Ontology Term Enrichment Analysis
2.6. Comparison with Drosophila melanogaster
3. Results
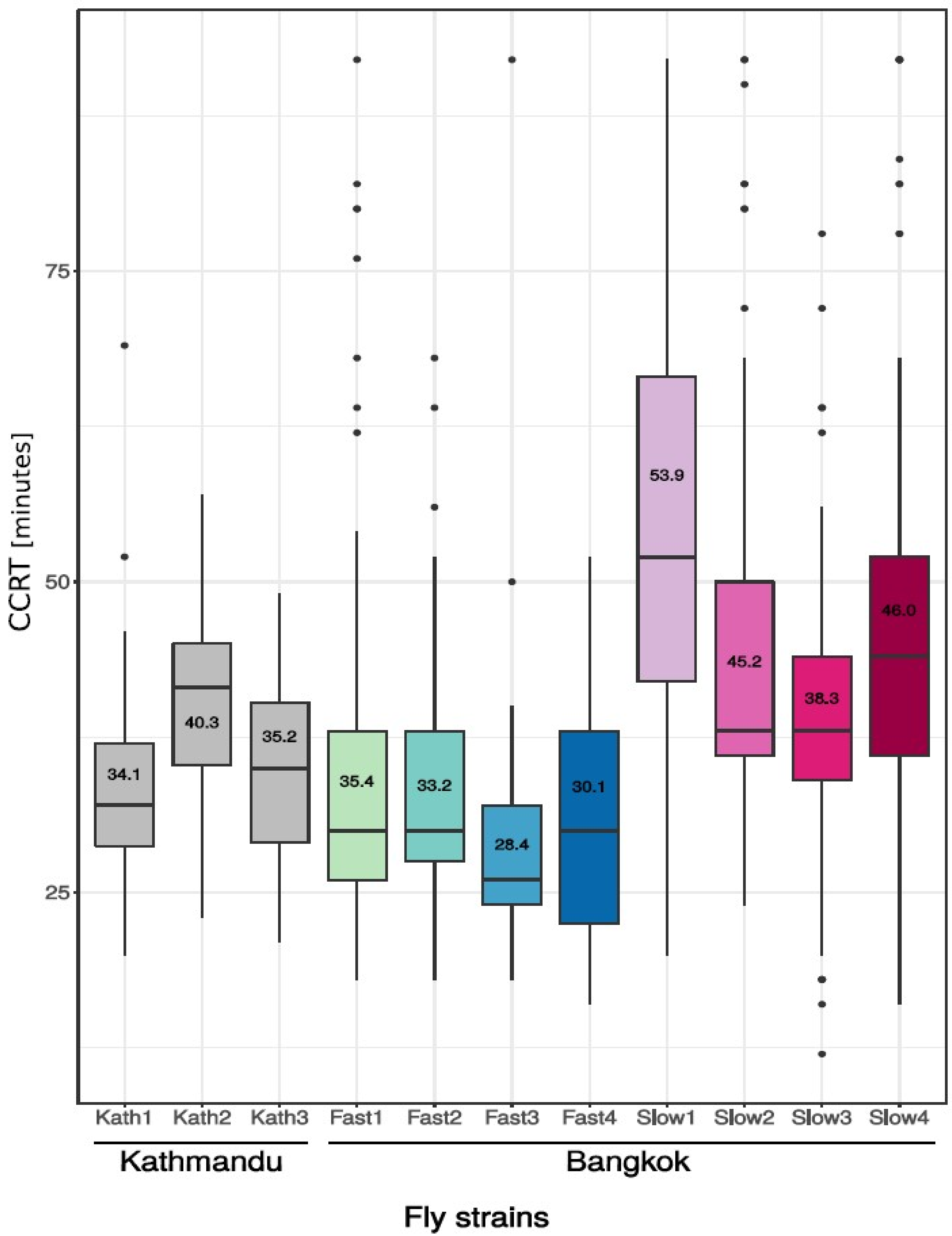
3.1. Phenotypic Differences in Cold Tolerance as Measured by Chill Coma Recovery Time
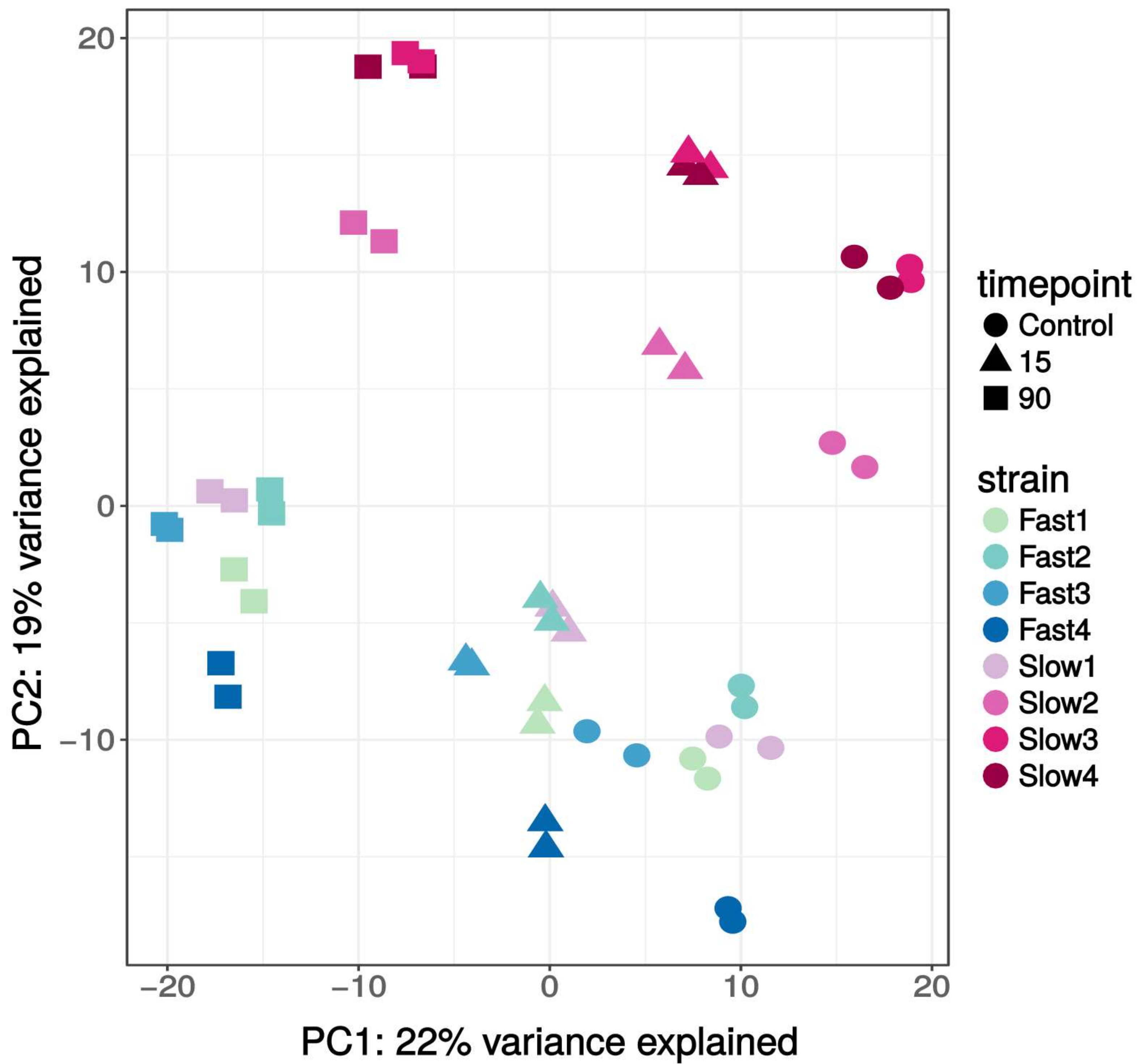
3.2. Transcriptome Overview
3.3. Analysis of Differential Gene Expression
3.4. Expression Differences before the Cold Shock
3.5. Expression Differences in the Recovery Phase
3.6. Comparison with Drosophila melanogaster
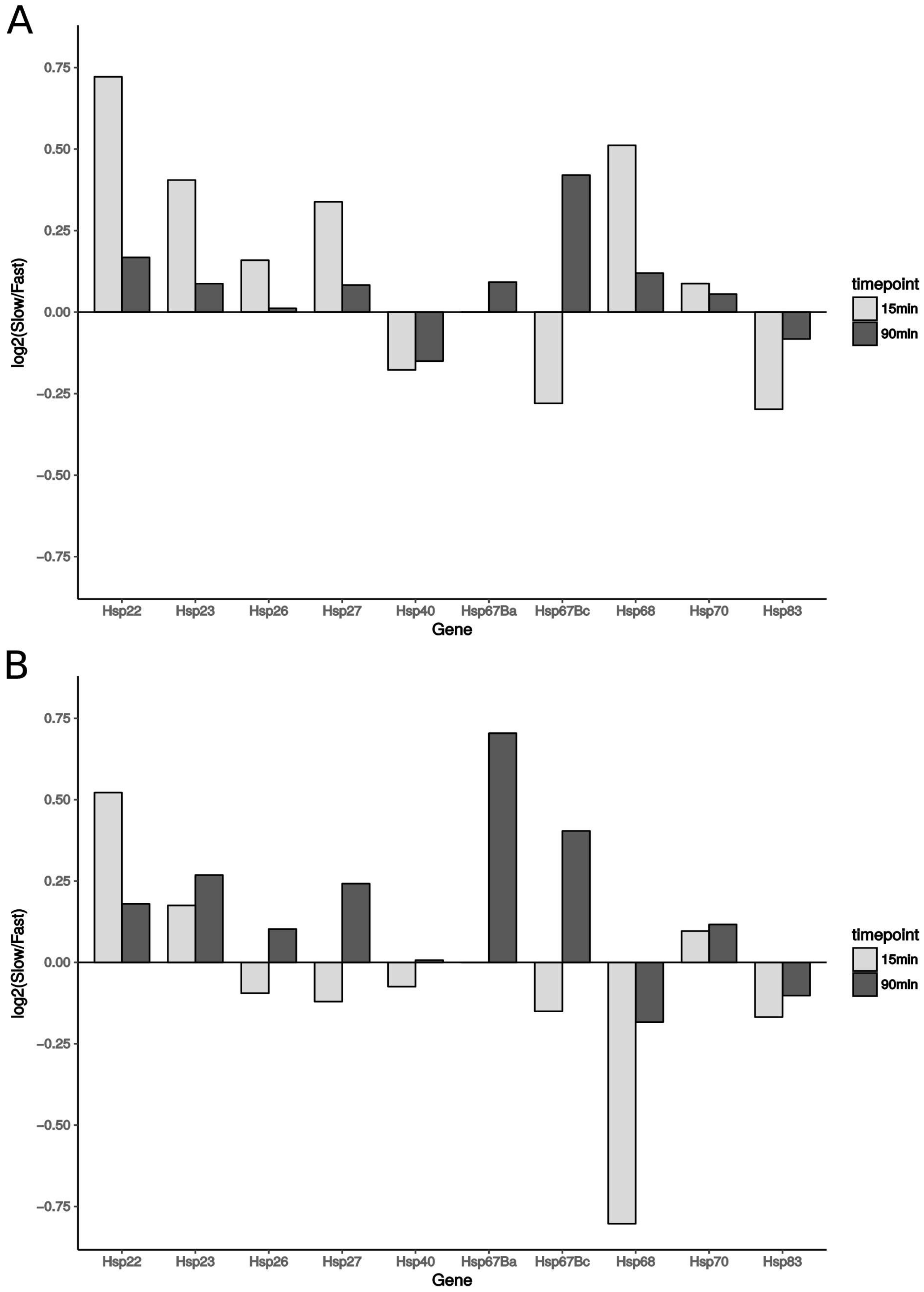
3.7. Expression of Heat-Shock Proteins
3.8. Identification of Candidate Genes Involved in Cold Tolerance in Drosophila ananassae
4. Discussion
4.1. Delayed Onset of Apoptosis May Improve Recovery from Chill Coma
4.2. Increased Expression of Hsps Is Unlikely to Improve Chill Coma Recovery Time in Drosophila
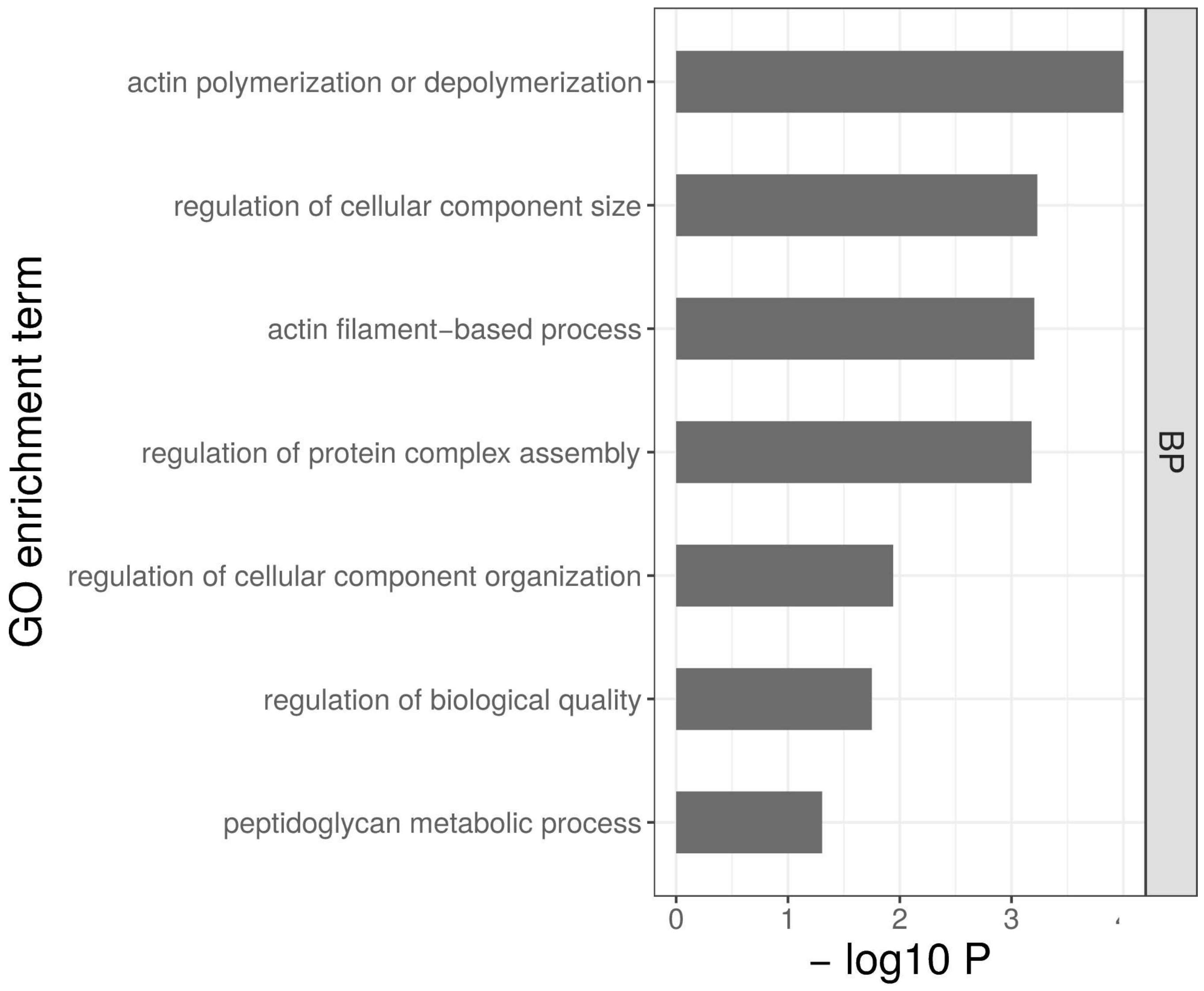
4.3. Genes Involved in Actin Polymerization May Have a Preemptive Role in Cold Tolerance
4.4. Strengths and Limitations of this Study and Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Overgaard, J.; MacMillan, H. The integrative physiology of insect chill tolerance. Annu. Rev. Physiol. 2016, 79, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, B.J.; Addo-Bediako, A.; Chown, S.L. Climatic variability and the evolution of insect freeze tolerance. Biol. Rev. Camb. Philos. Soc. 2003, 78, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Sformo, T.; Walters, K.; Jeannet, K.; Wowk, B.; Fahy, G.M.; Barnes, B.M.; Duman, J.G. Deep supercooling, vitrification and limited survival to −100 °C in the Alaskan beetle Cucujus clavipes puniceus (Coleoptera: Cucujidae) larvae. J. Exp. Biol. 2010, 213, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Throckmorton, L.H. The phylogeny, ecology, and geography of Drosophila. In Handbook of Genetics, Volume 3; Plenum Press: New York, NY, USA, 1975; pp. 421–469. [Google Scholar]
- Bale, J.S.; Hayward, S.A.L. Insect overwintering in a changing climate. J. Exp. Biol. 2010, 213, 980–994. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, B.J. Insect cold tolerance: How many kinds of frozen? Eur. J. Entomol. 1999, 96, 157–164. [Google Scholar]
- David, J.; Gibert, P.; Pla, E.; Petavy, G.; Karan, D.; Moreteau, B. Cold stress tolerance in Drosophila: Analysis of chill coma recovery in D. melanogaster. J. Therm. Biol. 1998, 23, 291–299. [Google Scholar] [CrossRef]
- MacMillan, H.A.; Sinclair, B.J. Mechanisms underlying insect chill-coma. J. Insect. Physiol. 2011, 57, 12–20. [Google Scholar] [CrossRef]
- Andersen, J.L.; Manenti, T.; Sørensen, J.G.; MacMillan, H.A.; Loeschcke, V.; Overgaard, J. How to assess Drosophila cold tolerance: Chill coma temperature and lower lethal temperature are the best predictors of cold distribution limits. Funct. Ecol. 2015, 29, 55–65. [Google Scholar] [CrossRef]
- Telonis-Scott, M.; Hallas, R.; McKechnie, S.W.; Wee, C.W.; Hoffmann, A.A. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. J. Insect. Physiol. 2009, 55, 549–555. [Google Scholar] [CrossRef]
- Von Heckel, K.; Stephan, W.; Hutter, S. Canalization of gene expression is a major signature of regulatory cold adaptation in temperate Drosophila melanogaster. BMC Genom. 2016, 17. [Google Scholar] [CrossRef]
- Gerken, A.R.; Eller, O.C.; Hahn, D.A.; Morgan, T.J. Constraints, independence, and evolution of thermal plasticity: Probing genetic architecture of long- and short-term thermal acclimation. Proc. Natl. Acad. Sci. USA 2015, 112, 4399–4404. [Google Scholar] [CrossRef] [PubMed]
- Poupardin, R.; Schöttner, K.; Korbelová, J.; Provazník, J.; Doležel, D.; Pavlinic, D.; Beneš, V.; Koštál, V. Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Des Marteaux, L.E.; McKinnon, A.H.; Udaka, H.; Toxopeus, J.; Sinclair, B.J. Effects of cold-acclimation on gene expression in Fall field cricket (Gryllus pennsylvanicus) ionoregulatory tissues. BMC Genom. 2017, 18, 357. [Google Scholar] [CrossRef] [PubMed]
- Des Marteaux, L.E.; Stinziano, J.R.; Sinclair, B.J. Effects of cold acclimation on rectal macromorphology, ultrastructure, and cytoskeletal stability in Gryllus pennsylvanicus crickets. J. Insect. Physiol. 2018, 104, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Torson, A.S.; Yocum, G.D.; Rinehart, J.P.; Kemp, W.P.; Bowsher, J.H. Transcriptional responses to fluctuating thermal regimes underpinning differences in survival in the solitary bee Megachile rotundata. J. Exp. Biol. 2015, 218, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Ragland, G.J.; Denlinger, D.L.; Hahn, D.A. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc. Natl. Acad. Sci. USA 2010, 107, 14909–14914. [Google Scholar] [CrossRef] [PubMed]
- Kvist, J.; Wheat, C.W.; Kallioniemi, E.; Saastamoinen, M.; Hanski, I.; Frilander, M.J. Temperature treatments during larval development reveal extensive heritable and plastic variation in gene expression and life history traits. Mol. Ecol. 2013, 22, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Tobari, Y.N. Drosophila ananassae: Genetical and Biological Aspects; Japan Scientific Societies Press: Tokyo, Japan; Karger: Basel, Switzerland, 1993. [Google Scholar]
- Das, A.; Mohanty, S.; Stephan, W. Inferring the population structure and demography of Drosophila ananassae from multilocus data. Genetics 2004, 168, 1975–1985. [Google Scholar] [CrossRef]
- MacMillan, H.A.; Andersen, J.L.; Davies, S.A.; Overgaard, J. The capacity to maintain ion and water homeostasis underlies interspecific variation in Drosophila cold tolerance. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Gibert, P.; Moreteau, B.; Pétavy, G.; Karan, D.; David, J.R. Chill-coma tolerance, a major climatic adaptation among Drosophila species. Evolution 2001, 55, 1063–1068. [Google Scholar] [CrossRef]
- David, J.R.; Gibert, P.; Moreteau, B.; Gilchrist, G.W.; Huey, R.B. The fly that came in from the cold: Geographic variation of recovery time from low-temperature exposure in Drosophila subobscura. Funct. Ecol. 2003, 17, 425–430. [Google Scholar] [CrossRef]
- Sisodia, S.; Singh, B.N. Resistance to environmental stress in Drosophila ananassae: Latitudinal variation and adaptation among populations. J. Evol. Biol. 2010, 23, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Grath, S. Molecular evolution of sex-biased genes in Drosophila ananassae. Ph.D. Thesis, LMU München, München, Germany, 2010. [Google Scholar]
- Grath, S.; Baines, J.F.; Parsch, J. Molecular evolution of sex-biased genes in the Drosophila ananassae subgroup. BMC Evol. Biol. 2009, 9, 291. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Richards, S.; Carbone, M.A.; Zhu, D.; Anholt, R.R.H.; Ayroles, J.F.; Duncan, L.; Jordan, K.W.; Lawrence, F.; Magwire, M.M.; et al. Epistasis dominates the genetic architecture of Drosophila quantitative traits. PNAS 2012, 109, 15553–15559. [Google Scholar] [CrossRef] [PubMed]
- Drosophila 12 Genomes Consortium Evolution of Genes Genomes on the Drosophila Phylogeny. Available online: https://wwwuniprotorg/citations/17994087 (accessed on 25 September 2018).
- Müller, L.; Grath, S.; von Heckel, K.; Parsch, J. Inter- and intraspecific variation in Drosophila genes with sex-biased expression. Int. J. Evol. Biol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Attrill, H.; Falls, K.; Goodman, J.L.; Millburn, G.H.; Antonazzo, G.; Rey, A.J.; Marygold, S.J. FlyBase: Establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016, 44, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef] [PubMed]
- Huylmans, A.K.; Parsch, J. Population- and Sex-Biased Gene Expression in the Excretion Organs of Drosophila melanogaster. G3 2014, 4, 2307–2315. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 25 September 2018).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e0021800. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 4 April 2016).
- Coulter, D.E.; Swaykus, E.A.; Beran-Koehn, M.A.; Goldberg, D.; Wieschaus, E.; Schedl, P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990, 9, 3795–3804. [Google Scholar] [CrossRef] [PubMed]
- Gramates, L.S.; Marygold, S.J.; dos Santos, G.; Urbano, J.-M.; Antonazzo, G.; Matthews, B.B.; Rey, A.J.; Tabone, C.J.; Crosby, M.A.; Emmert, D.B.; et al. FlyBase at 25: Looking to the future. Nucleic Acids Res. 2017, 45, D663–D671. [Google Scholar] [CrossRef] [PubMed]
- Castrillon, D.H.; Gonczy, P.; Alexander, S.; Rawson, R.; Eberhart, C.G.; Viswanathan, S.; DiNardo, S.; Wasserman, S.A. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: Characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 1993, 135, 489–505. [Google Scholar] [PubMed]
- Goto, S. Expression of Drosophila homologue of senescence marker protein-30 during cold acclimation. J. Insect Physiol. 2000, 46, 1111–1120. [Google Scholar] [CrossRef]
- The FlyBase Consortium. FlyBase: Enhancing Drosophila Gene Ontology Annotations. Available online: https://europepmc.org/articles/PMC2686450/ (accessed on 25 September 2018).
- Chae, H.-J.; Ke, N.; Kim, H.-R.; Chen, S.; Godzik, A.; Dickman, M.; Reed, J.C. Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 2003, 323, 101–113. [Google Scholar] [CrossRef]
- Yuan, S.; Yu, X.; Topf, M.; Dorstyn, L.; Kumar, S.; Ludtke, S.J.; Akey, C.W. Structure of the Drosophila apoptosome at 6.9Å resolution. Structure 2011, 19, 128–140. [Google Scholar] [CrossRef]
- Stephan, W.; Li, H. The recent demographic and adaptive history of Drosophila melanogaster. Heredity 2007, 98, 65–68. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef]
- Sinclair, B.J.; Gibbs, A.G.; Roberts, S.P. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Mol. Biol. 2007, 16, 435–443. [Google Scholar] [CrossRef]
- Sejerkilde, M.; Sørensen, J.G.; Loeschcke, V. Effects of cold- and heat hardening on thermal resistance in Drosophila melanogaster. J. Insect Physiol. 2003, 49, 719–726. [Google Scholar] [CrossRef]
- Kimura, M.T.; Yoshida, K.M.; Goto, S.G. Accumulation of Hsp70 mRNA under environmental stresses in diapausing and nondiapausing adults of Drosophila triauraria. J. Insect Physiol. 1998, 44, 1009–1015. [Google Scholar] [PubMed]
- Storey, K.B.; Storey, J.M. Insect cold hardiness: Metabolic, gene, and protein adaptation. Can. J. Zool. 2012, 90, 456–475. [Google Scholar] [CrossRef]
- Liefting, M.; Hoffmann, A.A.; Ellers, J. Plasticity versus environmental canalization: Population differences in thermal responses along a latitudinal gradient in Drosophila serrata. Evolution 2009, 63, 1954–1963. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, H.A.; Knee, J.M.; Dennis, A.B.; Udaka, H.; Marshall, K.E.; Merritt, T.J.S.; Sinclair, B.J. Cold acclimation wholly reorganizes the Drosophila melanogaster transcriptome and metabolome. Sci. Rep. 2016, 6, 28999. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zeng, Z.; Liu, Q.; Zhang, R.; Ni, J. Se-methylselenocysteine inhibits apoptosis induced by clusterin knockdown in neuroblastoma N2a and SH-SY5Y cell lines. Int. J. Mol. Sci. 2014, 15, 21331–21347. [Google Scholar] [CrossRef]
- Walter Bock, K. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: Animal-plant arms-race and co-evolution. Biochem. Pharmacol. 2015, 99, 11–17. [Google Scholar] [CrossRef]
- Yi, S.-X.; Moore, C.W.; Lee, R.E. Rapid cold-hardening protects Drosophila melanogaster from cold-induced apoptosis. Apoptosis 2007, 12, 1183–1193. [Google Scholar] [CrossRef]
- Teets, N.; Denlinger, D.; Teets, N.M.; Denlinger, D.L. Physiological mechanisms of seasonal and rapid cold-hardening in insects. Physiol. Entomol. 2013, 38, 105–116. [Google Scholar] [CrossRef]
- Yi, S.-X.; Lee, R.E. Rapid cold-hardening blocks cold-induced apoptosis by inhibiting the activation of pro-caspases in the flesh fly Sarcophaga crassipalpis. Apoptosis 2011, 16, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, J.P.; Hartl, F.-U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, F.; Koonce, N.L.; Guo, L.; Fatima, S.; Qiu, C.; Moon, M.T.; Zheng, Y.; Ordway, R.W. Small heat shock protein-mediated cell-autonomous and nonautonomous protection in a Drosophila model for environmental stress-induced degeneration. Dis. Models Mech. 2016, 9, dmm–026385. [Google Scholar] [CrossRef] [PubMed]
- Morrow, G.; Tanguay, R.M. Drosophila melanogaster Hsp22: A mitochondrial small heat shock protein influencing the aging process. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Colinet, H.; Lee, S.F.; Hoffmann, A. Knocking down expression of Hsp22 and Hsp23 by RNA interference affects recovery from chill coma in Drosophila melanogaster. J. Exp. Biol. 2010, 213, 4146–4150. [Google Scholar] [CrossRef] [PubMed]
- Colinet, H.; Lee, S.F.; Hoffmann, A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 2010, 277, 174–185. [Google Scholar] [CrossRef]
- Štětina, T.; Koštál, V.; Korbelová, J. The role of inducible Hsp70, and other heat shock proteins, in adaptive complex of cold tolerance of the fruit fly (Drosophila melanogaster). PLoS ONE 2015, 10, e0128976. [Google Scholar] [CrossRef]
- Rinehart, J.P.; Li, A.; Yocum, G.D.; Robich, R.M.; Hayward, S.A.L.; Denlinger, D.L. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. USA 2007, 104, 11130–11137. [Google Scholar] [CrossRef]
- Udaka, H.; Ueda, C.; Goto, S.G. Survival rate and expression of Heat-shock protein 70 and Frost genes after temperature stress in Drosophila melanogaster lines that are selected for recovery time from temperature coma. J. Insect Physiol. 2010, 56, 1889–1894. [Google Scholar] [CrossRef]
- Nielsen, M.M.; Overgaard, J.; Sørensen, J.G.; Holmstrup, M.; Justesen, J.; Loeschcke, V. Role of HSF activation for resistance to heat, cold and high-temperature knock-down. J. Insect Physiol. 2005, 51, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.A.; Sept, D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 2008, 267, 183–206. [Google Scholar] [PubMed]
- Mullins, R.D.; Heuser, J.A.; Pollard, T.D. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. PNAS 1998, 95, 6181–6186. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.M.; Keshavmurthy, S.; Fontana, S.; Takuma, M.; Chou, W.-H.; Chen, C.A. Transcriptomic response in Acropora muricata under acute temperature stress follows preconditioned seasonal temperature fluctuations. BMC Res. Notes 2018, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Garland, M.A.; Stillman, J.H.; Tomanek, L. The proteomic response of cheliped myofibril tissue in the eurythermal porcelain crab Petrolisthes cinctipes to heat shock following acclimation to daily temperature fluctuations. J. Exp. Biol. 2015, 218 Pt 3, 388–403. [Google Scholar] [CrossRef]
- Cottam, D.M.; Tucker, J.B.; Rogers-Bald, M.M.; Mackie, J.B.; Macintyre, J.; Scarborough, J.A.; Ohkura, H.; Milner, M.J. Non-centrosomal microtubule-organising centres in cold-treated cultured Drosophila cells. Cell Motil. 2006, 63, 88–100. [Google Scholar] [CrossRef]
- Kim, M.; Robich, R.M.; Rinehart, J.P.; Denlinger, D.L. Upregulation of two actin genes and redistribution of actin during diapause and cold stress in the northern house mosquito, Culex pipiens. J. Insect Physiol. 2006, 52, 1226–1233. [Google Scholar] [CrossRef]
- Bowman, L.L.; Kondrateva, E.S.; Timofeyev, M.A.; Yampolsky, L.Y. Temperature gradient affects differentiation of gene expression and SNP allele frequencies in the dominant Lake Baikal zooplankton species. Mol. Ecol. 2018, 27, 2544–2559. [Google Scholar] [CrossRef]
- Chen, Y.-R.; Jiang, T.; Zhu, J.; Xie, Y.-C.; Tan, Z.-C.; Chen, Y.-H.; Tang, S.-M.; Hao, B.-F.; Wang, S.-P.; Huang, J.-S.; et al. Transcriptome sequencing reveals potential mechanisms of diapause preparation in bivoltine silkworm Bombyx mori (Lepidoptera: Bombycidae). Comp. Biochem. Physiol. Part D 2017, 24, 68–78. [Google Scholar] [CrossRef]
- Örvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef]
- Wu, J.-Y.; Jin, C.; Qu, H.-Y.; Tao, S.-T.; Xu, G.-H.; Wu, J.; Wu, H.-Q.; Zhang, S.-L. Low temperature inhibits pollen viability by alteration of actin cytoskeleton and regulation of pollen plasma membrane ion channels in Pyrus pyrifolia. Environ. Exp. Bot. 2012, 78, 70–75. [Google Scholar] [CrossRef]
- Sinclair, B.J.; Coello, L.A.; Ferguson, L.V. An invitation to measure insect cold tolerance: Methods, approaches, and workflow. J. Therm. Biol. 2015, 53, 180–197. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Gonda, R.L.; Garlena, R.A.; Stronach, B. Drosophila heat shock response requires the JNK pathway and phosphorylation of mixed lineage kinase at a conserved serine-proline motif. PLoS ONE 2012, 7, e0042369. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef]
- Teets, N.M.; Peyton, J.T.; Ragland, G.J.; Colinet, H.; Renault, D.; Hahn, D.A.; Denlinger, D.L. Combined transcriptomic and metabolomic approach uncovers molecular mechanisms of cold tolerance in a temperate flesh fly. Physiol. Genom. 2012, 44, 764–772. [Google Scholar] [CrossRef] [PubMed]

| Timepoint | Direction | Fast CCR | Slow CCR | Fast vs. Slow |
|---|---|---|---|---|
| control | up | - | - | 552 (3.87%) |
| down | - | - | 557 (3.91%) | |
| 15 min vs control | up | 46 (0.32%) | 82 (0.67%) | 0 |
| down | 11 (0.08%) | 96 (0.78%) | 0 | |
| 90 min vs control | up | 1096 (8.00%) | 1086 (7.80%) | 3 (0.02%) |
| down | 653 (4.80%) | 986 (7.10%) | 0 | |
| 90 min vs 15 min | up | 1114 (8.00%) | 1145 (8.20%) | - |
| down | 733 (5.20%) | 666 (4.80%) | - |
| Timepoint | Direction | Fast CCR | Slow CCR | Fast vs. Slow |
|---|---|---|---|---|
| control | up | - | - | 1080 (8.10%) |
| down | - | - | 1075 (8.10%) | |
| 15 min vs control | up | 96 (0.73%) | 142 (1.00%) | 0 |
| down | 86 (0.65%) | 97 (0.72%) | 1 (0.01%) | |
| 90 min vs control | up | 1258 (9.30%) | 1257 (9.46%) | 10 (0.08%) |
| down | 1342 (9.90%) | 1401 (11.00%) | 13 (0.10%) | |
| 90 min vs 15 min | up | 1060 (7.80%) | 1086 (8.00%) | - |
| down | 582 (4.30%) | 837 (6.20%) | - |
| Log2 Fold Change 15 min vs. Control | Log2 Fold Change 90 min vs. Control | Significance of Interaction a | |||
|---|---|---|---|---|---|
| Gene | Cold-Tolerant | Cold-Sensitive | Cold-Tolerant | Cold-Sensitive | Corrected P-Value b |
| GF14647 | 0.9597 | 0.6600 | 1.7301 | 0.2751 | 7.79 × 10−3 |
| GF15058 | −0.2726 | −0.7023 | 0.2298 | −0.5815 | 9.35 × 10−3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Königer, A.; Grath, S. Transcriptome Analysis Reveals Candidate Genes for Cold Tolerance in Drosophila ananassae. Genes 2018, 9, 624. https://doi.org/10.3390/genes9120624
Königer A, Grath S. Transcriptome Analysis Reveals Candidate Genes for Cold Tolerance in Drosophila ananassae. Genes. 2018; 9(12):624. https://doi.org/10.3390/genes9120624
Chicago/Turabian StyleKöniger, Annabella, and Sonja Grath. 2018. "Transcriptome Analysis Reveals Candidate Genes for Cold Tolerance in Drosophila ananassae" Genes 9, no. 12: 624. https://doi.org/10.3390/genes9120624
APA StyleKöniger, A., & Grath, S. (2018). Transcriptome Analysis Reveals Candidate Genes for Cold Tolerance in Drosophila ananassae. Genes, 9(12), 624. https://doi.org/10.3390/genes9120624





