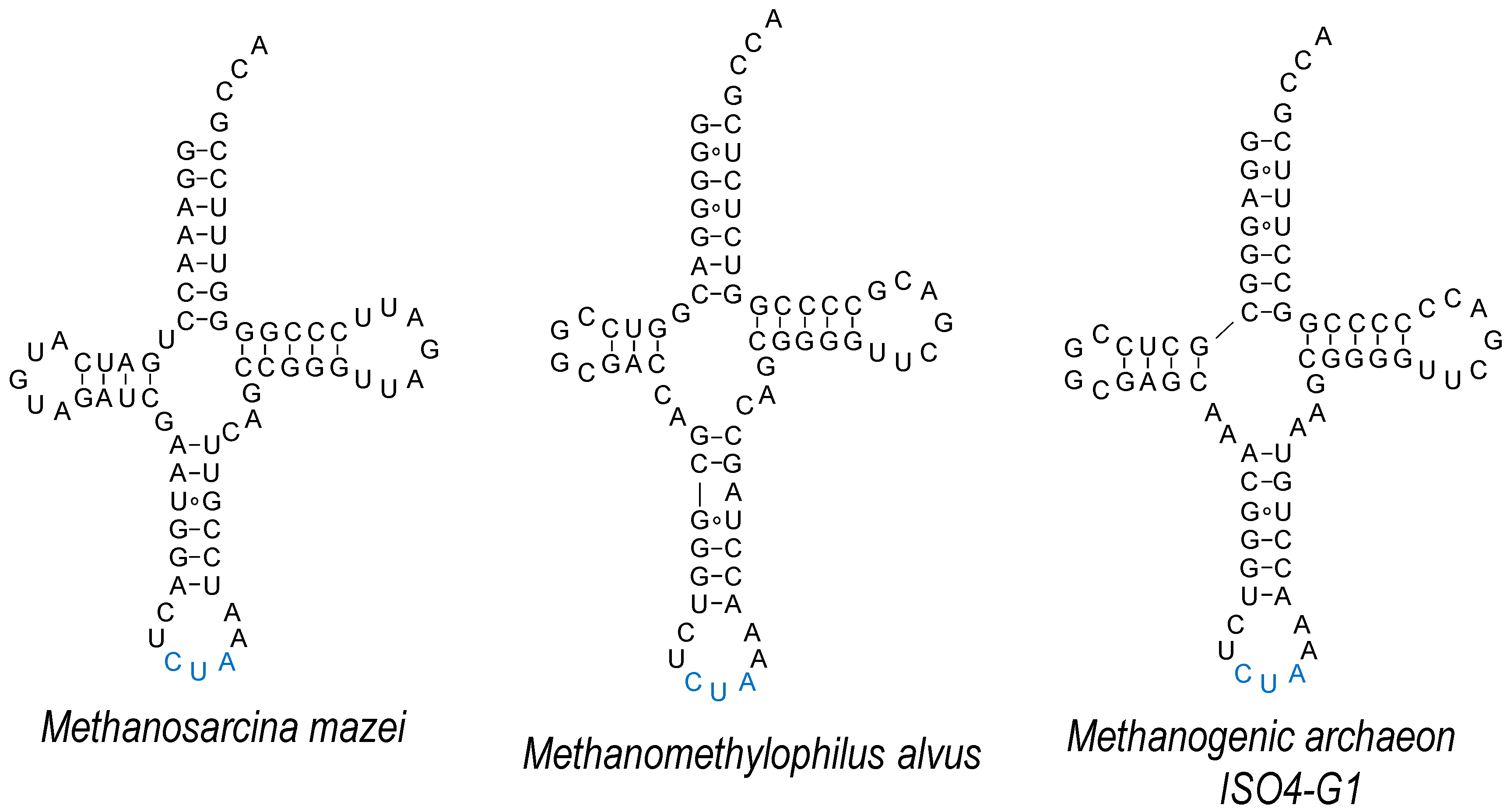Versatility of Synthetic tRNAs in Genetic Code Expansion
Abstract
1. Introduction
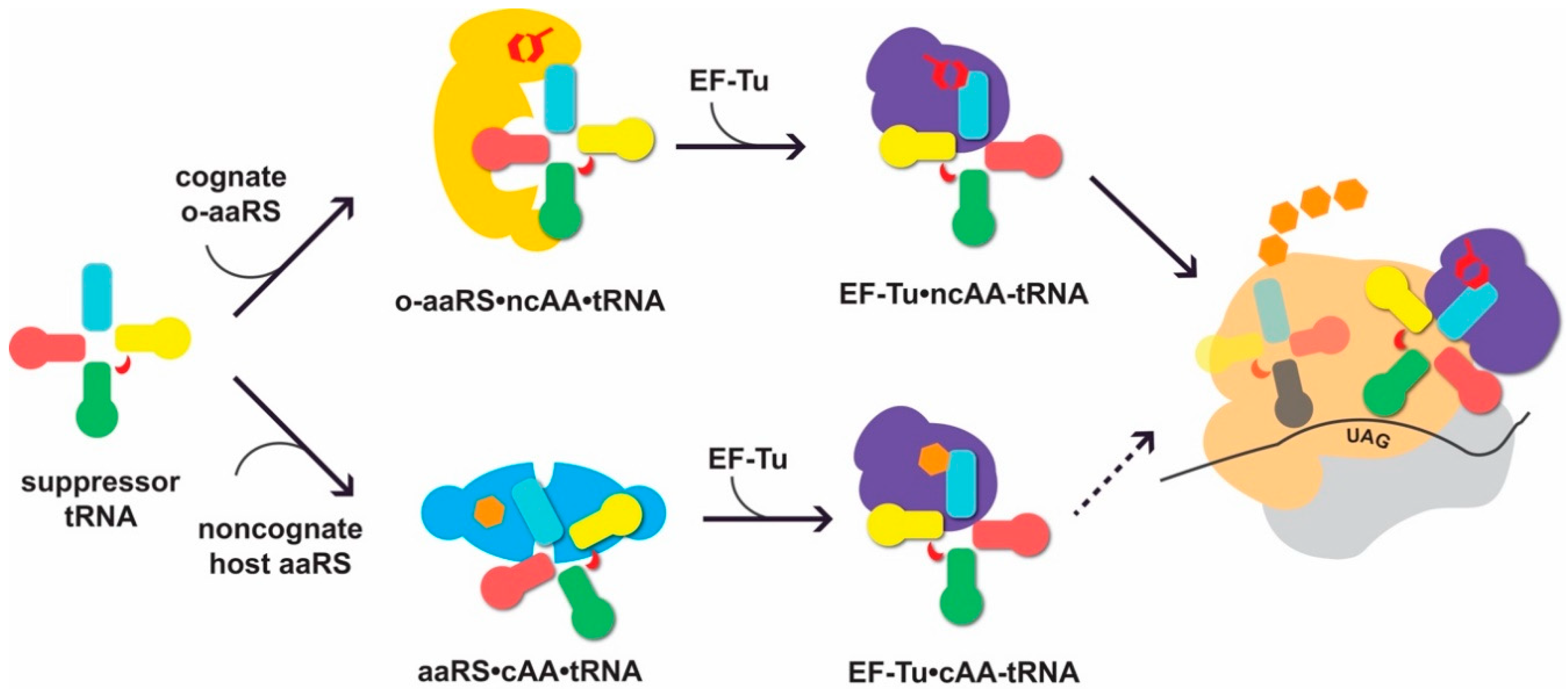
2. Aspects of Heterologous tRNA Expression
2.1. Identity Elements and Recognition
2.2. Heterologous tRNA Modification and Maturation
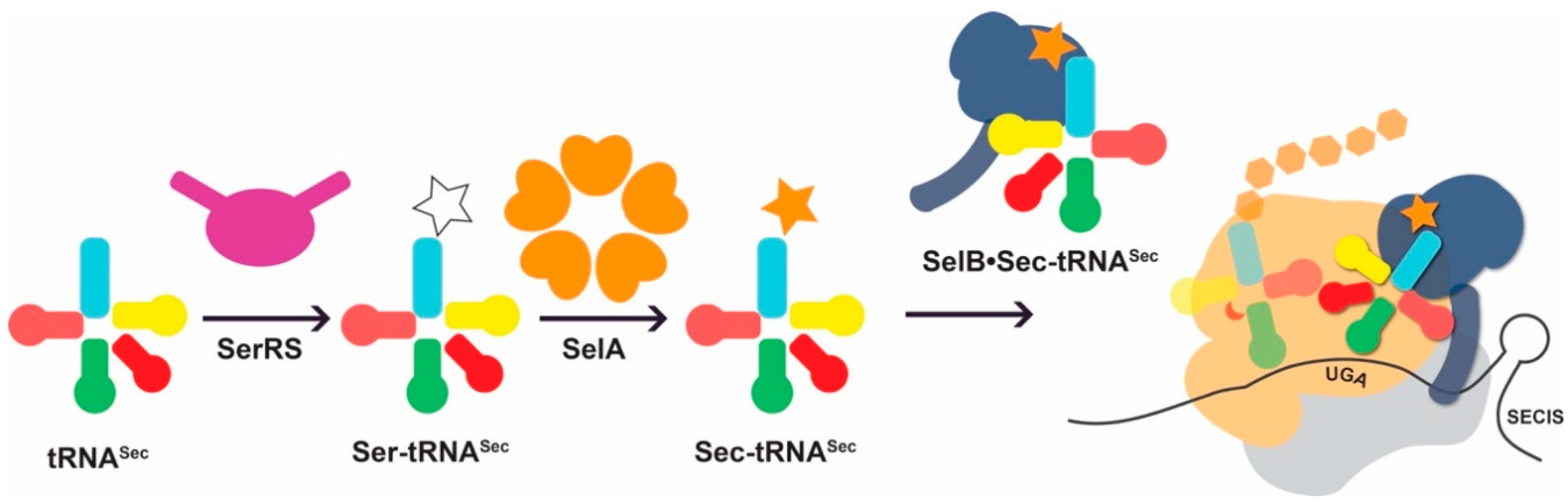
3. When Amino Acid Biosynthesis is o-tRNA-Dependent: Challenges in tRNASec Engineering
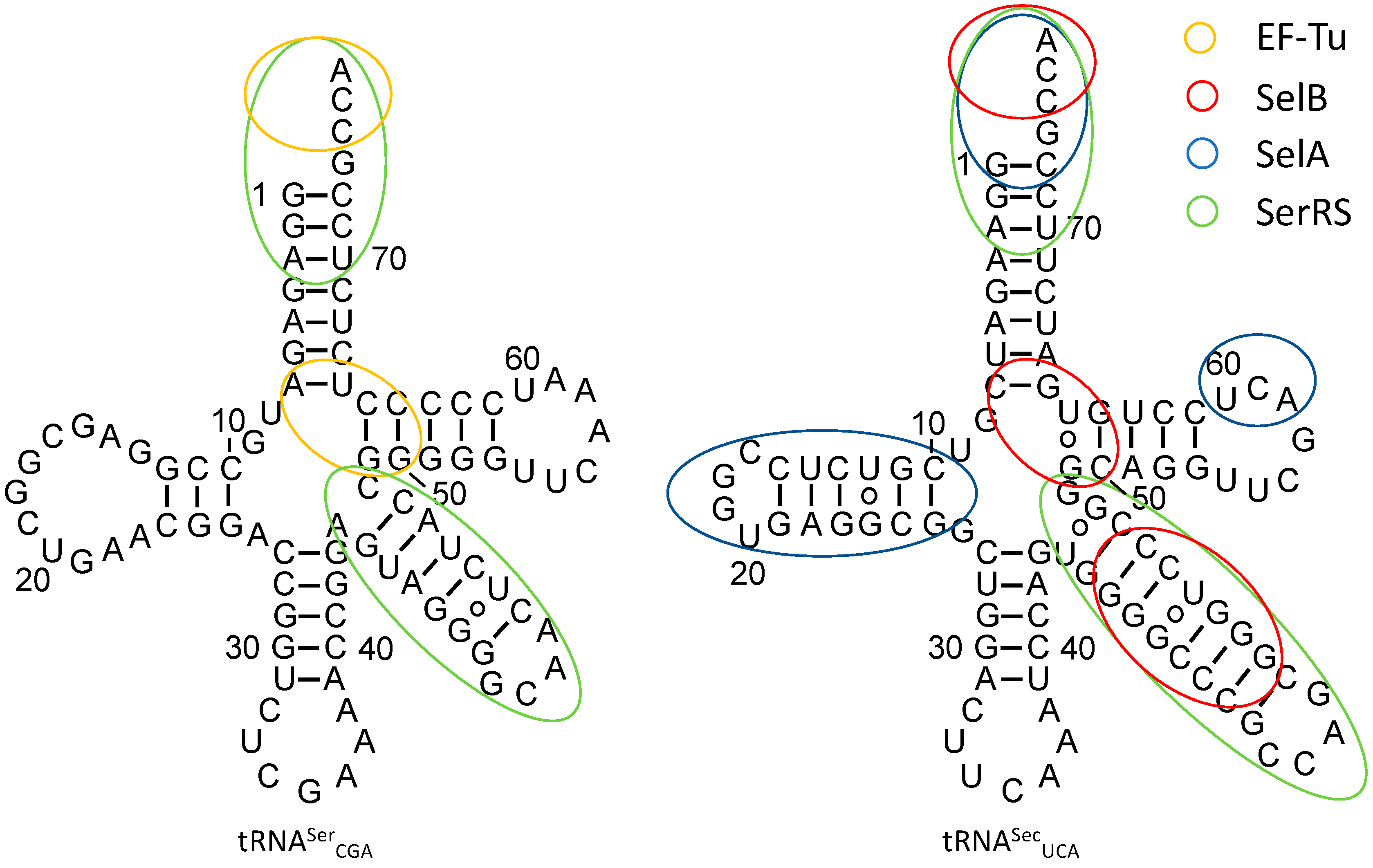
3.1. tRNASec Interactions
3.2. Converting tRNASec Recognition from SelB to EF-Tu
3.3. Improving Ser-to-Sec Conversion
3.4. Different tRNASec Structures for the Optimization of Selenoprotein Production
4. Absolutely Orthogonal? Unique Features of tRNAPyl
5. Conclusions/Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chin, J.W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014, 83, 379–408. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Lajoie, M.J.; Englert, M.; Soll, D. Rewriting the Genetic Code. Annu. Rev. Microbiol. 2017, 71, 557–577. [Google Scholar] [CrossRef] [PubMed]
- Young, D.D.; Schultz, P.G. Playing with the molecules of life. ACS Chem. Biol. 2018, 13, 854–870. [Google Scholar] [CrossRef] [PubMed]
- Herring, S.; Ambrogelly, A.; Gundllapalli, S.; O’Donoghue, P.; Polycarpo, C.R.; Söll, D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007, 581, 3197–3203. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Krzycki, J.A. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J. Biol. Chem. 2012, 287, 32738–32746. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W.; Cropp, T.A.; Anderson, J.C.; Mukherji, M.; Zhang, Z.; Schultz, P.G. An expanded eukaryotic genetic code. Science 2003, 301, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Greiss, S.; Chin, J.W. Expanding the genetic code of an animal. J. Am. Chem. Soc. 2011, 133, 14196–14199. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Xu, H.; Zhou, X.; Zhang, Z.; Tian, Z.; Wang, Y.; Wu, Y.; Zhang, B.; Niu, Z.; Zhang, C.; et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 2016, 354, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Townsley, F.M.; Greiss, S.; Lang, K.; Chin, J.W. Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 2012, 8, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Reich, H.J.; Hondal, R.J. Why nature chose selenium. ACS Chem. Biol. 2016, 11, 821–841. [Google Scholar] [CrossRef] [PubMed]
- Arai, K.; Takei, T.; Okumura, M.; Watanabe, S.; Amagai, Y.; Asahina, Y.; Moroder, L.; Hojo, H.; Inaba, K.; Iwaoka, M. Preparation of Selenoinsulin as a Long-Lasting Insulin Analogue. Angew. Chem. Int. Ed. Engl. 2017, 56, 5522–5526. [Google Scholar] [CrossRef] [PubMed]
- Metanis, N.; Hilvert, D. Natural and synthetic selenoproteins. Curr. Opin. Chem. Biol. 2014, 22, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Shchedrina, V.A.; Novoselov, S.V.; Malinouski, M.Y.; Gladyshev, V.N. Identification and characterization of a selenoprotein family containing a diselenide bond in a redox motif. Proc. Natl. Acad. Sci. USA 2007, 104, 13919–13924. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.A.; Xu, X.M.; Kryukov, G.V.; Rao, M.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 12848–12853. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Carlson, B.A.; Mix, H.; Zhang, Y.; Saira, K.; Glass, R.S.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007, 5, e4. [Google Scholar] [CrossRef] [PubMed]
- Palioura, S.; Sherrer, R.L.; Steitz, T.A.; Söll, D.; Simonovic, M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science 2009, 325, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Palioura, S.; Salazar, J.C.; Su, D.; O’Donoghue, P.; Hohn, M.J.; Cardoso, A.M.; Whitman, W.B.; Soll, D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA 2006, 103, 18923–18927. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Leinfelder, W.; Böck, A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 1989, 342, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Fagegaltier, D.; Hubert, N.; Yamada, K.; Mizutani, T.; Carbon, P.; Krol, A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000, 19, 4796–4805. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Berry, M.J. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life 2008, 60, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.; Copeland, P.R. Threading the needle: Getting selenocysteine into proteins. Antioxid. Redox Signal. 2010, 12, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Giege, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.T.; Wang, Y.S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef] [PubMed]
- Proc Natl Acad Sci U S AFEBS LettOhtake, K.; Mukai, T.; Iraha, F.; Takahashi, M.; Haruna, K.-i.; Date, M.; Yokoyama, K.; Sakamoto, K. Engineering an automaturing transglutaminase with enhanced thermostability by genetic code expansion with two codon reassignments. ACS Synth. Biol. 2018, 7, 2170–2176. [Google Scholar] [CrossRef]
- Fan, Z.; Song, J.; Guan, T.; Lv, X.; Wei, J. Efficient expression of glutathione peroxidase with chimeric tRNA in amber-less Escherichia coli. ACS Synth. Biol. 2018, 7, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Bröcker, M.J.; Prat, L.; Ip, K.; Chirathivat, N.; Feiock, A.; Veszpremi, M.; Söll, D. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015, 589, 2194–2199. [Google Scholar] [CrossRef] [PubMed]
- Thyer, R.; Robotham, S.A.; Brodbelt, J.S.; Ellington, A.D. Evolving tRNASec for efficient canonical incorporation of selenocysteine. J. Am. Chem. Soc. 2015, 137, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Haruna, K.; Alkazemi, M.H.; Liu, Y.; Söll, D.; Englert, M. Engineering the elongation factor Tu for efficient selenoprotein synthesis. Nucleic Acids Res. 2014, 42, 9976–9983. [Google Scholar] [CrossRef] [PubMed]
- Aldag, C.; Bröcker, M.J.; Hohn, M.J.; Prat, L.; Hammond, G.; Plummer, A.; Söll, D. Rewiring translation for elongation factor Tu-dependent selenocysteine incorporation. Angew. Chem. Int. Ed. Engl. 2013, 52, 1441–1445. [Google Scholar] [CrossRef] [PubMed]
- Baron, C.; Böck, A. The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNASec of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J. Biol. Chem. 1991, 266, 20375–20379. [Google Scholar] [PubMed]
- Li, W.T.; Mahapatra, A.; Longstaff, D.G.; Bechtel, J.; Zhao, G.; Kang, P.T.; Chan, M.K.; Krzycki, J.A. Specificity of pyrrolysyl-tRNA synthetase for pyrrolysine and pyrrolysine analogs. J. Mol. Biol. 2009, 385, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Bryson, D.I.; Fan, C.; Guo, L.T.; Miller, C.; Söll, D.; Liu, D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017, 13, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miller, C.; Guo, L.T.; Ho, J.M.L.; Bryson, D.I.; Wang, Y.S.; Liu, D.R.; Söll, D. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017, 13, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S.; Schimmel, P. Switching recognition of two tRNA synthetases with an amino acid swap in a designed peptide. Science 1995, 267, 1994–1996. [Google Scholar] [CrossRef] [PubMed]
- Perret, V.; Garcia, A.; Grosjean, H.; Ebel, J.P.; Florentz, C.; Giege, R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature 1990, 344, 787–789. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.D.; Giege, R.; Kern, D. Identity of prokaryotic and eukaryotic tRNAAsp for aminoacylation by aspartyl-tRNA synthetase from Thermus thermophilus. Biochemistry 1996, 35, 7447–7458. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; RajBhandary, U.L. Mutants of Escherichia coli initiator tRNA that suppress amber codons in Saccharomyces cerevisiae and are aminoacylated with tyrosine by yeast extracts. Proc. Natl. Acad. Sci. USA 1991, 88, 11378–11382. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.; Perez, J.G.; Carlson, E.D.; Ntai, I.; Isaacs, F.J.; Kelleher, N.L.; Jewett, M.C. Translation system engineering in Escherichia coli enhances non-canonical amino acid incorporation into proteins. Biotechnol. Bioeng. 2017, 114, 1074–1086. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oh, S.; Yang, A.; Kim, J.; Soll, D.; Lee, D.; Park, H.S. A facile strategy for selective incorporation of phosphoserine into histones. Angew. Chem. Int. Ed. Engl. 2013, 52, 5771–5775. [Google Scholar] [CrossRef] [PubMed]
- Rogerson, D.T.; Sachdeva, A.; Wang, K.; Haq, T.; Kazlauskaite, A.; Hancock, S.M.; Huguenin-Dezot, N.; Muqit, M.M.; Fry, A.M.; Bayliss, R.; et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat. Chem. Biol. 2015, 11, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Xiao, H.; Schultz, P.G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, 14841–14846. [Google Scholar] [CrossRef] [PubMed]
- Bröcker, M.J.; Ho, J.M.; Church, G.M.; Söll, D.; O’Donoghue, P. Recoding the genetic code with selenocysteine. Angew. Chem. Int. Ed. Engl. 2014, 53, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Baron, C.; Heider, J.; Böck, A. Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA-species in E. coli: Effects on in vivo function. Nucleic Acids Res. 1990, 18, 6761–6766. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, J.B.; Aerni, H.R.; Rogulina, S.; Liu, Y.; Rinehart, J. Expanded cellular amino acid pools containing phosphoserine, phosphothreonine, and phosphotyrosine. ACS Chem. Biol. 2014, 9, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Galli, G.; Hofstetter, H.; Birnstiel, M.L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 1981, 294, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Sharp, S.; DeFranco, D.; Dingermann, T.; Farrell, P.; Soll, D. Internal control regions for transcription of eukaryotic tRNA genes. Proc. Natl. Acad. Sci. USA 1981, 78, 6657–6661. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L. Genetic incorporation of unnatural amino acids into proteins in yeast. Methods Mol. Biol. 2012, 794, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Schultz, P.G.; Brock, A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J. Mol. Biol. 2007, 371, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Otter, C.A.; Straby, K.B. Transcription of eukaryotic genes with impaired internal promoters: The use of a yeast tRNA gene as promoter. J. Biotechnol. 1991, 21, 289–293. [Google Scholar] [CrossRef]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Munch, R.; Hohn, M.J.; Jahn, D.; Söll, D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 2005, 433, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Lünse, E.C.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Väre, Y.V.; Eruysal, R.E.; Narendran, A.; Sarachan, L.K.; Agris, F.P. Chemical and conformational diversity of modified nucleosides affects trna structure and function. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Wang, X.; Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 2006, 12, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Dewe, J.M.; Whipple, J.M.; Chernyakov, I.; Jaramillo, L.N.; Phizicky, E.M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012, 18, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.D.; Hoffman, K.S.; Genereaux, J.; Mian, S.; Trussler, R.S.; Haniford, D.B.; O’Donoghue, P.; Brandl, C.J. Evolving mistranslating tRNAs through a phenotypically ambivalent intermediate in Saccharomyces cerevisiae. Genetics 2017, 206, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef] [PubMed]
- Gefter, M.L.; Russell, R.L. Role modifications in tyrosine transfer RNA: A modified base affecting ribosome binding. J. Mol. Biol. 1969, 39, 145–157. [Google Scholar] [CrossRef]
- Laten, H.; Gorman, J.; Böck, R.M. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1978, 5, 4329–4342. [Google Scholar] [CrossRef] [PubMed]
- Crnkovic, A.; Vargas-Rodriguez, O.; Merkuryev, A.; Söll, D. Effects of heterologous tRNA modifications on the production of proteins containing noncanonical amino acids. Bioengineering 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Schaffrath, R. Collaboration of tRNA modifications and elongation factor eEF1A in decoding and nonsense suppression. Sci. Rep. 2018, 8, 12749. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; Saks, M.E. Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: A kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993, 21, 4467–4475. [Google Scholar] [CrossRef] [PubMed]
- Biou, V.; Yaremchuk, A.; Tukalo, M.; Cusack, S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 1994, 263, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Ollick, T.; Abelson, J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl. Acad. Sci. USA 1992, 89, 5680–5684. [Google Scholar] [CrossRef] [PubMed]
- Himeno, H.; Hasegawa, T.; Ueda, T.; Watanabe, K.; Shimizu, M. Conversion of aminoacylation specificity from tRNATyr to tRNASer in vitro. Nucleic Acids Res. 1990, 18, 6815–6819. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.J.; Söll, D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc. Natl. Acad. Sci. USA 1988, 85, 6627–6631. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Ogden, R.C.; Horvath, S.J.; Abelson, J. Changing the identity of a transfer RNA. Nature 1986, 321, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Sekine, S.; Suetsugu, S.; Yokoyama, S. Tertiary structure of bacterial selenocysteine tRNA. Nucleic Acids Res. 2013, 41, 6729–6738. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, Y.; Tian, Q.; Jia, Q.; Gao, Y.; Zhang, Q.; Zhou, C.; Xie, W. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015, 43, 10534–10545. [Google Scholar] [CrossRef] [PubMed]
- Himeno, H.; Yoshida, S.; Soma, A.; Nishikawa, K. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeu in vitro. J. Mol. Biol. 1997, 268, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Bröcker, M.J.; Sekine, S.; Hammond, G.; Suetsugu, S.; Söll, D.; Yokoyama, S. Decameric SelA•tRNASec ring structure reveals mechanism of bacterial selenocysteine formation. Science 2013, 340, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Selmer, M.; Su, X.D. Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J. 2002, 21, 4145–4153. [Google Scholar] [CrossRef] [PubMed]
- Kromayer, M.; Wilting, R.; Tormay, P.; Böck, A. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol. 1996, 262, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Neumann, P.; Bock, L.V.; Maracci, C.; Wang, Z.; Paleskava, A.; Konevega, A.L.; Schroder, G.F.; Grubmuller, H.; Ficner, R.; et al. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature 2016, 540, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.M.; Chapman, S.J.; Uhlenbeck, O.C. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J. Mol. Biol. 2009, 386, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Rudinger, J.; Hillenbrandt, R.; Sprinzl, M.; Giege, R. Antideterminants present in minihelixSec hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996, 15, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Vargas-Rodriguez, O.; Englert, M.; Tripp, H.J.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2017, 45, 2776–2785. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Englert, M.; Tripp, H.J.; Miller, C.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Facile Recoding of Selenocysteine in Nature. Angew. Chem. Int. Ed. Engl. 2016, 55, 5337–5341. [Google Scholar] [CrossRef] [PubMed]
- Tharp, J.M.; Ehnbom, A.; Liu, W.R. tRNAPyl: Structure, function, and applications. RNA Biol. 2018, 15, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Polycarpo, C.; Ambrogelly, A.; Berube, A.; Winbush, S.M.; McCloskey, J.A.; Crain, P.F.; Wood, J.L.; Söll, D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA 2004, 101, 12450–12454. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Ohtsuki, T.; Shimizu, Y.; Ueda, T.; Sisido, M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J. Am. Chem. Soc. 2007, 129, 14458–14462. [Google Scholar] [CrossRef] [PubMed]
- Uhlenbeck, O.C.; Schrader, J.M. Evolutionary tuning impacts the design of bacterial tRNAs for the incorporation of unnatural amino acids by ribosomes. Curr. Opin. Chem. Biol. 2018, 46, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xiong, H.; Reynolds, N.M.; Söll, D. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 2015, 43, e156. [Google Scholar] [CrossRef] [PubMed]
- LaRiviere, F.J.; Wolfson, A.D.; Uhlenbeck, O.C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 2001, 294, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Shepotinovskaya, I.; Uhlenbeck, O.C. tRNA residues evolved to promote translational accuracy. RNA 2013, 19, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Sun, S.B.; Furman, J.L.; Xiao, H.; Schultz, P.G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 2013, 52, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Schmied, W.H.; Elsässer, S.J.; Uttamapinant, C.; Chin, J.W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 2014, 136, 15577–15583. [Google Scholar] [CrossRef] [PubMed]
- Serfling, R.; Lorenz, C.; Etzel, M.; Schicht, G.; Bottke, T.; Mörl, M.; Coin, I. Designer tRNAs for efficient incorporation of non-canonical amino acids by the pyrrolysine system in mammalian cells. Nucleic Acids Res. 2018, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ambrogelly, A.; Gundllapalli, S.; Herring, S.; Polycarpo, C.; Frauer, C.; Söll, D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. USA 2007, 104, 3141–3146. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, K.; O’Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Fladischer, P.; Weingartner, A.; Blamauer, J.; Darnhofer, B.; Birner-Gruenberger, R.; Kardashliev, T.; Ruff, A.J.; Schwaneberg, U.; Wiltschi, B. A semi-rationally engineered bacterial pyrrolysyl-tRNA synthetase genetically encodes phenyl azide chemistry. Biotechnol. J. 2018, e1800125. [Google Scholar] [CrossRef] [PubMed]
- Meineke, B.; Heimgärtner, J.; Lafranchi, L.; Elsässer, S.J. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. ACS Chem. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.C.W.; Chin, J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018, 10, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; Gaci, N.; Peyret, P.; O’Toole, P.W.; Gribaldo, S.; Brugere, J.F. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: Implications for the evolution of a genetic code expansion cassette. Archaea 2014, 2014, 374146. [Google Scholar] [CrossRef] [PubMed]
- Beranek, V.; Willis, J.C.W.; Chin, J.W. An evolved Methanomethylophilus alvus pyrrolysyl-tRNA synthetase/tRNA pair is highly active and orthogonal in mammalian cells. Biochemistry 2018. [Google Scholar] [CrossRef] [PubMed]
- Venkat, S.; Sturges, J.; Stahman, A.; Gregory, C.; Gan, Q.; Fan, C. Genetically incorporating two distinct post-translational modifications into one protein simultaneously. ACS Synth. Biol. 2018, 7, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Prat, L.; Aerni, H.R.; Ling, J.; Merryman, C.; Glass, J.I.; Rinehart, J.; Söll, D. Transfer RNA misidentification scrambles sense codon recoding. Chembiochem 2013, 14, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffman, K.S.; Crnković, A.; Söll, D. Versatility of Synthetic tRNAs in Genetic Code Expansion. Genes 2018, 9, 537. https://doi.org/10.3390/genes9110537
Hoffman KS, Crnković A, Söll D. Versatility of Synthetic tRNAs in Genetic Code Expansion. Genes. 2018; 9(11):537. https://doi.org/10.3390/genes9110537
Chicago/Turabian StyleHoffman, Kyle S., Ana Crnković, and Dieter Söll. 2018. "Versatility of Synthetic tRNAs in Genetic Code Expansion" Genes 9, no. 11: 537. https://doi.org/10.3390/genes9110537
APA StyleHoffman, K. S., Crnković, A., & Söll, D. (2018). Versatility of Synthetic tRNAs in Genetic Code Expansion. Genes, 9(11), 537. https://doi.org/10.3390/genes9110537