Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress
Abstract
1. Introduction
2. Materials and Methods
Expression Analysis of Heat Shock Proteins 70 Genes Under Thermal and Hypoxia Stress
3. Results
3.1. Genome-Wide Identification of the Heat Shock Proteins 70 Gene Family in the Large Yellow Croaker
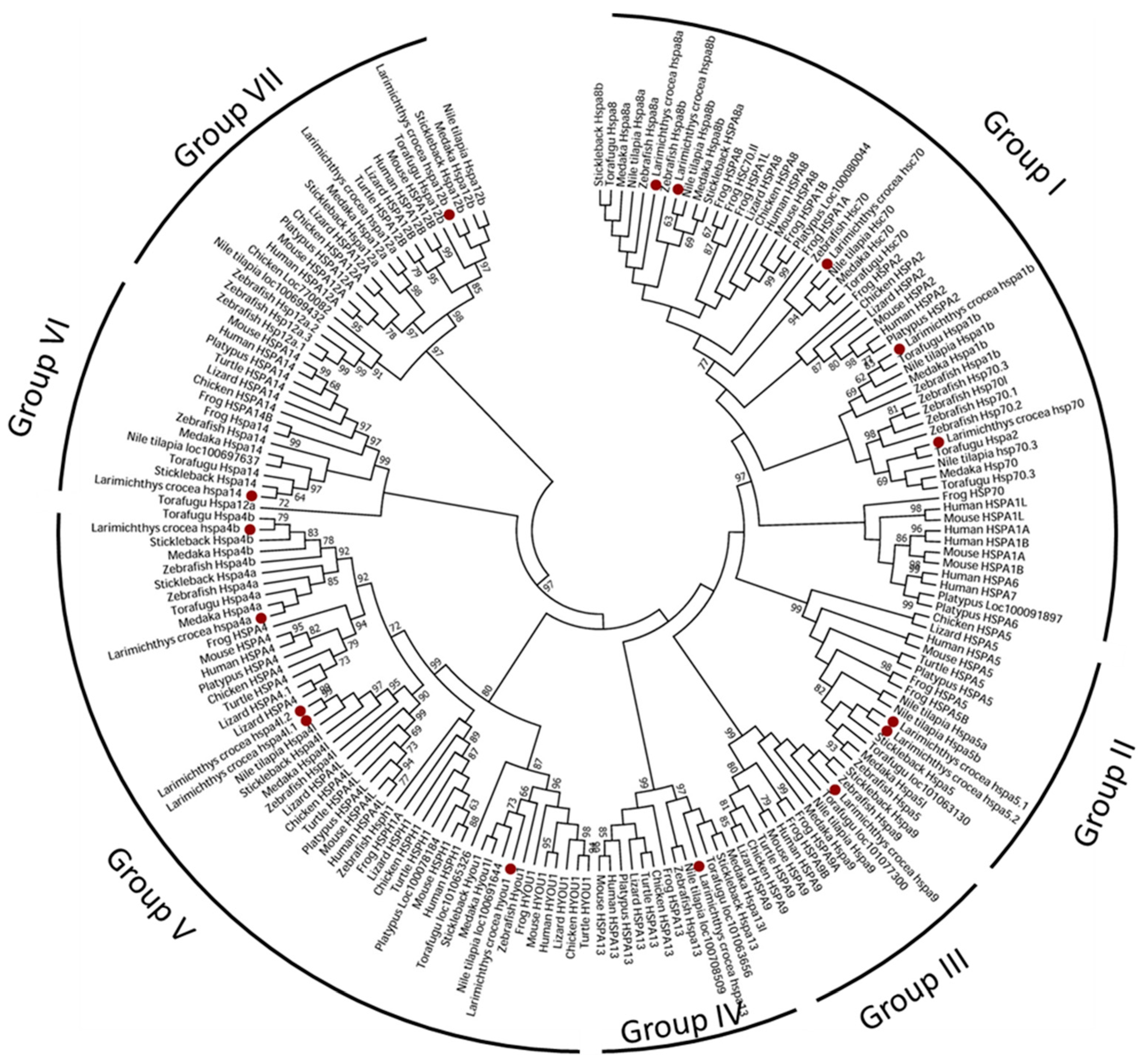
3.2. Phylogenetic Relationships of the Heat Shock Proteins 70 Genes Among Species
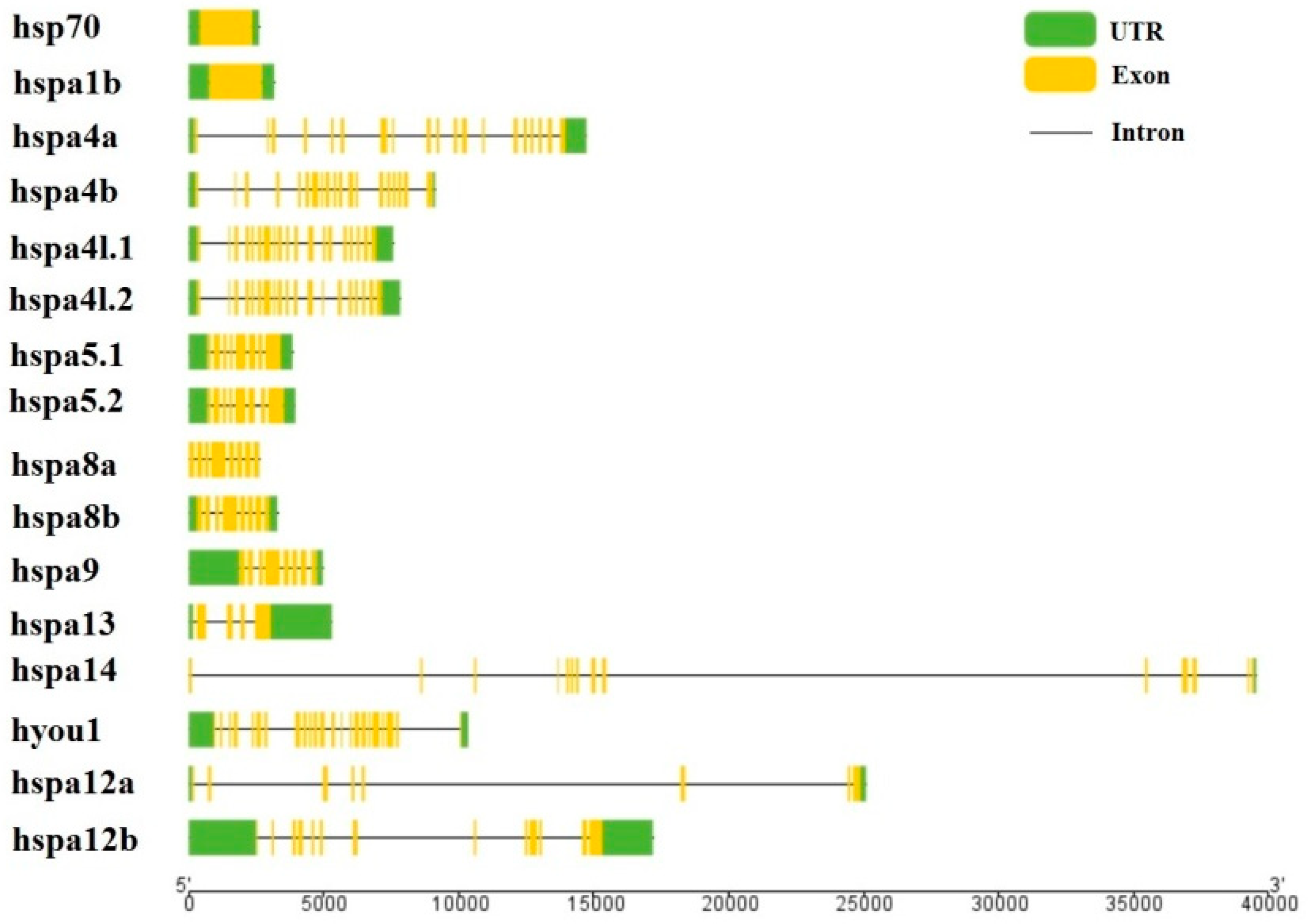
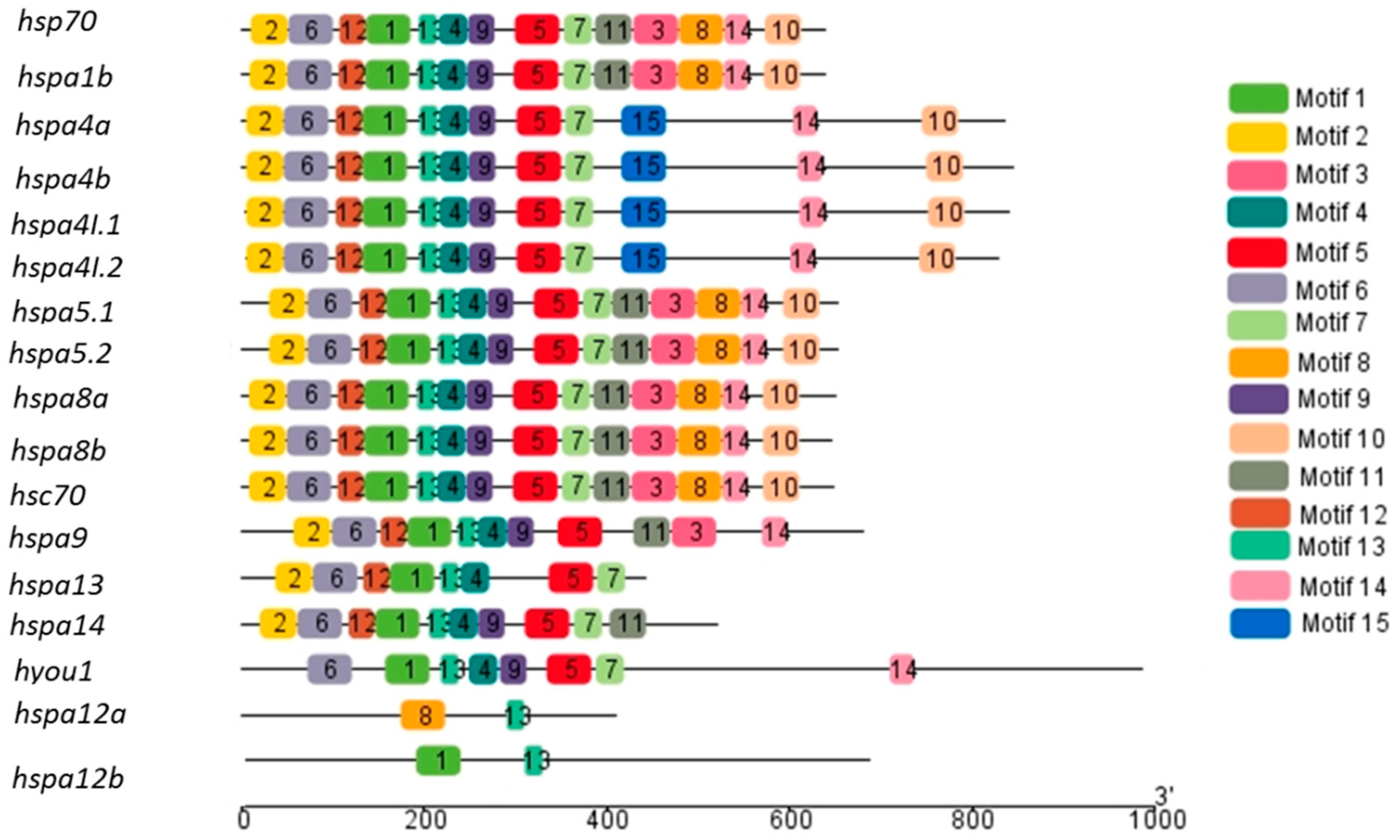
3.3. Gene Structure and Motif Analysis of the Heat Shock Proteins 70 in the Large Yellow Croaker
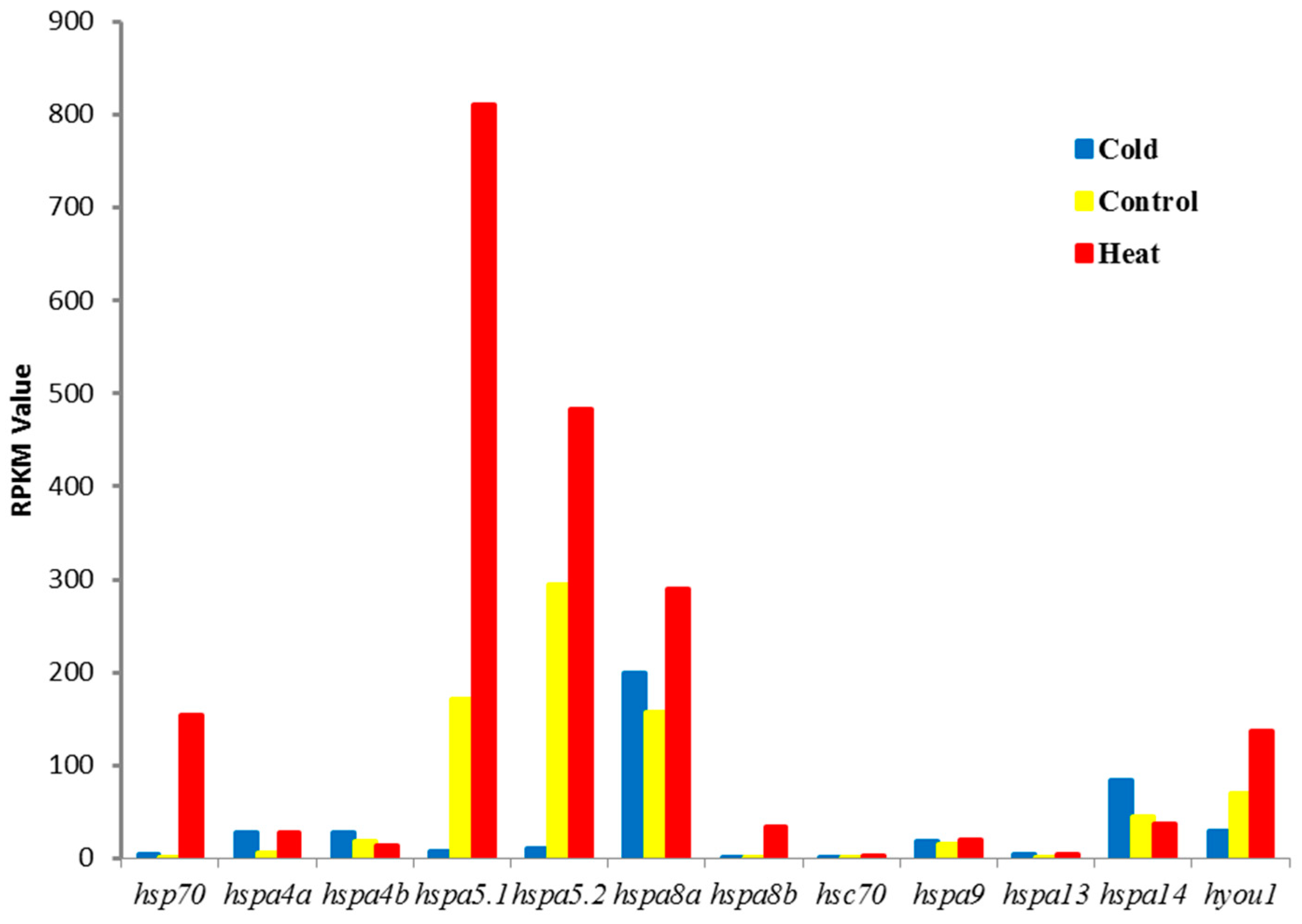
3.4. Expression Regulation of the Heat Shock Proteins 70Gene Under Thermal Stress Treatment
3.5. Validation of RNA-Seq Results by Quantitative Polymerase Chain Reaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Song, L.; Li, C.; Xie, Y.J.; Liu, S.K.; Zhang, J.R.; Yao, J.; Jiang, C.; Li, Y.; Liu, Z.J. Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immun. 2016, 49, 154–162. [Google Scholar]
- Metzger, D.C.; Hemmerhansen, J.; Schulte, P.M. Conserved structure and expression of hsp70 paralogs in teleost fishes. Comp. Biochem. Phys. D 2016, 18, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, D.; Chen, Q.; Ma, L. Genome-wide survey and characterization of the small heat shock protein gene family in Bursaphelenchus xylophilus. Gene 2016, 579, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Brocchieri, L.; Conway, D.M.E.; Macario, A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar]
- Liu, J.; Wang, R.; Liu, W.; Zhang, H.; Guo, Y.; Wen, R. Genome-wide characterization of heat-shock protein 70s from Chenopodium quinoa and expression analyses of Cqhsp70s in response to drought stress. Genes 2018, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Brocchieri, L. Heat shock protein 70 family: Multiple sequence comparisons, function, and evolution. J. Mol. Evol. 1998, 47, 565–577. [Google Scholar] [CrossRef]
- Ritossa, F. Discovery of the heat shock response. Cell Stress Chaperon 1996, 1, 97–98. [Google Scholar] [CrossRef]
- Giri, S.S.; Sen, S.S.; Sukumaran, V. Role of HSP70 in cytoplasm protection against thermal stress in rohu, Labeo rohita. Fish Shellfish Immun. 2014, 41, 294–299. [Google Scholar] [CrossRef]
- Yamashita, M.; Yabu, T.; Ojima, N. Stress protein HSP70 in fish. ABSM 2010, 3, 111–141. [Google Scholar] [CrossRef]
- Cheng, J.; Xun, X.G.; Kong, Y.F.; Wang, S.Y.; Yang, Z.H.; Li, Y.J.; Kong, D.X.; Wang, S.; Zhang, L.L.; Hu, X.L.; et al. Hsp70 gene expansions in the scallop (Patinopecten yessoensis) genome and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immun. 2016, 58, 266–273. [Google Scholar]
- Thanaphum, S.; Haymer, D.S. A member of the hsp70 gene family from the Mediterranean fruit fly, Ceratitis capitata. Insect Mol. Biol. 1998, 7, 63–72. [Google Scholar] [CrossRef]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Mu, Y.; Xiang, L.X.; Fan, D.D.; Feng, M.J.; Zhang, S.; Shi, Q.; Zhu, L.Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Q.; Xu, H.X.; Shui, B.N.; Zhou, Y.D.; Gao, T.X. Lack of genetic structure in endangered large yellow croaker Larimichthys crocea, from China inferred from mitochondrial control region sequence data. Biochem. Syst. Ecol. 2015, 61, 1–7. [Google Scholar] [CrossRef]
- Liu, M.; Mitcheson, S. Profile of a fishery collapse: Why mariculture failed to save the large yellow croaker. Fish Fish. 2008, 9, 219–242. [Google Scholar] [CrossRef]
- He, J.Y.; Wang, J.R.; Xu, M.S.; Wu, C.W.; Liu, H.H. The cooperative expression of heat shock protein 70 KD and 90 KD gene in juvenile Larimichthys crocea under Vibrio alginolyticus stress. Fish Shellfish Immun. 2016, 58, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Y.; Xue, L.Y.; Huang, H.L. Liver transcriptome analysis of the large yellow croaker (Larimichthys crocea) during fasting by using RNA-seq. PLoS ONE 2016, 11, 0150240. [Google Scholar] [CrossRef]
- Zhang, C.L.; Liu, J.F.; Li, Y.C.; Chen, Z. Analyzing the present condition and countermeasure of cultured large yellow croaker Pseudosciaena crocea in Fujian Province. Shanghai Fish. Univ. 2002, 11, 77–83. [Google Scholar]
- Qian, B.Y.; Xue, L.Y. Liver transcriptome sequencing and de novo annotation of the large yellow croaker (Larimichthys crocea) under heat and cold stress. Mar. Genom. 2016, 25, 95–102. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information Home Page. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 29 November 2018).
- Pfam Home Page. Available online: https://pfam.xfam.org/ (accessed on 15 November 2018).
- PANTHER Classification System Home Page. Available online: http://www.pantherdb.org/ (accessed on 5 October 2018).
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Simple Modular Architecture Research Tool Home Page. Available online: http://smart.embl-heidelberg.de/ (accessed on 2 November 2018).
- National Center for Biotechnology Information conserved domain database. Available online: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/ (accessed on 12 November 2018).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; AAAI Press: Menlo Park, CA, USA, 1994; pp. 28–36. [Google Scholar]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free; open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Sonna, L.A.; Fujita, J.; Gaffin, S.L.; Lilly, C.M. Molecular biology of thermoregulation invited review: Effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 2002, 92, 1725–1742. [Google Scholar] [CrossRef]
- Dana, R.C.; Welch, W.J.; Deftos, L.J. Heat shock proteins bind calcitonin. Endocrinology 1990, 126, 672–674. [Google Scholar] [CrossRef]
- Xia, B.P.; Liu, Z.; Zhou, Y.J.; Wang, Y.J.; Huang, J.Q.; Li, Y.J.; Kang, Y.J.; Wang, J.F.; Liu, X.X. Effects of heat stress on biochemical parameters and heat shock protein family a (Hsp70) member 5 (HSPA5) mRNA expression in rainbow trout (Oncorhynchus mykiss). Mar. Freshw. Res. 2018. [Google Scholar] [CrossRef]
- Huang, J.Q.; Li, Y.J.; Liu, Z.; Kang, Y.J.; Wang, J.F. Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish Shellfish Immun. 2018, 82, 32–40. [Google Scholar] [CrossRef]
- Otterson, G.A.; Gregory, C.F.; Kratzke, R.A.; Coxon, A.; Johnston, P.G.; Kaye, J.F. Stch encodes the ‘ATPase core’ of a microsomal stress 70 protein. EMBO J. 1994, 13, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Airaksinen, S.; Jokilehto, T.; Rabergh, C.M.; Nikinmaa, M. Heat-and-cold inducible regulation of HSP70 expression in zebrafish ZF4 cells. Comp. Biochem. Physiol. B 2003, 136, 275–282. [Google Scholar] [CrossRef]
- Bertotto, D.; Poltronieri, C.; Negrato, E.; Richrad, J.; Pascoli, F.; Simontacchi, C.; Radaelli, G. Whole body cortisol and expression of HSP70, IGF-I and MSTN in early development of sea bass subjected to heat shock. Gen. Comp. Endocrinol. 2011, 174, 44–50. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene Accession Number | Protein Accession Number | CDS Length (bp) | Protein Length (aa) | Hsp70 Domain Location (aa) | Domain Feature | WT (kDa) | |
|---|---|---|---|---|---|---|---|---|
| 1 | hsp70 | XM_010755062.2 | XP_010753364.1 | 1920 | 639 | 8–614 | HSPA1-2-6-8-like_NBD | 70.3 |
| 2 | hspa1b | XM_010740692.2 | XP_010738994.1 | 1917 | 638 | 7–613 | HSPA1-2-6-8-like_NBD | 70.3 |
| 3 | hspa4a | XM_010729327.2 | XP_010727629.1 | 2514 | 837 | 3–586 | HSPA4_NBD | 93.6 |
| 4 | hspa4b | XM_010754752.2 | XP_010753054.2 | 2535 | 844 | 3–598 | HSPA4_NBD | 94.6 |
| 5 | hspa4l.1 | XM_019277085.1 | XP_019132630.1 | 2523 | 840 | 3–696 | HSPA4_NBD | 94.4 |
| 6 | hspa4l.2 | XM_019279519.1 | XP_019135064.1 | 2493 | 830 | 3–557 | HSPA4_NBD | 93.5 |
| 7 | hspa5.1 | XM_010738795.2 | XP_010737097.1 | 1965 | 654 | 28–634 | HSPA5-like_NBD | 72.2 |
| 8 | hspa5.2 | XM_019274093.1 | XP_019129638.1 | 1965 | 654 | 28–634 | HSPA5-like_NBD | 72.2 |
| 9 | hspa8a | XM_019279481.1 | XP_019135026.1 | 1953 | 650 | 6–612 | HSPA1-2-6-8-like_NBD | 71.1 |
| 10 | hspa8b | XM_019270840 | XP_019126385.1 | 1941 | 646 | 6–612 | HSPA1-2-6-8-like_NBD | 70.9 |
| 11 | hsc70 | XM_010747566.2 | XP_010745868.1 | 1950 | 649 | 6–612 | HSPA1-2-6-8-like_NBD | 71.07 |
| 12 | hspa9 | XM_010734059.2 | XP_010732361.2 | 2043 | 680 | 55–653 | HSPA9-like_NBD | 73.4 |
| 13 | hspa12a | XM_019257914.1 | XP_019113459.1 | 1236 | 411 | 40–389 | HSPA12A-like_NBD | 46.4 |
| 14 | hspa12b | XM_019255647.1 | XP_019111192.1 | 2064 | 687 | 51–584 | HSPA12B-like_NBD | 76.5 |
| 15 | hspa13 | XM_010757137.2 | XP_010755439.1 | 1329 | 442 | 33–431 | HSPA13-like_NBD | 48.3 |
| 16 | hspa14 | XM_019266323.1 | XP_019121868.1 | 1566 | 521 | 18–521 | HSPA14-like_NBD | 56.4 |
| 17 | hyou1 | XM_019279502.1 | XP_019135047.1 | 2961 | 986 | 28–690 | HYOU1-like_NBD | 110.2 |
| Gene Name | hsp70 | hspa4a | hspa4b | hspa5.1 | hspa5.2 | hspa8a | hspa8b | hsc70 | hspa9 | hspa13 | hspa14 | hyou1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold | 2.17 * | 2.03 * | 0.64 | −4.49 * | −4.85 * | 0.33 | −0.92 | 0.68 | 0.28 | 1.66 | 0.90 | −1.27 |
| Heat | 7.32 * | 2.01 * | −0.44 | 2.24 * | 0.71 | 0.88 | 5.33 * | 2.13 * | 0.44 | 1.82 | −0.25 | 0.95 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Xu, H.; Han, Z. Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress. Genes 2018, 9, 590. https://doi.org/10.3390/genes9120590
Xu K, Xu H, Han Z. Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress. Genes. 2018; 9(12):590. https://doi.org/10.3390/genes9120590
Chicago/Turabian StyleXu, Kaida, Hanxiang Xu, and Zhiqiang Han. 2018. "Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress" Genes 9, no. 12: 590. https://doi.org/10.3390/genes9120590
APA StyleXu, K., Xu, H., & Han, Z. (2018). Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress. Genes, 9(12), 590. https://doi.org/10.3390/genes9120590




