UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans
Abstract
1. Introduction
2. Materials and Methods
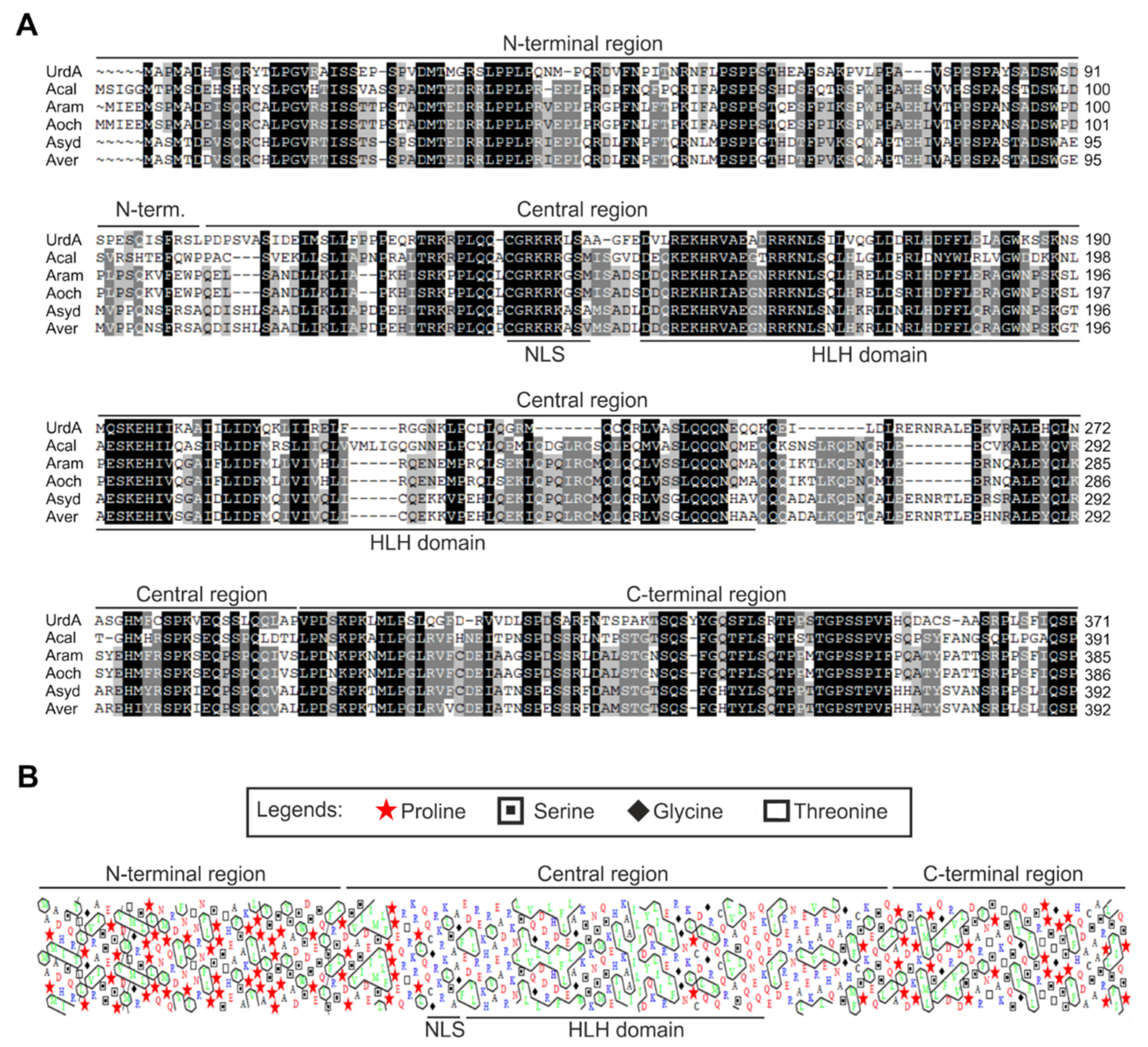
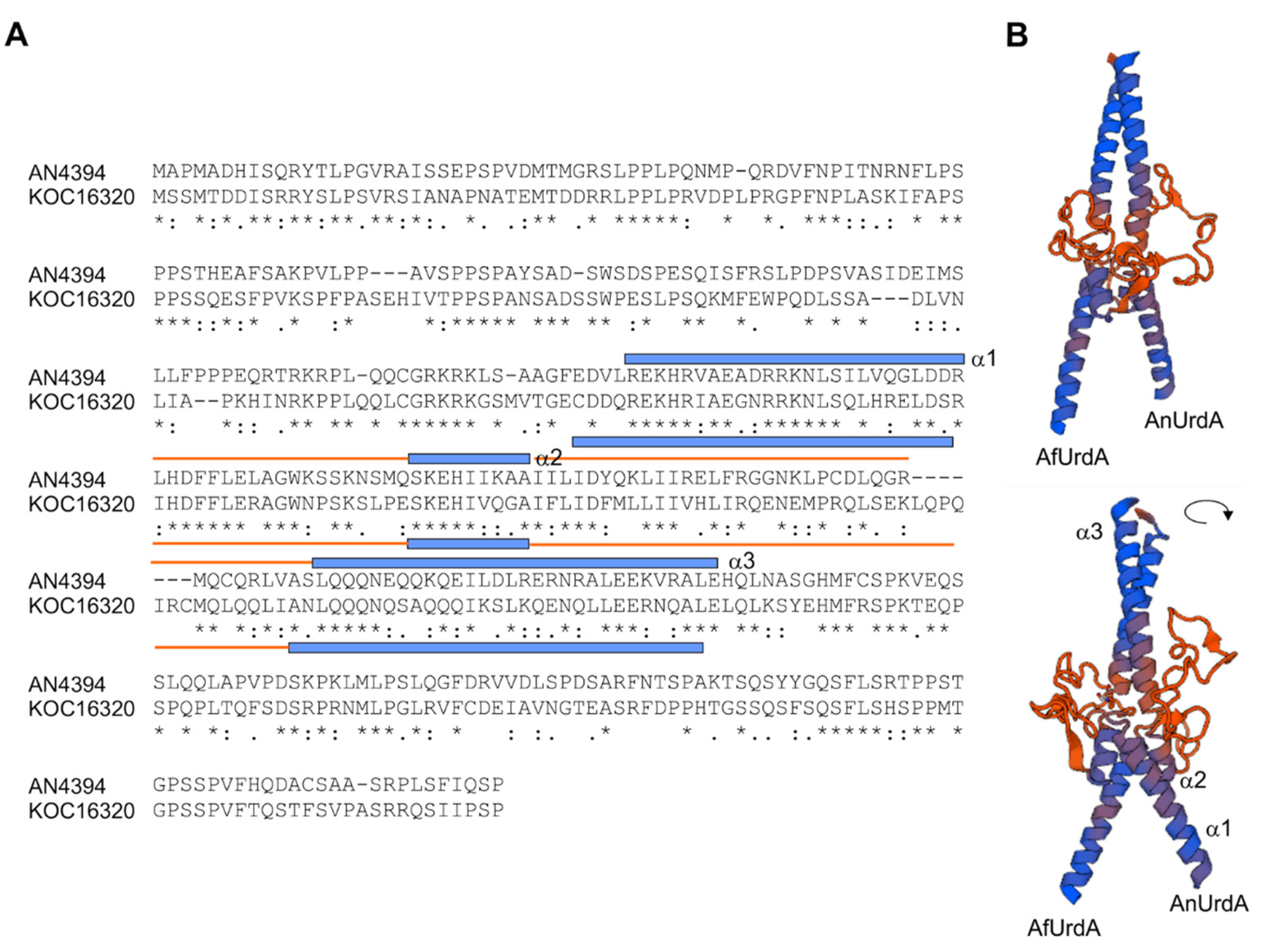
2.1. Sequence Search, Alignment and Phylogenetic Analyses
2.2. Fungal Strains and Culture Conditions
2.3. Generation of the urdA Deletion Strain (∆urdA)
2.4. Generation of the Aspergillus nidulans urdA Complementation Strain
2.5. Heterologous Complementation
2.6. Morphological Studies
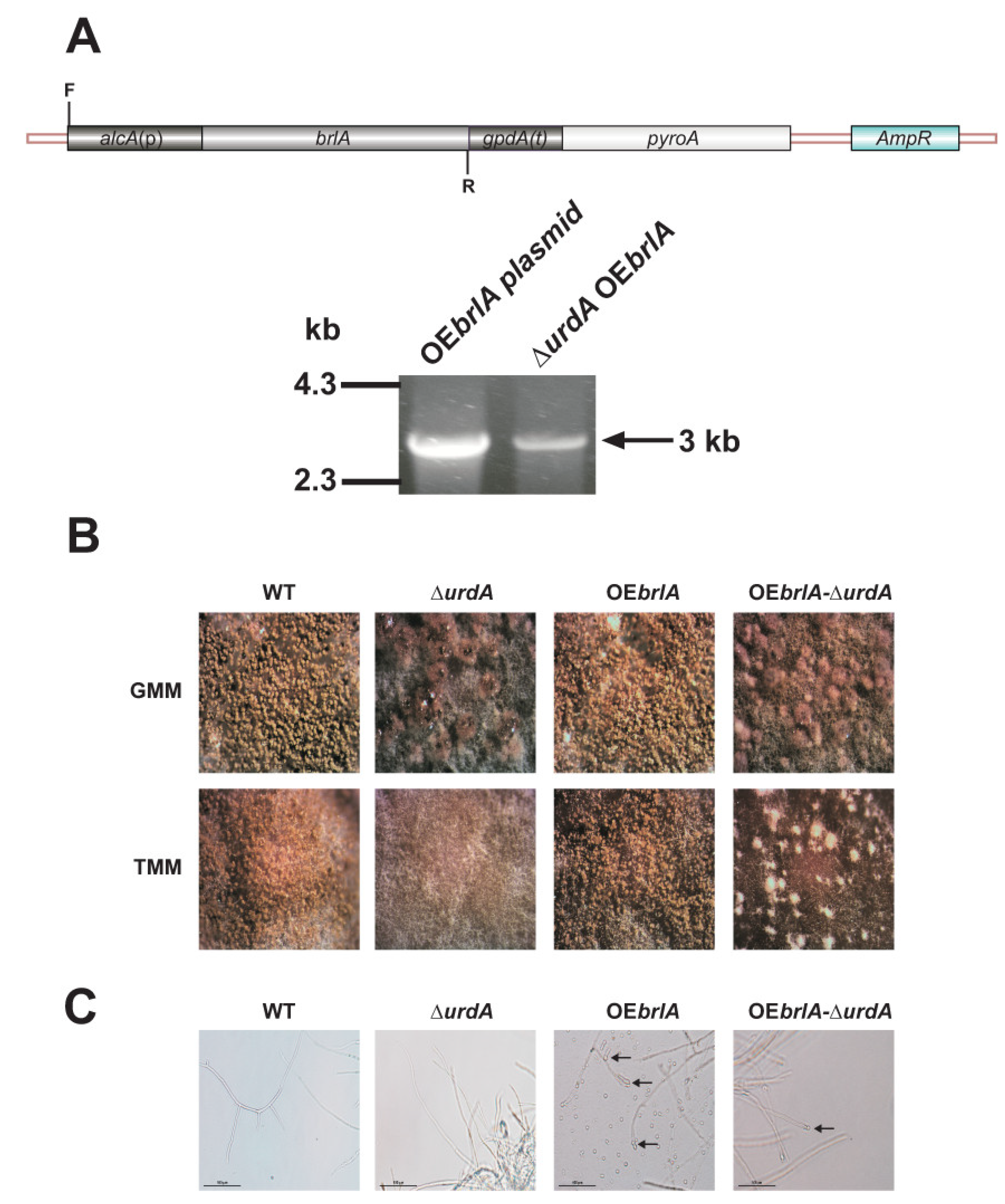
2.7. Overexpression of brlA in a ∆urdA Background
2.8. Toxin Analysis
2.9. Penicillin Analysis
2.10. Fluorescence Microscopy
2.11. Construction of a Double ∆urdA∆veA Strain
2.12. Gene Expression Analysis
2.13. Statistical Analysis
3. Results
3.1. UrdA Orthologs Are Found Exclusively in the Order Eurotiales
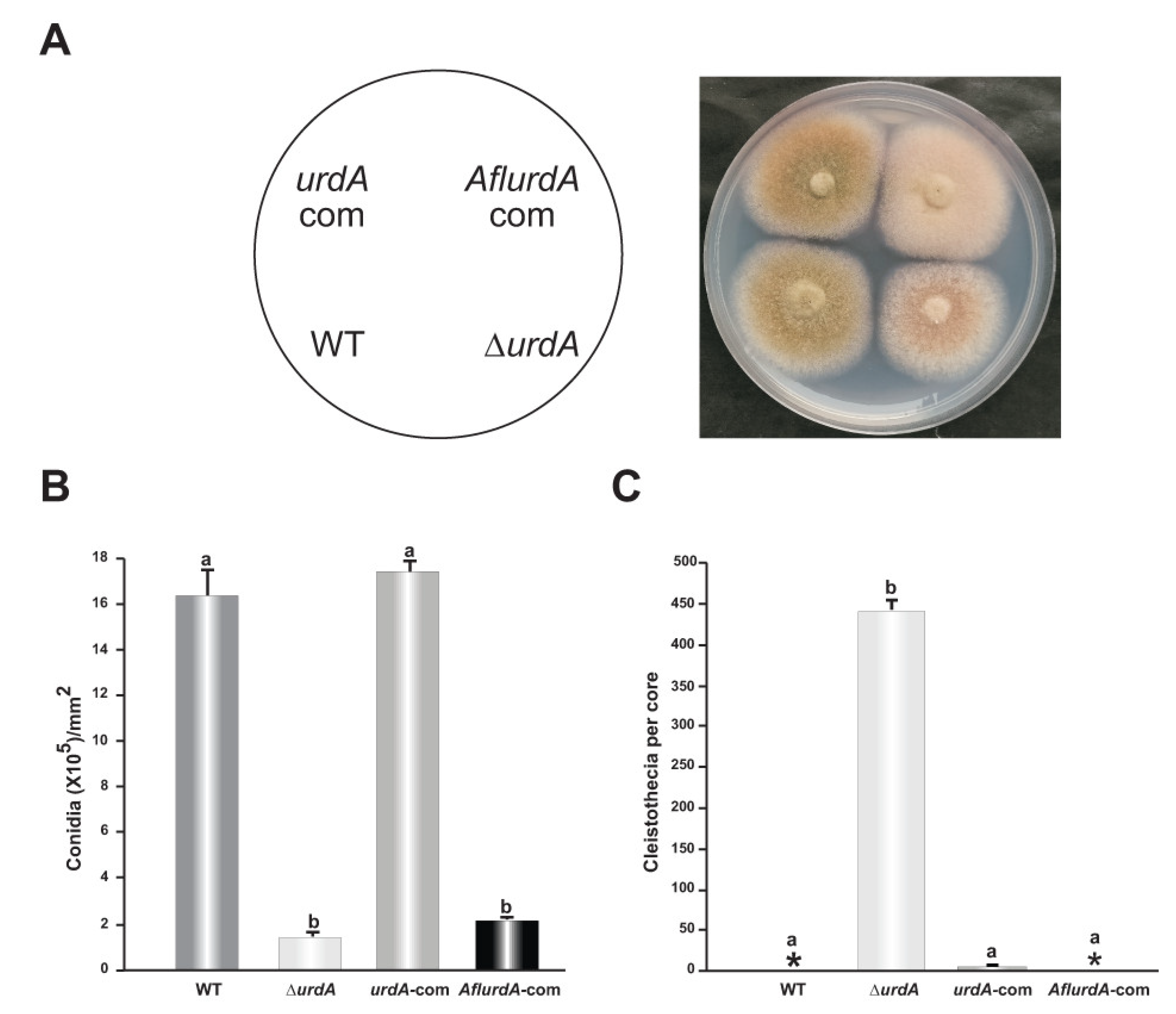
3.2. urdA Is Required for Normal Conidiation in A. nidulans
3.3. Overexpression of brlA Is Not Sufficient to Induce Conidiation in the Absence of urdA
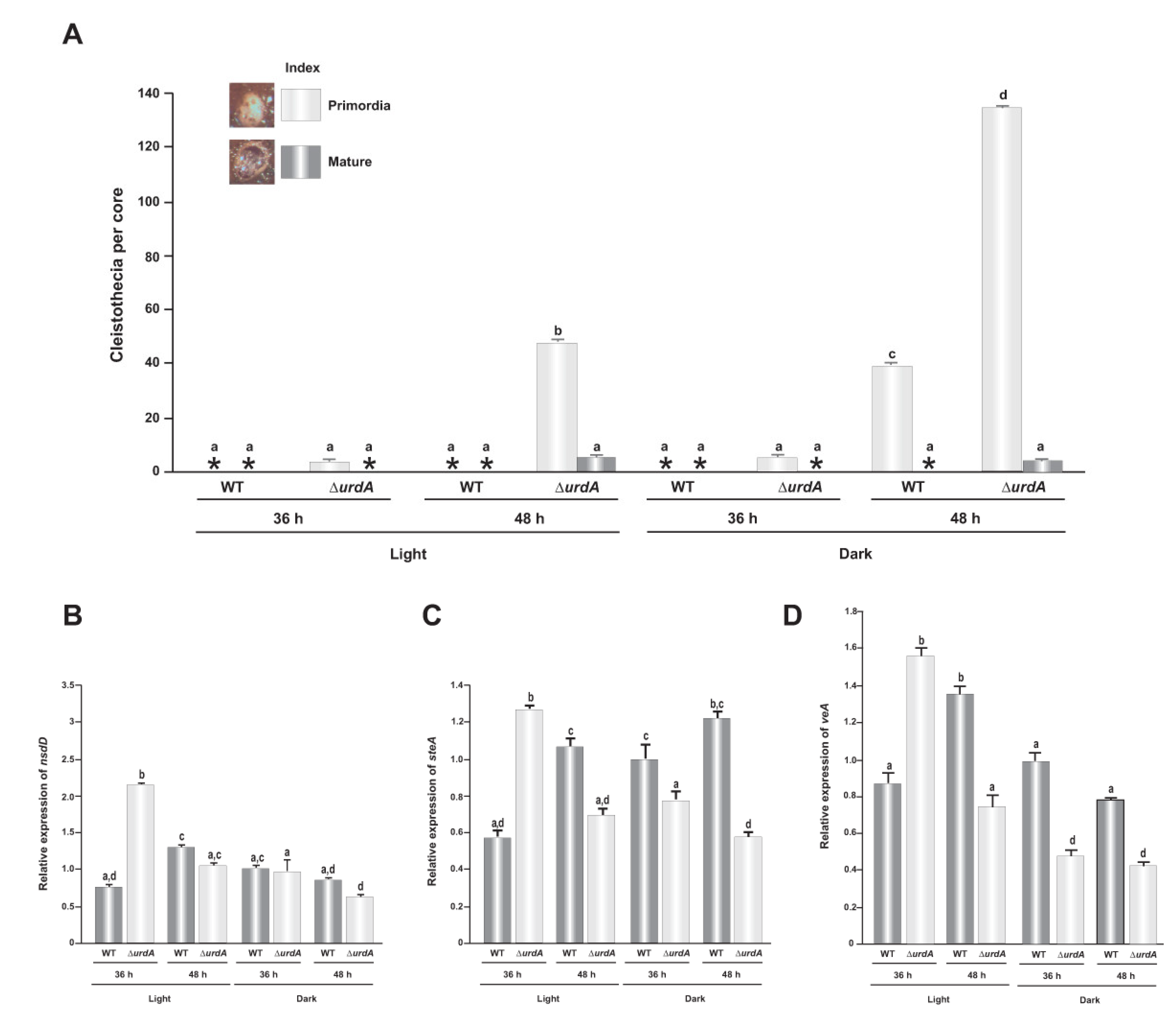
3.4. Effect of urdA on Sexual Development
3.5. Heterologous Complementation of A. nidulans ∆urdA with the Putative urdA Ortholog of A. flavus Does Not Fully Rescue Wild-Type Morphological Phenotype
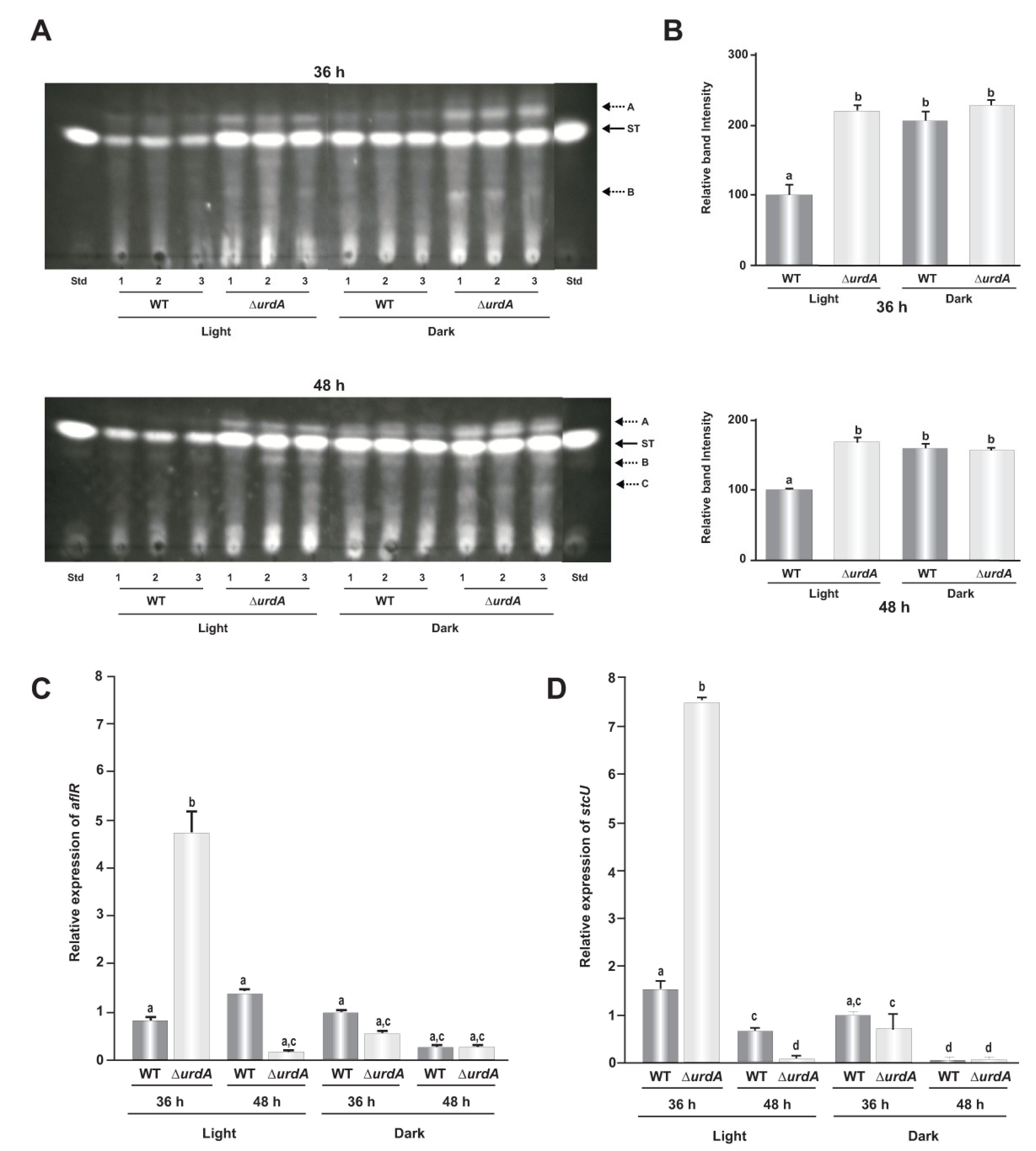
3.6. UrdA Is a Negative Regulator of Sterigmatocystin Biosynthesis and Production of Several Unknown Metabolites in a Light-Dependent Manner
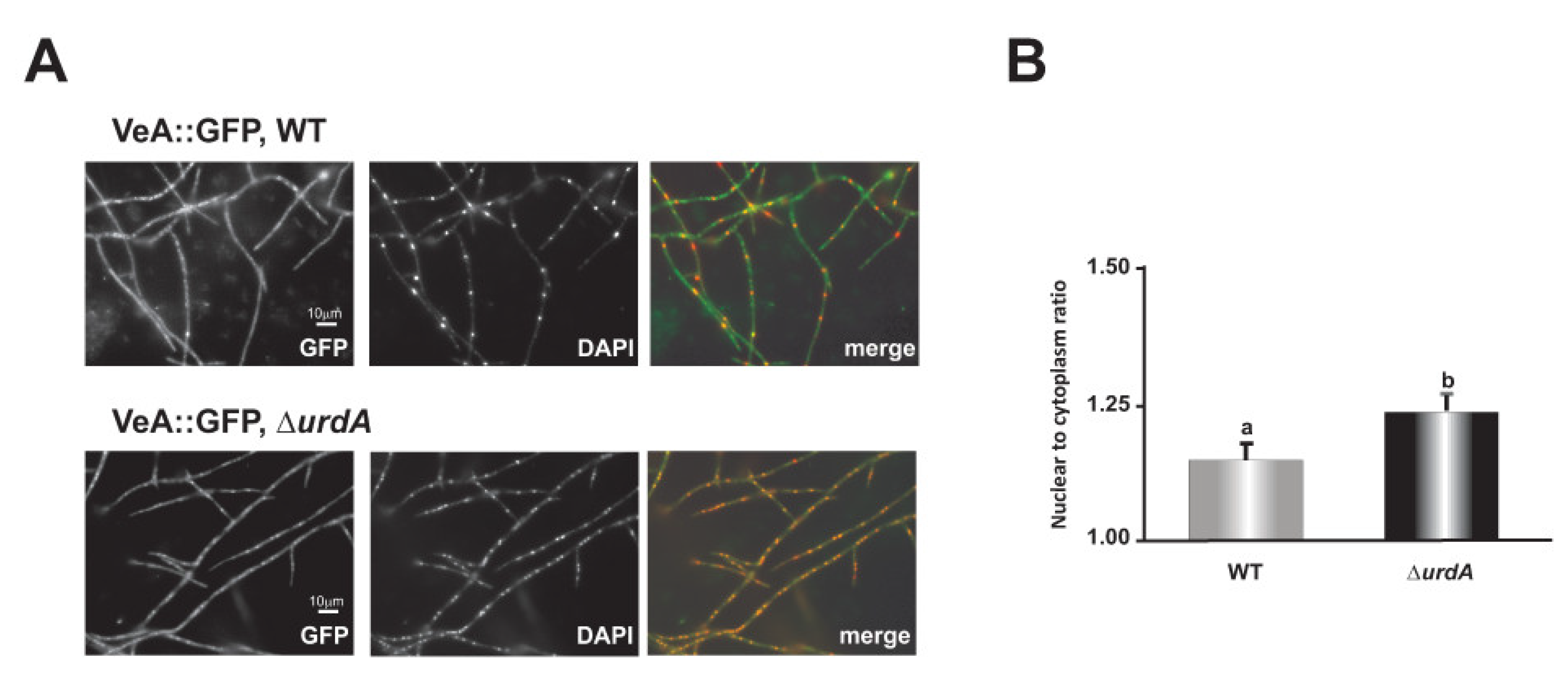
3.7. UrdA Affects the Level of VeA in Fungal Cells
3.8. Epistatic Relationship between urdA and veA
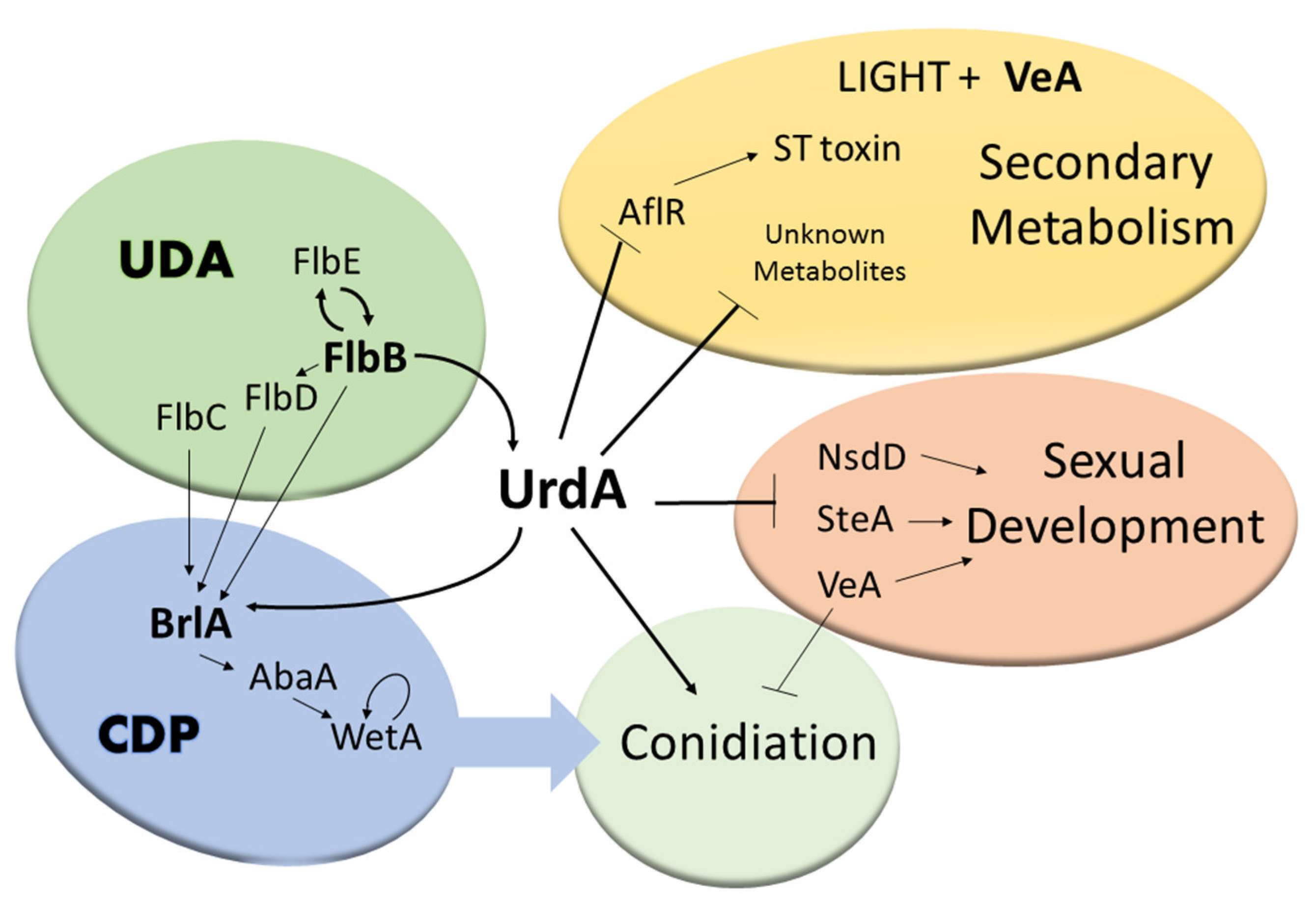
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix
| Strain | Pertinent Genotype | Source |
|---|---|---|
| FGSC4 | Wild type | FGSC* |
| BD834 | ∆urdA::pyrGA. fum; pyrG89; pyroA4 | This study |
| TRV50.2 | Wild type | [73] |
| TRV50 | pyroA4, ∆nkuA::argB; veA+ | [73] |
| T-17 | veA::gfp:: pyrGA.fum; pyrG89; pyroA4 | [30] |
| RJMP1.49 | pyrG89; argB2; ∆nku::argB; pyroA4 | [74] |
| TXF3.1 | pyrG89; argB2; ∆nku::argB; ∆veA::pyro; pyroA4 | [75] |
| TXFp2.1 | ∆veA::pyrGA.fum; pyrG89; argB2; ∆nku::argB; pyroA4, | [75] |
| TSSP1.1 | ∆urdA::pyrGA.fum; pyrG89; argB2; ∆nku::argB; pyroA4; | This study |
| TSSP3.1 | veA::gfp::pyrGA.fum; pyrG89, ∆nku::argB; argB2; | This study |
| TSSP4.1 | pyrG89, argB2; ∆nku::argB; ∆urdA::pyroA pyroA4 | This study |
| TSSP6.1 | pyrG89; veA::gfp::pyrGA.fum; ∆nku::argB; argB2; ∆urdA::pyroA; pyroA4; | This study |
| TSSP7.1 | pyrG89; argB2; ∆nkuA::argB | [75] |
| TSSP13.1 | pyrG89; ∆urdA::pyrGA.fum argB2; ∆nku::argB; ∆veA::pyroA; pyroA4 | This study |
| TSSP23.1 | pyrG89; argB2; ∆nkuA::argB; alcA::brlA::gpdA::pyroA; pyroA4, | This study |
| TSSP25.1 | ∆urdA::pyrG A. fum;pyrG89;argB2; ∆nkuA::argB | This study |
| TSSP26.1 | ∆urdA::pyrG A. fum; pyrG89; ∆nkuA::argB; argB2;; alcA::brlA::gpdA::pyroA; pyroA4 | This study |
| CA14 | niaD-;pyrG-; Δku70 | USDA** |
| TSSP27.1 | ∆urdA::pyrGA.fum; pyrG89; ∆nkuA::argB: argB2; urdA::pyro; pyroA4 | This Study |
| TSSP28.1 | ∆urdA::pyrG A. fum; pyrG89; ∆nkuA::argB; argB2; urdAA.flavus::pyroA;pyroA4 | This Study |
References
- Adrio, J.L.; Demain, A.L. Fungal biotechnol. Int. Microbiol. 2003, 6, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Ricelli, A.; Zjalic, S.; Fabbri, A.A.; Fanelli, C. Natural functions of mycotoxin and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 2010, 87, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites-strategies to activate silent gene clusters. Fungal Gen. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Woloshuk, C.P.; Shim, W.B. Aflatoxins, fumonisins, and trichothecenes: A convergence of knowledge. FEMS Microbiol. Rev. 2013, 37, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Pontecorvo, G.; Roper, J.A.; Chemmons, L.M.; MacDonald, K.D.; Bufton, A.W.J. The genetics of Aspergillus nidulans. Adv. Genet. 1953, 5, 141–238. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.A.; Nierman, W.C.; Wortman, J.R.; Pritchard, B.L.; Brown, D. Whole genome comparison of Aspergillus flavus and A. Oryzae. Med. Mycol. 2006, 44 (Suppl. 1), S9–S11. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.J.; Dobson, A.D. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol. Lett. 1999, 175, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.A.; Yu, J. Ecology, development and gene regulation in Aspergillus flavus. In Aspergillus: Molecular Biology and Genomics; Machida, M., Gomi, K., Eds.; Caister Academic Press: Norfolk, UK, 2010; pp. 157–171. [Google Scholar]
- Kew, M.C. Aflatoxins as a cause of hepatocellular carcinoma. J. Gastrointest. Liver Dis. 2013, 22, 301–310. [Google Scholar] [PubMed]
- Cole, R.J.; Cox, R.H. Handbook of Toxic Fungal Metabolites; Academic Press: New York, NY, USA, 1981; ISBN 0121797600. [Google Scholar]
- Keller, N.P.; Kantz, N.J.; Adams, T.H. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 1994, 60, 1444–1450. [Google Scholar] [PubMed]
- Yu, J.H.; Butchko, R.A.; Fernandes, M.; Keller, N.P.; Leonard, T.J.; Adams, T.H. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 1996, 29, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P.; Hohn, T.M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997, 21, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.; Keller, N.P.; Adams, T.H. Sequence-specific binding by Aspergillus nidulans AflR, a C-6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 1998, 28, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Balibar, C.J.; Howard-Jones, A.R.; Walsh, C.T. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat. Chem. Biol. 2007, 3, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Thön, M.; Spröte, P.; Scharf, D.H.; Al-Abdallah, Q.; Wolke, S.M.; Hortschansky, P. Aspects on evolution of fungal beta-lactam biosynthesis gene clusters and recruitment of trans-acting factors. Phytochemistry 2009, 70, 1801–1811. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Lohmar, J.M.; Ibarra, B.; Satterlee, T. Velvet Regulation of Fungal Development. In Growth, Differentiation and Sexuality, 3rd ed.; Wedland, J., Ed.; Mycota Series; Springer International Publishing: Cham, Switzerland, 2016; Volume I, Chapter 18; pp. 475–497. [Google Scholar] [CrossRef]
- Adams, T.H.; Boylan, M.T.; Timberlake, W.E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 1988, 54, 353–362. [Google Scholar] [CrossRef]
- Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans asexual development: Making the most of cellular modules. Trends Microbiol. 2010, 18, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Gao, N.; Kwon, N.J.; Shin, K.S.; Yu, J.H. Regulation of Aspergillus conidiation. In Cellular and Molecular Biology of Filamentous Fungi; Borkovich, K.A., Ebbole, D.J., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 557–576. [Google Scholar]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Kües, U. Asexual sporulation in mycelial fungi. In Growth, Differentiation, and Sexuality; Kües, U., Fischer, R., Eds.; Springer: Berlin, Germany, 2006; Volume 1, pp. 263–292. [Google Scholar]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [PubMed]
- Wieser, J.; Lee, B.N.; Fondon, J., III; Adams, T.H. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 1994, 27, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Oiartzabal-Arano, E.; Garzia, A.; Gorostidi, A.; Ugalde, U.; Espeso, E.A.; Etxebeste, O. Beyond asexual development: Modifications in the gene expression profile caused by the absence of the Aspergillus nidulans transcription factor FlbB. Genetics 2015, 199, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Gerke, J.; Bayram, Ö.; Feussner, K.; Landesfeind, M.; Shelest, E.; Ivo, F.; Braus, G.H. Breaking the silence: Protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl. Environ. Microbiol. 2012, 78, 8234–8244. [Google Scholar] [CrossRef] [PubMed]
- Purschwitz, J.; Müller, S.; Kastner, C.; Schöser, M.; Haas, H.; Espeso, E.A.; Atoui, A.; Calvo, A.M.; Fischer, R. Functional and physical interaction of blue-and red-light sensors in Aspergillus nidulans. Curr. Boil. 2008, 18, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Stinnett, S.M.; Espeso, E.A.; Cobeno, L.; Araujo-Bazan, L.; Calvo, A.M. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 2007, 63, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Bazan, L.; Dhingra, S.; Chu, J.; Fernandez-Martinez, J.J.; Calvo, A.M.; Espeso, E.A. Importin alpha is an essential nuclear import carrier adaptor required for proper sexual and asexual development and secondary metabolism in Aspergillus nidulans. Fungal Genet. Biol. 2009, 46, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, A.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008, 45, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 2010, 38 (Suppl. 2), W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Käfer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977, 19, 33–131. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Shantappa, S.; Dhingra, S.; Calvo, A.M. veA-dependent RNA-pol II transcription elongation factor-like protein, RtfA, is associated with secondary metabolism and morphological development in Aspergillus nidulans. Mol. Microbiol. 2012, 85, 795–814. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2007, 1, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ukil, L.; Osmani, A.; Nahm, F.; Davies, J.; De Souza, C.P.; Dou, X.; Perez-Balaguer, A.; Osmani, S.A. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryote Cell 2004, 3, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Etxebeste, O.; Herrero-García, E.; Araújo-Bazán, L.; Rodríguez-Urra, A.B.; Garzia, A.; Ugalde, U.; Espeso, E.A. The bZIP-type transcription factor FlbB regulates distinct morphogenetic stages of colony formation in Aspergillus nidulans. Mol. Microbiol. 2009, 73, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, T.E.; Bhatnagar, D.; Foell, C.J.; McCormick, S.P. Conversion of a new metabolite to aflatoxin B2 by Aspergillus parasiticus. Appl. Environ. Microbiol. 1987, 53, 2804–2807. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Browne, P.; Turner, G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J. Bacteriol. 1992, 174, 3789–3799. [Google Scholar] [CrossRef] [PubMed]
- Peñalva, M.A. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 2005, 42, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Han, K.Y.; Yu, J.H.; Chae, K.S.; Jahng, K.Y.; Dong-Min, H. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 2001, 41, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Vallim, M.A.; Miller, K.Y.; Miller, B.L. Aspergillus SteA (Sterile12-like) is a homeodomain-C2/H2-Zn+ 2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 2000, 36, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Atoui, A.; Kastner, C.; Larey, C.M.; Thokala, R.; Etxebeste, O.; Espeso, E.A.; Fischer, R.; Calvo, A.M. Cross-talk between light and glucose regulation controls toxin production and morphogenesis in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Yu, J.H.; Keller, K.P.; Adams, T.H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 1997, 16, 4916–4923. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. “NIH Image to ImageJ: 25 years of image analysis”. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Brooks, W.; Calvo, A.M. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryote Cell 2003, 2, 1178–1186. [Google Scholar] [CrossRef]
- Cortese, M.S.; Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Elucidation of functional markers from Aspergillus nidulans developmental regulator FlbB and their phylogenetic distribution. PLoS ONE 2011, 6, e17505. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Asadollahi, M.; Askin, M.; Barry, K.; Battaglia, E.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Krizsan, K.; Almasi, E.; Merenyi, Z.; Sahu, N.; Viragh, M.; Tamas, K.; Mondo, S.; Kiss, B.; Balint, B.; Kues, U.; et al. Transcriptomic atlas of mushroom development highlights an independent origin of complex multicellularity. bioRxiv 2018. [Google Scholar] [CrossRef]
- Mooney, J.L.; Yager, L.N. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 1990, 4, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Tokuok, M.; Jin, F.J.; Takahashi, T.; Koyama, Y. Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Nishida, M.; Hara, S.; Koyama, Y. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet. Biol. 2011, 48, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Coley-Smith, J.R.; Cooke, R.C. Survival and germination of fungal sclerotia. Annu. Rev. Phytopathol. 1971, 9, 65–92. [Google Scholar] [CrossRef]
- Malloch, D.; Cain, R.F. The Trichocomataceae: Ascomycetes with Aspergillus, Paeciloyces and Penicillium imperfect states. Can. J. Bot. 1972, 50, 2613–2628. [Google Scholar] [CrossRef]
- Wicklow, D.T. Survival of Aspergillus flavus sclerotia in soil. Mycol. Res. 1987, 89, 131–134. [Google Scholar] [CrossRef]
- Horn, B.W.; Sorensen, R.B.; Lamb, M.C.; Sobolev, V.S.; Olarte, R.A.; Worthington, C.J.; Carbone, I. Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 2014, 104, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Moore, G.G.; Carbone, I. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 2011, 103, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Moore, G.G.; Carbone, I. Sexual reproduction in Aspergillus flavus. Mycologia 2009, 101, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.H.; Timberlake, W.E. Developmental repression of growth and gene expression in Aspergillus. Proc. Natl. Acad. Sci. USA 1990, 87, 5405–5409. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Bok, J.; Brooks, W.; Keller, N.P. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004, 70, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Myung, K.; Li, S.; Butchko, R.A.; Busman, M.; Proctor, R.H.; Abbas, H.K.; Calvo, A.M. FvVE1 regulates biosynthesis of the mycotoxins fumonisins and fusarins in Fusarium verticillioides. J. Agric. Food Chem. 2009, 57, 5089–5094. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, S.; Andes, D.; Calvo, A.M. VeA regulates conidiation, gliotoxin production and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryote Cell 2012, 11, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Laskowski-Peak, M.C.; Calvo, A.M.; Rohrssen, J.; Smulian, A.G. VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum. Fungal Genet. Biol. 2012, 49, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Chettri, P.; Calvo, A.M.; Cary, J.W.; Dhingra, S.; Guo, Y.; McDougal, R.L.; Bradshaw, R.E. The veA gene of the pine needle pathogen Dothistroma septosporum regulates sporulation and secondary metabolism. Fung Gen. Biol. 2012, 49, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8, e74122. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, M.; Palmer, J.; EL-Naggar, W.A.; EL-Sokkary, M.A.; Habib, E.E.; Keller, N.P. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet. Biol. 2010, 47, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Ramamoorthy, V.; Pandit, S.S.; Prieto, A.; Espeso, E.A.; Calvo, A.M. cpsA regulates mycotoxin production, morphogenesis and cell wall biosynthesis in the fungus Aspergillus nidulans. Mol. Microbiol. 2017, 105, 1–24. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandit, S.S.; Lohmar, J.M.; Ahmed, S.; Etxebeste, O.; Espeso, E.A.; Calvo, A.M. UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans. Genes 2018, 9, 570. https://doi.org/10.3390/genes9120570
Pandit SS, Lohmar JM, Ahmed S, Etxebeste O, Espeso EA, Calvo AM. UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans. Genes. 2018; 9(12):570. https://doi.org/10.3390/genes9120570
Chicago/Turabian StylePandit, Sandesh S., Jessica M. Lohmar, Shawana Ahmed, Oier Etxebeste, Eduardo A. Espeso, and Ana M. Calvo. 2018. "UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans" Genes 9, no. 12: 570. https://doi.org/10.3390/genes9120570
APA StylePandit, S. S., Lohmar, J. M., Ahmed, S., Etxebeste, O., Espeso, E. A., & Calvo, A. M. (2018). UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans. Genes, 9(12), 570. https://doi.org/10.3390/genes9120570






