Serum and Lipoprotein Particle miRNA Profile in Uremia Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. microRNA in Serum and Lipoprotein Particles
2.3. TaqMan™ Arrays
2.4. Synthetic miRNA
2.5. Lipoprotein Particle Isolation
2.6. Reconstitution of HDL Particles
2.7. Labeling of LDL Particles
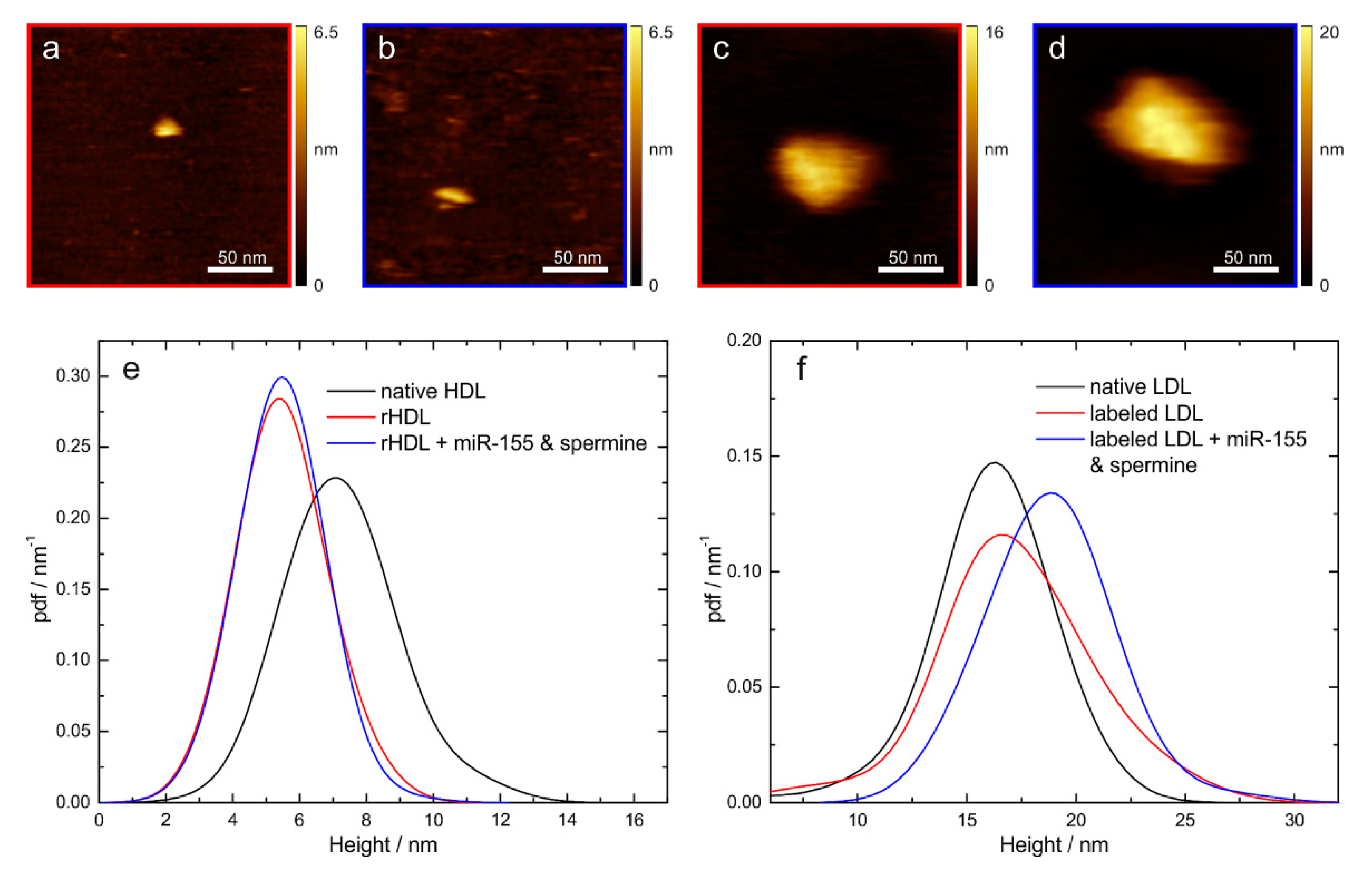
2.8. Quality Control of Reconstituted/Labeled Lipoprotein Particles
2.9. Cells
2.10. miRNA-Extraction
2.11. Reverse Transcription
2.12. qPCR
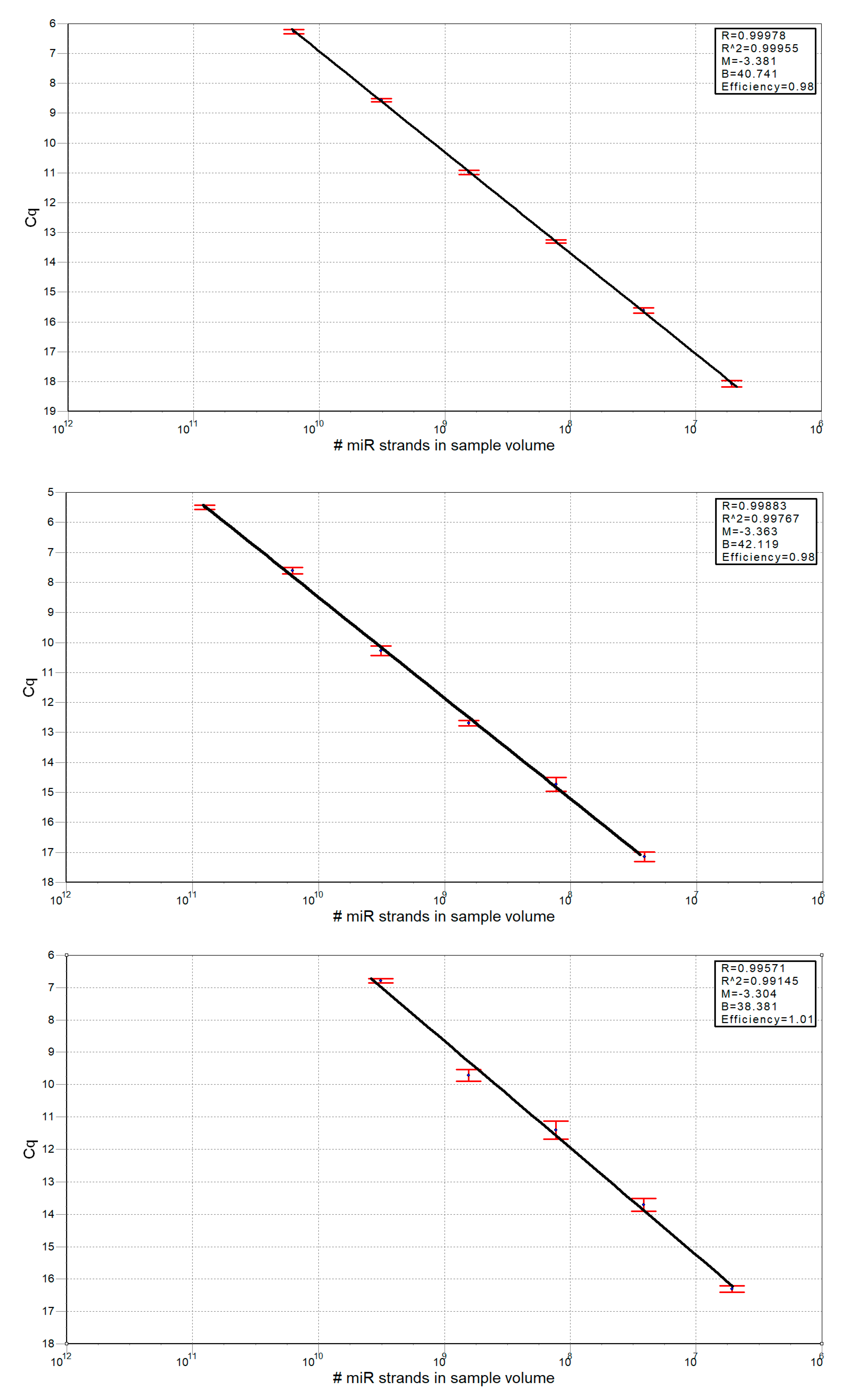
2.13. Calculation of miRNA Strands Number
3. Results
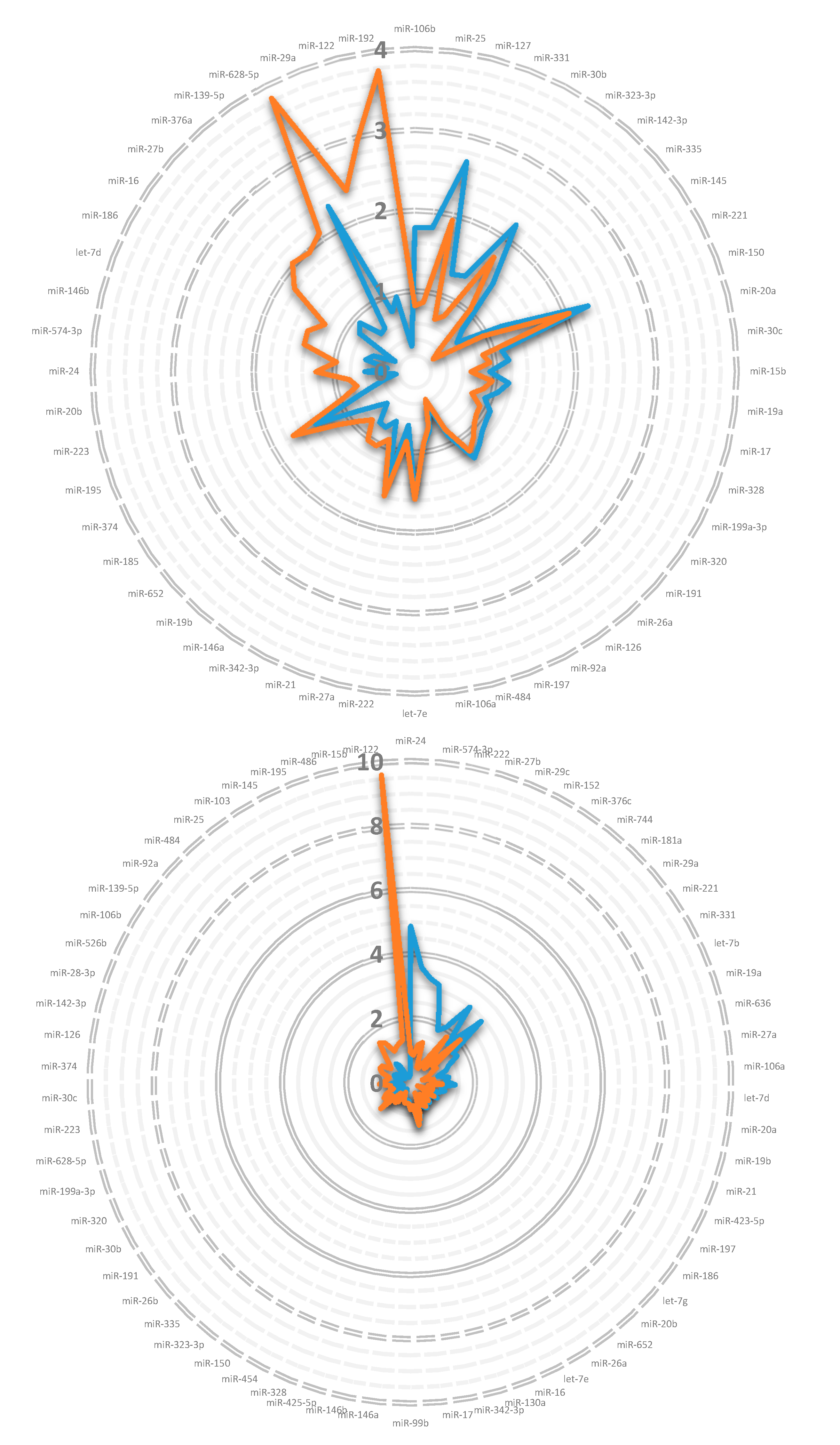
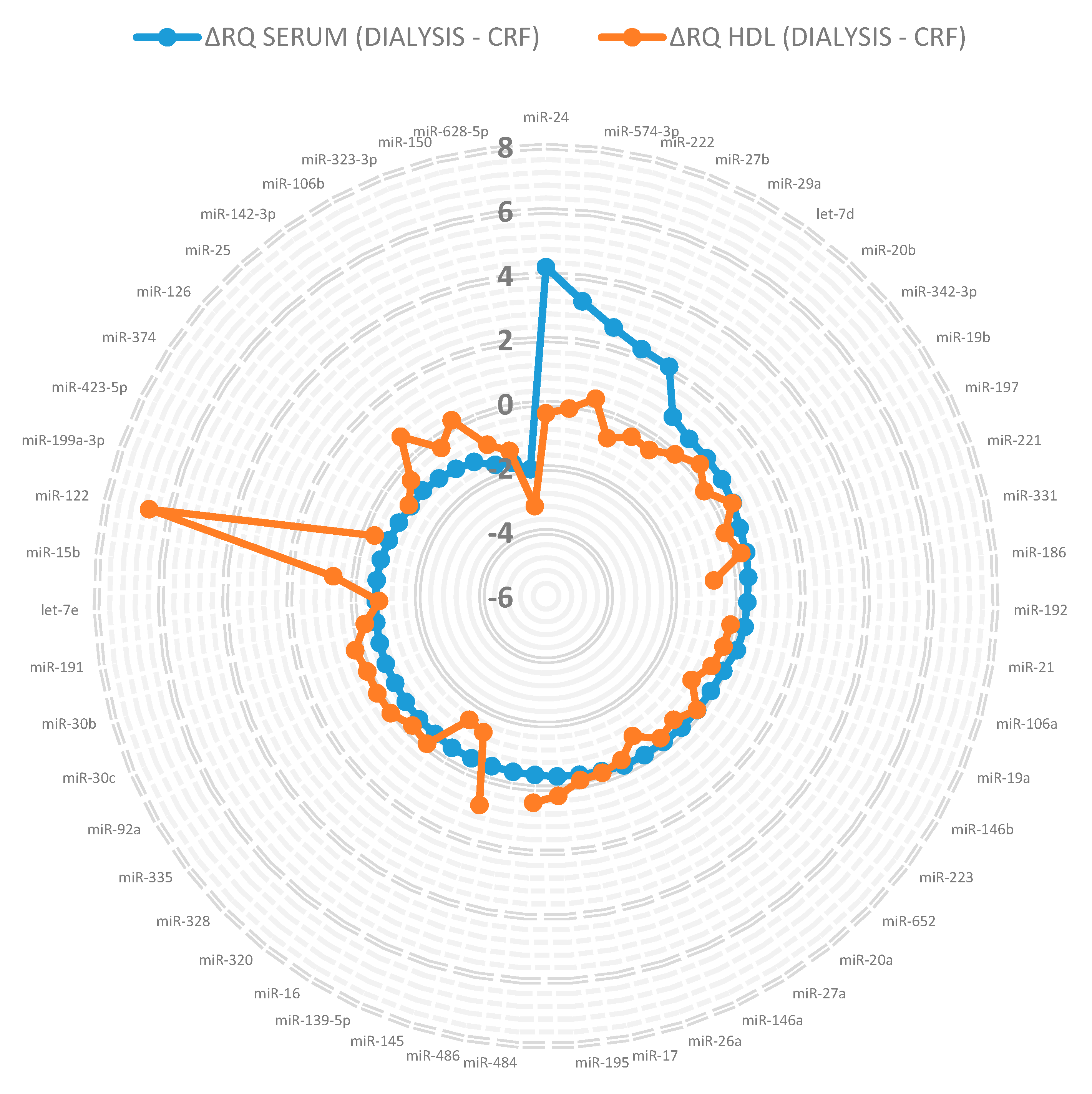
3.1. miRNA in Lipoprotein Particle Fractions
3.2. miRNA Associated to Lipoprotein Particles
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Chamber Conditions | < # miRNA > | SD < # miRNA > |
|---|---|---|
| Blocking with 5% BSA for 1 h + medium for 16 h | 2600 | 5 |
| Blocking with 500 µg/mL native HDL for 1 h + medium for 16 h | 2200 | 90 |
| Blocking with 5% BSA for 1 h + 50 µg/mL rHDL + miR-155 & spermine for 16 h | 14500 | 2800 |
| Blocking with 500 µg/mL native HDL for 1 h + 50 µg/mL rHDL + miR-155 & spermine for 16 h | 11000 | 2800 |
| 50 µg/mL rHDL + miR-155 & spermine for 16 h | 11000 | 2100 |
| 5 µg/mL rHDL + miR-155 & spermine for 16 h | 2800 | 140 |
| Assay | RQSERUM | RQHDL | Assay | RQSERUM | RQHDL | Assay | RQSERUM | RQHDL |
|---|---|---|---|---|---|---|---|---|
| miR-106b | 1.78 | 0.82 | miR-19b | 0.52 | 0.80 | miR-425-5p | N.D. | 0.83 |
| miR-25 | 1.80 | 0.87 | miR-652 | 0.64 | 0.93 | miR-148b | N.D. | 0.85 |
| miR-127 | 2.68 | 1.94 | miR-185 | 0.86 | 1.16 | miR-485-3p | N.D. | 0.87 |
| miR-331 | 1.29 | 0.70 | miR-374 | 1.39 | 1.71 | miR-99b | N.D. | 0.88 |
| miR-30b | 1.34 | 0.78 | miR-195 | 0.62 | 0.97 | miR-28-3p | N.D. | 0.96 |
| miR-323-3p | 2.21 | 1.73 | miR-223 | 0.38 | 0.74 | miR-376c | N.D. | 0.97 |
| miR-142-3p | 1.46 | 1.04 | miR-20b | 0.23 | 0.82 | let-7b | N.D. | 1.12 |
| miR-335 | 0.79 | 0.39 | miR-24 | 0.62 | 1.23 | let-7g | N.D. | 1.34 |
| miR-145 | 0.63 | 0.27 | miR-574-3p | 0.35 | 0.98 | miR-29c | N.D. | 1.39 |
| miR-221 | 1.22 | 0.95 | miR-146b | 0.63 | 1.41 | miR-26b | N.D. | 1.45 |
| miR-150 | 2.30 | 2.05 | let-7d | 0.55 | 1.42 | miR-155 | N.D. | 1.66 |
| miR-20a | 1.11 | 0.87 | miR-186 | 0.28 | 1.26 | miR-494 | N.D. | 1.85 |
| miR-30c | 1.17 | 0.94 | miR-16 | 0.83 | 1.82 | miR-152 | N.D. | 1.97 |
| miR-15b | 0.93 | 0.70 | miR-27b | 0.91 | 2.03 | miR-454 | N.D. | 2.05 |
| miR-19a | 1.17 | 0.97 | miR-376a | 0.63 | 1.97 | miR-133a | N.D. | 2.12 |
| miR-17 | 1.07 | 0.88 | miR-139-5p | 0.65 | 2.09 | miR-539 | N.D. | 2.37 |
| miR-328 | 0.93 | 0.75 | miR-628-5p | 2.32 | 3.83 | miR-148a | N.D. | 2.85 |
| miR-199a-3p | 1.06 | 0.91 | miR-29a | 0.80 | 2.40 | miR-518f | N.D. | 2.99 |
| miR-320 | 1.07 | 0.93 | miR-122 | 0.96 | 2.97 | miR-627 | N.D. | 3.02 |
| miR-191 | 1.12 | 0.98 | miR-192 | 0.32 | 3.76 | miR-636 | N.D. | 3.73 |
| miR-26a | 1.20 | 1.09 | miR-142-5p | N.D. | 0.35 | miR-301b | N.D. | 3.83 |
| miR-126 | 1.30 | 1.20 | miR-526b | N.D. | 0.38 | miR-107 | N.D. | 5.30 |
| miR-92a | 0.90 | 0.81 | miR-103 | N.D. | 0.45 | miR-410 | N.D. | 5.77 |
| miR-197 | 0.46 | 0.37 | miR-744 | N.D. | 0.45 | miR-424 | N.D. | 6.91 |
| miR-484 | 0.75 | 0.71 | miR-340 | N.D. | 0.48 | miR-487b | N.D. | 6.92 |
| miR-106a | 0.92 | 0.90 | let-7a | N.D. | 0.50 | let-7c | N.D. | 13.26 |
| let-7e | 1.53 | 1.58 | miR-130b | N.D. | 0.56 | miR-486 | 0.61 | N.D. |
| miR-222 | 0.67 | 0.87 | miR-130a | N.D. | 0.58 | miR-532 | 1.11 | N.D. |
| miR-27a | 1.38 | 1.59 | miR-301 | N.D. | 0.65 | miR-375 | 1.20 | N.D. |
| miR-21 | 0.66 | 0.90 | miR-128a | N.D. | 0.72 | miR-423-5p | 1.24 | N.D. |
| miR-342-3p | 0.78 | 1.04 | miR-28 | N.D. | 0.75 | miR-212 | 1.63 | N.D. |
| miR-146a | 0.77 | 1.03 | miR-345 | N.D. | 0.82 | |||
| miR-24 | 4.86 | 0.92 | miR-99b | 0.82 | 0.84 | miR-483-5p | 0.43 | N.D. |
| miR-574-3p | 3.59 | 0.86 | miR-146a | 0.58 | 0.65 | miR-192 | 0.59 | N.D. |
| miR-222 | 3.29 | 1.20 | miR-146b | 0.55 | 0.65 | miR-99a | 2.80 | N.D. |
| miR-27b | 3.16 | 1.30 | miR-425-5p | 0.56 | 0.66 | miR-660 | 3.94 | N.D. |
| miR-29c | 2.36 | 0.52 | miR-328 | 0.44 | 0.54 | miR-193a-5p | 6.52 | N.D. |
| miR-152 | 1.84 | 0.50 | miR-454 | 0.23 | 0.34 | miR-125b | 16.31 | N.D. |
| miR-376c | 2.05 | 0.73 | miR-150 | 0.57 | 0.71 | miR-193b | 17.65 | N.D. |
| miR-744 | 2.98 | 1.81 | miR-323-3p | 0.59 | 0.78 | miR-200c | 90.15 | N.D. |
| miR-181a | 1.67 | 0.61 | miR-335 | 0.26 | 0.45 | miR-203 | 1626.14 | N.D. |
| miR-29a | 2.90 | 2.03 | miR-26b | 1.03 | 1.24 | miR-532 | N.D. | 0.39 |
| miR-221 | 1.61 | 0.86 | miR-191 | 0.46 | 0.69 | miR-370 | N.D. | 0.47 |
| miR-331 | 1.67 | 0.92 | miR-30b | 0.72 | 0.96 | miR-339-3p | N.D. | 0.52 |
| let-7b | 1.30 | 0.68 | miR-320 | 0.58 | 0.83 | miR-301 | N.D. | 0.54 |
| miR-19a | 1.17 | 0.56 | miR-199a-3p | 0.24 | 0.57 | miR-125a-5p | N.D. | 0.54 |
| miR-636 | 0.87 | 0.37 | miR-628-5p | 0.29 | 0.65 | miR-148b | N.D. | 0.55 |
| miR-27a | 1.20 | 0.72 | miR-223 | 0.27 | 0.63 | miR-590-5p | N.D. | 0.58 |
| miR-106a | 1.09 | 0.64 | miR-30c | 0.59 | 0.98 | miR-142-5p | N.D. | 0.62 |
| let-7d | 1.38 | 0.99 | miR-374 | 0.45 | 0.83 | miR-597 | N.D. | 0.64 |
| miR-20a | 0.94 | 0.55 | miR-126 | 0.34 | 0.73 | miR-133a | N.D. | 0.65 |
| miR-19b | 1.09 | 0.71 | miR-142-3p | 0.30 | 0.69 | miR-539 | N.D. | 0.70 |
| miR-21 | 0.91 | 0.71 | miR-28-3p | 0.52 | 0.93 | let-7a | N.D. | 0.73 |
| miR-423-5p | 0.36 | 0.23 | miR-526b | 0.57 | 1.05 | miR-127 | N.D. | 0.79 |
| miR-197 | 0.95 | 0.85 | miR-106b | 0.52 | 1.03 | miR-18a | N.D. | 0.87 |
| miR-186 | 0.60 | 0.51 | miR-139-5p | 0.22 | 0.76 | miR-411 | N.D. | 0.92 |
| let-7g | 0.68 | 0.60 | miR-92a | 0.32 | 0.88 | miR-324-3p | N.D. | 1.10 |
| miR-20b | 0.85 | 0.78 | miR-484 | 0.34 | 1.15 | miR-185 | N.D. | 1.28 |
| miR-652 | 0.53 | 0.46 | miR-25 | 0.75 | 1.58 | miR-148a | N.D. | 1.31 |
| miR-26a | 0.93 | 0.87 | miR-103 | 0.63 | 1.46 | miR-130b | N.D. | 1.35 |
| let-7e | 0.83 | 0.78 | miR-145 | 0.20 | 1.11 | miR-155 | N.D. | 1.65 |
| miR-16 | 0.39 | 0.35 | miR-195 | 0.25 | 1.20 | miR-132 | N.D. | 2.02 |
| miR-130a | 1.07 | 1.04 | miR-486 | 0.18 | 1.28 | miR-487b | N.D. | 2.11 |
| miR-342-3p | 1.37 | 1.35 | miR-15b | 0.23 | 1.35 | miR-28 | N.D. | 2.22 |
| miR-17 | 0.73 | 0.71 | miR-122 | 0.23 | 9.61 |
References
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mrnas and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Cuesta Torres, L.F.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. Hdl-transferred microRNA-223 regulates icam-1 expression in endothelial cells. Nat. Commun. 2014, 5, 3292. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by micrornas. Ann. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: Cell-cell communication function? Front. Genet. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Vaisar, T.; Pennathur, S.; Green, P.S.; Gharib, S.A.; Hoofnagle, A.N.; Cheung, M.C.; Byun, J.; Vuletic, S.; Kassim, S.; Singh, P.; et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 2007, 117, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T.; Kopecky, C.; Kubicek, M.; Haidinger, M.; Doller, D.; Katholnig, K.; Suarna, C.; Eller, P.; Tolle, M.; Gerner, C.; et al. Serum amyloid a in uremic HDL promotes inflammation. J. Am. Soc. Nephrol. 2012, 23, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Wolf, P.; Curcic, S.; Birner-Gruenberger, R.; Weger, W.; Inzinger, M.; El-Gamal, D.; Wadsack, C.; Heinemann, A.; Marsche, G. Psoriasis alters HDL composition and cholesterol efflux capacity. J. Lipid Res. 2012, 53, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Birner-Gruenberger, R.; Stojakovic, T.; El-Gamal, D.; Binder, V.; Wadsack, C.; Heinemann, A.; Marsche, G. Uremia alters HDL composition and function. J. Am. Soc. Nephrol. 2011, 22, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.A.; Andraski, A.B.; Pieper, B.; Goh, W.; Mendivil, C.O.; Sacks, F.M.; Aikawa, M. Multiple apolipoprotein kinetics measured in human HDL by high-resolution/accurate mass parallel reaction monitoring. J. Lipid Res. 2016, 57, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.A.; Aikawa, M. Unbiased and targeted mass spectrometry for the HDL proteome. Curr. Opin. Lipidol. 2017, 28, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.Y. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune Netw. 2011, 11, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, M.S.; Weakley, S.M.; Zhang, L.; Kougias, P.; Lin, P.H.; Yao, Q.; Chen, C. miRNAs: Roles and clinical applications in vascular disease. Expert Rev. Mol. Diagn. 2011, 11, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Zhang, C. MicroRNAs in vascular disease. J. Cardiovasc. Pharm. 2011, 57, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Remaley, A.T. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr. Opin. Endocrinol. 2010, 17, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Akkina, S.; Becker, B.N. MicroRNAs in kidney function and disease. Transl. Res. 2011, 157, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, K.; Mi, Q.S.; Dong, Z. MicroRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am. J. Physiol. Renal Physiol. 2011, 300, F602–F610. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Yong, T.Y.; Michael, M.Z.; Gleadle, J.M. Review: The role of microRNAs in kidney disease. Nephrology 2010, 15, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Neal, C.S.; Michael, M.Z.; Pimlott, L.K.; Yong, T.Y.; Li, J.Y.; Gleadle, J.M. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol. Dial. Transplant. 2011, 26, 3794–3802. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci. Transl. Med. 2014, 6, 239ps3. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.M.; Wultsch, A.; Hollaus, M.; Ammann, M.; Pemberger, E.; Liebscher, F.; Lambers, B.; Fruhwurth, S.; Stojakovic, T.; Scharnagl, H.; et al. Effect of chronic kidney disease on macrophage cholesterol efflux. Life Sci. 2015, 136, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Schumaker, V.N.; Puppione, D.L. Sequential flotation ultracentrifugation. Method Enzymol. 1986, 128, 155–170. [Google Scholar]
- Jonas, A. Reconstitution of high-density lipoproteins. Method Enzymol. 1986, 128, 553–582. [Google Scholar]
- Krieger, M. Reconstitution of the hydrophobic core of low-density lipoprotein. Method Enzymol. 1986, 128, 608–613. [Google Scholar]
- Stangl, H.; Cao, G.; Wyne, K.L.; Hobbs, H.H. Scavenger receptor, class b, type I-dependent stimulation of cholesterol esterification by high density lipoproteins, low density lipoproteins, and nonlipoprotein cholesterol. J. Biol. Chem. 1998, 273, 31002–31008. [Google Scholar] [CrossRef] [PubMed]
- Stangl, H.; Hyatt, M.; Hobbs, H.H. Transport of lipids from high- and low-density lipoproteins via scavenger receptor-bi. J. Biol. Chem. 1999, 274, 32692–32698. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.B.; Jiang, Z.P. Role of miR-21 and its signaling pathways in renal diseases. J. Recept. Signal Transduct. 2014, 34, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, X.; Chen, C.; Zhou, J.; Han, M. Bone mesenchymal stem cells ameliorate ischemia/reperfusion-induced damage in renal epithelial cells via microRNA-223. Stem Cell Res. Ther. 2017, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Roxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. 2013, 33, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Kaucsar, T.; Schauerte, C.; Schmitt, R.; Rong, S.; Hubner, A.; Scherf, K.; Fiedler, J.; Martino, F.; Kumarswamy, R.; et al. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J. Am. Soc. Nephrol. 2014, 25, 2717–2729. [Google Scholar] [CrossRef] [PubMed]
- Desgagne, V.; Guerin, R.; Guay, S.P.; Corbin, F.; Couture, P.; Lamarche, B.; Bouchard, L. Changes in high-density lipoprotein-carried miRNA contribution to the plasmatic pool after consumption of dietary trans fat in healthy men. Epigenomics 2017, 9, 669–688. [Google Scholar] [CrossRef] [PubMed]
| Sample | miRNA | < ΔΔcq > | SD < ΔΔcq > |
|---|---|---|---|
| LPDS | 223 | 0.0 | 0.1 |
| 21 | 0.0 | 0.2 | |
| VLDL | 223 | 5.7 | 0.2 |
| 21 | 4.6 | 0.2 | |
| LDL | 223 | 2.8 | 0.2 |
| 21 | 1.8 | 0.2 | |
| HDL | 223 | −3.5 | 0.1 |
| 21 | −2.5 | 0.1 |
| Sample | Donor | miRNA | < # miRNA/particle > | SD < # miRNA/particle > |
|---|---|---|---|---|
| Native HDL | 1 | 223 | 6.10−9 | 7.10−10 |
| 155 | 3.10−9 | 9.10−10 | ||
| 2 | 223 | 7.10−9 | 4.10−9 | |
| 155 | 4.10−8 | 2.10−8 | ||
| rHDL | 1 | 223 | 4.10−9 | 8.10−10 |
| 155 | 5.10−9 | 3.10−9 | ||
| 2 | 223 | 7.10−9 | 4.10−9 | |
| 155 | 1.10−8 | 1.10−8 | ||
| rHDL + miR-155 | 1 | 155 | 1.10−4 | 1.10−4 |
| 2 | 155 | 8.10−5 | 5.10−5 | |
| rHDL + miR-155 & spermine | 1 | 155 | 1.10−4 | 5.10−5 |
| 2 | 155 | 7.10−5 | 2.10−5 |
| Sample | Donor | miRNA | < # miRNA/particle > | SD < # miRNA/particle > |
|---|---|---|---|---|
| Native LDL | 1 | 223 | 3.10−8 | 2.10−8 |
| 155 | 9.10−8 | 5.10−8 | ||
| 2 | 223 | 5.10−9 | 3.10−9 | |
| 155 | 9.10−8 | 2.10−8 | ||
| Labeled LDL | 1 | 223 | 5.10−8 | 3.10−8 |
| 155 | 9.10−8 | 5.10−8 | ||
| 2 | 223 | 4.10−9 | 3.10−9 | |
| 155 | 9.10−8 | 2.10−8 | ||
| Labeled LDL + miR-155 | 1 | 155 | 1.10−4 | 6.10−5 |
| 2 | 155 | 2.10−3 | 3.10−4 | |
| Labeled LDL + miR-155 & spermine | 1 | 155 | 5.10−4 | 3.10−4 |
| 2 | 155 | 5.10−3 | 6.10−4 |
| Cell Line | HDL Conditions and Concentration | < # miRNA/cell > | SD < # miRNA/cell > |
|---|---|---|---|
| ldlA7 | no HDL addition | 240 | 35 |
| native, 50 µg/mL | 250 | 25 | |
| miR-155 and spermine, 50 µg/mL | 980 | 55 | |
| ldlA7-SR-B1 | no HDL addition | 260 | 20 |
| native, 50 µg/mL | 330 | 55 | |
| miR-155 and spermine, 50 µg/mL | 2500 | 190 |
| Cell Line | LDL Conditions and Concentration | < # miR/cell > | SD < # miR/cell > |
|---|---|---|---|
| ldlA7 | no LDL addition | 190 | 10 |
| native, 5 µg/mL | 240 | 20 | |
| miR-155 and spermine, 5 µg/mL | 210 | 40 | |
| CHOK1 | no LDL addition | 120 | 10 |
| native, 5 µg/mL | 130 | 20 | |
| miR-155 and spermine, 5 µg/mL | 160 | 70 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Axmann, M.; Meier, S.M.; Karner, A.; Strobl, W.; Stangl, H.; Plochberger, B. Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes 2018, 9, 533. https://doi.org/10.3390/genes9110533
Axmann M, Meier SM, Karner A, Strobl W, Stangl H, Plochberger B. Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes. 2018; 9(11):533. https://doi.org/10.3390/genes9110533
Chicago/Turabian StyleAxmann, Markus, Sabine M. Meier, Andreas Karner, Witta Strobl, Herbert Stangl, and Birgit Plochberger. 2018. "Serum and Lipoprotein Particle miRNA Profile in Uremia Patients" Genes 9, no. 11: 533. https://doi.org/10.3390/genes9110533
APA StyleAxmann, M., Meier, S. M., Karner, A., Strobl, W., Stangl, H., & Plochberger, B. (2018). Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes, 9(11), 533. https://doi.org/10.3390/genes9110533