Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly
Abstract
1. Introduction
2. Effects of Donor’s Age on Cell Reprogramming
3. Epigenetic Remodelling during Reprogramming and Differentiation
4. Epigenetic Age Changes upon Cellular Reprogramming and Differentiation
5. Impact of Reprogramming-Associated Alterations in the Study of Age-Related Diseases
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kirkwood, T.B.; Kowald, A. Network theory of aging. Exp. Gerontol. 1997, 32, 395–399. [Google Scholar] [CrossRef]
- Franceschi, C.; Valensin, S.; Bonafè, M.; Paolisso, G.; Yashin, A.I.; Monti, D.; De Benedictis, G. The network and the remodeling theories of aging: Historical background and new perspectives. Exp. Gerontol. 2000, 35, 879–896. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14234–14239. [Google Scholar] [CrossRef] [PubMed]
- Raab, S.; Klingenstein, M.; Liebau, S.; Linta, L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells (Dayton Ohio) 2009, 27, 2667–2674. [Google Scholar] [CrossRef] [PubMed]
- Somers, A.; Jean, J.-C.; Sommer, C.A.; Omari, A.; Ford, C.C.; Mills, J.A.; Ying, L.; Sommer, A.G.; Jean, J.M.; Smith, B.W.; et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells (Dayton Ohio) 2010, 28, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Martín-Maestro, P.; Gargini, R.; Sproul, A.A.; García, E.; Antón, L.C.; Noggle, S.; Arancio, O.; Avila, J.; García-Escudero, V. Mitophagy Failure in Fibroblasts and iPSC-Derived Neurons of Alzheimer’s Disease-Associated Presenilin 1 Mutation. Front. Mol. Neurosci. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Swistowski, A.; Peng, J.; Liu, Q.; Mali, P.; Rao, M.S.; Cheng, L.; Zeng, X. Efficient Generation of Functional Dopaminergic Neurons from Human Induced Pluripotent Stem Cells Under Defined Conditions. Stem Cells 2010, 28, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Santiago, R.; Carballo-Carbajal, I.; Castellano, G.; Torrent, R.; Richaud, Y.; Sanchez-Danes, A.; Vilarrasa-Blasi, R.; Sanchez-Pla, A.; Mosquera, J.L.; Soriano, J.; et al. Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. EMBO Mol. Med. 2015, 7, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Kawser Hossain, M.; Abdal Dayem, A.; Han, J.; Kumar Saha, S.; Yang, G.-M.; Choi, H.; Cho, S.-G. Recent Advances in Disease Modeling and Drug Discovery for Diabetes Mellitus Using Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2016, 17, 256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lian, Q.; Zhu, G.; Zhou, F.; Sui, L.; Tan, C.; Mutalif, R.A.; Navasankari, R.; Zhang, Y.; Tse, H.-F.; et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 2011, 8, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, A.; Yokote, K.; Tahara, H. Werner Syndrome-specific induced pluripotent stem cells: Recovery of telomere function by reprogramming. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Lo Sardo, V.; Ferguson, W.; Erikson, G.A.; Topol, E.J.; Baldwin, K.K.; Torkamani, A. Influence of donor age on induced pluripotent stem cells. Nat. Biotechnol. 2016, 35, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.R.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Miyagoe-Suzuki, Y.; Yada, E.; Ito, N.; Nishiyama, T.; Nakamura, M.; Ono, Y.; Motohashi, N.; Segawa, M.; Masuda, S.; et al. Reprogramming efficiency and quality of induced Pluripotent Stem Cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr. 2011, 3, RRN1274. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Ito, S.; Nishio, N.; Xiao, H.; Zhang, R.; Suzuki, H.; Okawa, Y.; Murohara, T.; Isobe, K.-i. Establishment of induced pluripotent stem cells from aged mice using bone marrow-derived myeloid cells. J. Mol. Cell Biol. 2011, 3, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Trokovic, R.; Weltner, J.; Noisa, P.; Raivio, T.; Otonkoski, T. Combined negative effect of donor age and time in culture on the reprogramming efficiency into induced pluripotent stem cells. Stem Cell Res. 2015, 15, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Banito, A.; Rashid, S.T.; Acosta, J.C.; Li, S.; Pereira, C.F.; Geti, I.; Pinho, S.; Silva, J.C.; Azuara, V.; Walsh, M.; et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009, 23, 2134–2139. [Google Scholar] [CrossRef] [PubMed]
- Boulting, G.L.; Kiskinis, E.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; Wainger, B.J.; Williams, D.J.; Kahler, D.J.; Yamaki, M.; Davidow, L.; et al. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Ganat, Y.M.; Kishinevsky, S.; Bowman, R.L.; Liu, B.; Tu, E.Y.; Mandal, P.K.; Vera, E.; Shim, J.; Kriks, S.; et al. Human iPSC-Based Modeling of Late-Onset Disease via Progerin-Induced Aging. Cell Stem Cell 2013, 13, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Lapasset, L.; Milhavet, O.; Prieur, A.; Besnard, E.; Babled, A.; Ait-Hamou, N.; Leschik, J.; Pellestor, F.; Ramirez, J.-M.; De Vos, J.; et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011, 25, 2248–2253. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Kosakai, A.; Ito, D.; Okada, Y.; Akamatsu, W.; Nihei, Y.; Nabetani, A.; Ishikawa, F.; Arai, Y.; Hirose, N.; et al. Establishment of Induced Pluripotent Stem Cells from Centenarians for Neurodegenerative Disease Research. PLoS ONE 2012, 7, e41572. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Wani, P.; Zhou, L.; Baer, T.; Phadnis, S.M.; Reijo Pera, R.A.; Chen, B. Reprogramming of fibroblasts from older women with pelvic floor disorders alters cellular behavior associated with donor age. Stem Cells Transl. Med. 2013, 2, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Frobel, J.; Hemeda, H.; Lenz, M.; Abagnale, G.; Joussen, S.; Denecke, B.; Šarić, T.; Zenke, M.; Wagner, W. Epigenetic Rejuvenation of Mesenchymal Stromal Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Rep. 2014, 3, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Ohmine, S.; Squillace, K.A.; Hartjes, K.A.; Deeds, M.C.; Armstrong, A.S.; Thatava, T.; Sakuma, T.; Terzic, A.; Kudva, Y.; Ikeda, Y. Reprogrammed keratinocytes from elderly type 2 diabetes patients suppress senescence genes to acquire induced pluripotency. Aging 2012, 4, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Pirazzini, C.; Delledonne, M.; Xumerle, L.; Descombes, P.; Marquis, J.; Mengozzi, G.; Monti, D.; Bellizzi, D.; Passarino, G.; et al. Centenarians as extreme phenotypes: An ecological perspective to get insight into the relationship between the genetics of longevity and age-associated diseases. Mech. Ageing Dev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007, 3, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Enroth, S.; Gyllensten, U. Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLoS ONE 2013, 8, e67378. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Ciccarone, F.; Calabrese, R.; Franceschi, C.; Bürkle, A.; Caiafa, P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015, 151, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Maleszewska, M.; Mawer, J.S.P.; Tessarz, P. Histone Modifications in Ageing and Lifespan Regulation. Curr. Mol. Biol. Rep. 2016, 2, 26–35. [Google Scholar] [CrossRef]
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Van Deursen, J.M. The role of senescent cells in ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Suhr, S.T.; Chang, E.A.; Rodriguez, R.M.; Wang, K.; Ross, P.J.; Beyhan, Z.; Murthy, S.; Cibelli, J.B. Telomere Dynamics in Human Cells Reprogrammed to Pluripotency. PLoS ONE 2009, 4, e8124. [Google Scholar] [CrossRef] [PubMed]
- Koche, R.P.; Smith, Z.D.; Adli, M.; Gu, H.; Ku, M.; Gnirke, A.; Bernstein, B.E.; Meissner, A. Reprogramming Factor Expression Initiates Widespread Targeted Chromatin Remodeling. Cell Stem Cell 2011, 8, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Kim, J.H.; Chung, M.K.; Hong, Y.J.; Jang, H.S.; Seo, B.J.; Jung, T.H.; Kim, J.S.; Chung, H.M.; Byun, S.J.; et al. Mitochondrial and metabolic remodeling during reprogramming and differentiation of the reprogrammed cells. Stem Cells Dev. 2015, 24, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- BI Ageing Clock Team; Stubbs, T.M.; Bonder, M.J.; Stark, A.-K.; Krueger, F.; von Meyenn, F.; Stegle, O.; Reik, W. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Kang, X.; Du, H.; Song, B.; Lu, Z.; Huang, Y.; Wang, D.; Sun, X.; Yu, Y.; Fan, Y. Defining Differentially Methylated Regions Specific for the Acquisition of Pluripotency and Maintenance in Human Pluripotent Stem Cells via Microarray. PLoS ONE 2014, 9, e108350. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Planello, A.C.; Ji, J.; Sharma, V.; Singhania, R.; Mbabaali, F.; Müller, F.; Alfaro, J.A.; Bock, C.; De Carvalho, D.D.; Batada, N.N. Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regen. 2014, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Toyoda, M.; Yamazaki-Inoue, M.; Fukawatase, Y.; Chikazawa, E.; Sakaguchi, H.; Akutsu, H.; Umezawa, A. DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time. PLoS Genet. 2011, 7, e1002085. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Koch, C.; Gupta, M.K.; Lin, Q.; Lenz, M.; Laufs, S.; Denecke, B.; Schmidt, M.; Linke, M.; Hennies, H.C.; et al. Induced pluripotent mesenchymal stromal cell clones retain donor-derived differences in DNA methylation profiles. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, K.; Ruan, W.; Bo, Z.; Liu, L.; Cao, Z.; Chai, L.; Cao, G. Higher methylation in genomic DNA indicates incomplete reprogramming in induced pluripotent stem cells. Cell. Reprogram. 2013, 15, 92–99. [Google Scholar] [CrossRef]
- Kim, K.; Zhao, R.; Doi, A.; Ng, K.; Unternaehrer, J.; Cahan, P.; Hongguang, H.; Loh, Y.-H.; Aryee, M.J.; Lensch, M.W.; et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Gao, S.; Hou, X.; Xu, Z.; Liu, Y.; Kang, L.; Tao, Y.; Liu, W.; Huang, B.; Kou, X.; et al. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014, 24, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Bar, S.; Schachter, M.; Eldar-Geva, T.; Benvenisty, N. Large-Scale Analysis of Loss of Imprinting in Human Pluripotent Stem Cells. Cell Rep. 2017, 19, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, J.D.; Jung, M.; Chen, C.; Lin, Z.; Ye, J.; Godatha, S.; Lizhar, E.; Wu, X.; Hsu, D.; Couture, L.A.; et al. Mapping Human Pluripotent-to-Cardiomyocyte Differentiation: Methylomes, Transcriptomes, and Exon DNA Methylation “Memories”. EBioMedicine 2016, 4, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Suelves, M.; Carrió, E.; Núñez-Álvarez, Y.; Peinado, M.A. DNA methylation dynamics in cellular commitment and differentiation. Brief. Funct. Genom. 2016. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, K.J.; Shamis, Y.; Hayman, R.B.; Margvelashvili, M.; Dong, S.; Carlson, M.W.; Garlick, J.A. Epigenetic and Phenotypic Profile of Fibroblasts Derived from Induced Pluripotent Stem Cells. PLoS ONE 2011, 6, e17128. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, M.; Chonabayashi, K.; Nomura, M.; Tanaka, A.; Nakamura, M.; Inagaki, A.; Nishikawa, M.; Takei, I.; Oishi, A.; Tanabe, K.; et al. Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell 2016. [Google Scholar] [CrossRef] [PubMed]
- Butcher, L.M.; Ito, M.; Brimpari, M.; Morris, T.J.; Soares, F.A.C.; Ährlund-Richter, L.; Carey, N.; Vallier, L.; Ferguson-Smith, A.C.; Beck, S. Non-CG DNA methylation is a biomarker for assessing endodermal differentiation capacity in pluripotent stem cells. Nat. Commun. 2016, 7, 10458. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Krassowska, A.; Gilbert, N.; Chevassut, T.; Forrester, L.; Ansell, J.; Ramsahoye, B. Severe Global DNA Hypomethylation Blocks Differentiation and Induces Histone Hyperacetylation in Embryonic Stem Cells. Mol. Cell. Biol. 2004, 24, 8862–8871. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Down, T.A.; Maslau, S.; Andrew, T.; Yang, T.P.; Beyan, H.; Whittaker, P.; McCann, O.T.; Finer, S.; Valdes, A.M.; et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010, 20, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Schwartz, J.; Wright, R.; Litonjua, A.; Tarantini, L.; Suh, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009, 130, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Sazzini, M.; Bacalini, M.G.; Pirazzini, C.; Marasco, E.; Fontanesi, E.; Franceschi, C.; Luiselli, D.; Garagnani, P. Epigenetic Variability across Human Populations: A Focus on DNA Methylation Profiles of the KRTCAP3, MAD1L1 and BRSK2 Genes. Genome Biol. Evol. 2016, 8, 2760–2773. [Google Scholar] [CrossRef] [PubMed]
- Pirazzini, C.; Giuliani, C.; Bacalini, M.G.; Boattini, A.; Capri, M.; Fontanesi, E.; Marasco, E.; Mantovani, V.; Pierini, M.; Pini, E.; et al. Space/population and time/age in DNA methylation variability in humans: A study on IGF2/H19 locus in different Italian populations and in mono- and di-zygotic twins of different age. Aging 2012, 4, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.; Horvath, S.; Raj, K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Bennett, D.A.; Horvath, S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging 2015, 7, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Ritz, B.R. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging 2015, 7, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Salvioli, S.; Gentilini, D.; Di Blasio, A.M.; Giuliani, C.; Tung, S.; Vinters, H.V.; et al. Accelerated epigenetic aging in Down syndrome. Aging Cell 2015, 14, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Durso, D.; Giulia Bacalini, M.; Sala, C.; Pirazzini, C.; Marasco, E.; Bonafé, M.; do Valle, Í.F.; Gentilini, D.; Castellani, G.; Caetano Faria, A.M.; et al. Acceleration of leukocytes’ epigenetic age as an early tumor and sex-specific marker of breast and colorectal cancer. Oncotarget 2017. [Google Scholar] [CrossRef]
- Fernandes Durso, D.; Giulia Bacalini, M.; do Valle, Í.F.; Pirazzini, C.; Bonafé, M.; Castellani, G.; Caetano Faria, A.M.; Franceschi, C.; Garagnani, P.; Nardini, C. Aberrant methylation patterns in colorectal cancer: A meta-analysis. Oncotarget 2017. [Google Scholar] [CrossRef]
- Horvath, S.; Pirazzini, C.; Bacalini, M.G.; Gentilini, D.; Di Blasio, A.M.; Delledonne, M.; Mari, D.; Arosio, B.; Monti, D.; Passarino, G.; et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging 2015, 7, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Weidner, C.; Lin, Q.; Koch, C.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.; Jöckel, K.-H.; Erbel, R.; Mühleisen, T.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef] [PubMed]
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D.; Di Blasio, A.M.; Gentilini, D.; Vitale, G.; Collino, S.; et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 2012, 11, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Bacalini, M.G.; Boattini, A.; Gentilini, D.; Giampieri, E.; Pirazzini, C.; Giuliani, C.; Fontanesi, E.; Remondini, D.; Capri, M.; Del Rio, A.; et al. A meta-analysis on age-associated changes in blood DNA methylation: Results from an original analysis pipeline for Infinium 450k data. Aging 2015, 7, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Roessler, R.; Smallwood, S.A.; Veenvliet, J.V.; Pechlivanoglou, P.; Peng, S.-P.; Chakrabarty, K.; Groot-Koerkamp, M.J.A.; Pasterkamp, R.J.; Wesseling, E.; Kelsey, G.; et al. Detailed Analysis of the Genetic and Epigenetic Signatures of iPSC-Derived Mesodiencephalic Dopaminergic Neurons. Stem Cell Rep. 2014, 2, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.-P.; Zhang, F.; Atkinson-Dell, R.; Rowe, C.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015, 62, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Byers, B.; Cord, B.; Shcheglovitov, A.; Byrne, J.; Gujar, P.; Kee, K.; Schüle, B.; Dolmetsch, R.E.; Langston, W.; et al. LRRK2 Mutant iPSC-Derived DA Neurons Demonstrate Increased Susceptibility to Oxidative Stress. Cell Stem Cell 2011, 8, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.Y.; Kishinevsky, S.; Mazzulli, J.R.; Graziotto, J.; Mrejeru, A.; Mosharov, E.V.; Puspita, L.; Valiulahi, P.; Sulzer, D.; Milner, T.A.; et al. Parkin and PINK1 Patient iPSC-Derived Midbrain Dopamine Neurons Exhibit Mitochondrial Dysfunction and α-Synuclein Accumulation. Stem Cell Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Noda, A.; Mishima, S.; Hirai, Y.; Hamasaki, K.; Landes, R.D.; Mitani, H.; Haga, K.; Kiyono, T.; Nakamura, N.; Kodama, Y. Progerin, the protein responsible for the Hutchinson-Gilford progeria syndrome, increases the unrepaired DNA damages following exposure to ionizing radiation. Genes Environ. 2015, 37. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P.; Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Vera, E.; Bosco, N.; Studer, L. Generating Late-Onset Human iPSC-Based Disease Models by Inducing Neuronal Age-Related Phenotypes through Telomerase Manipulation. Cell Rep. 2016, 17, 1184–1192. [Google Scholar] [CrossRef]
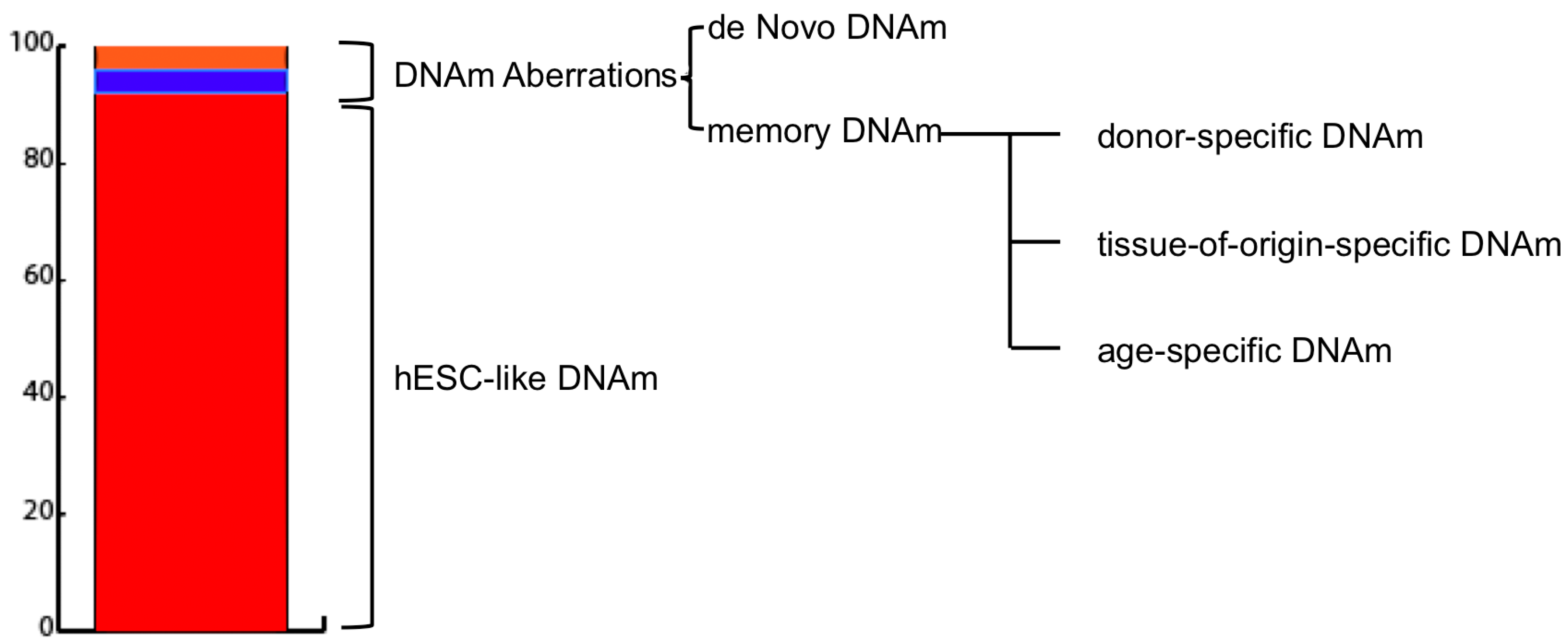
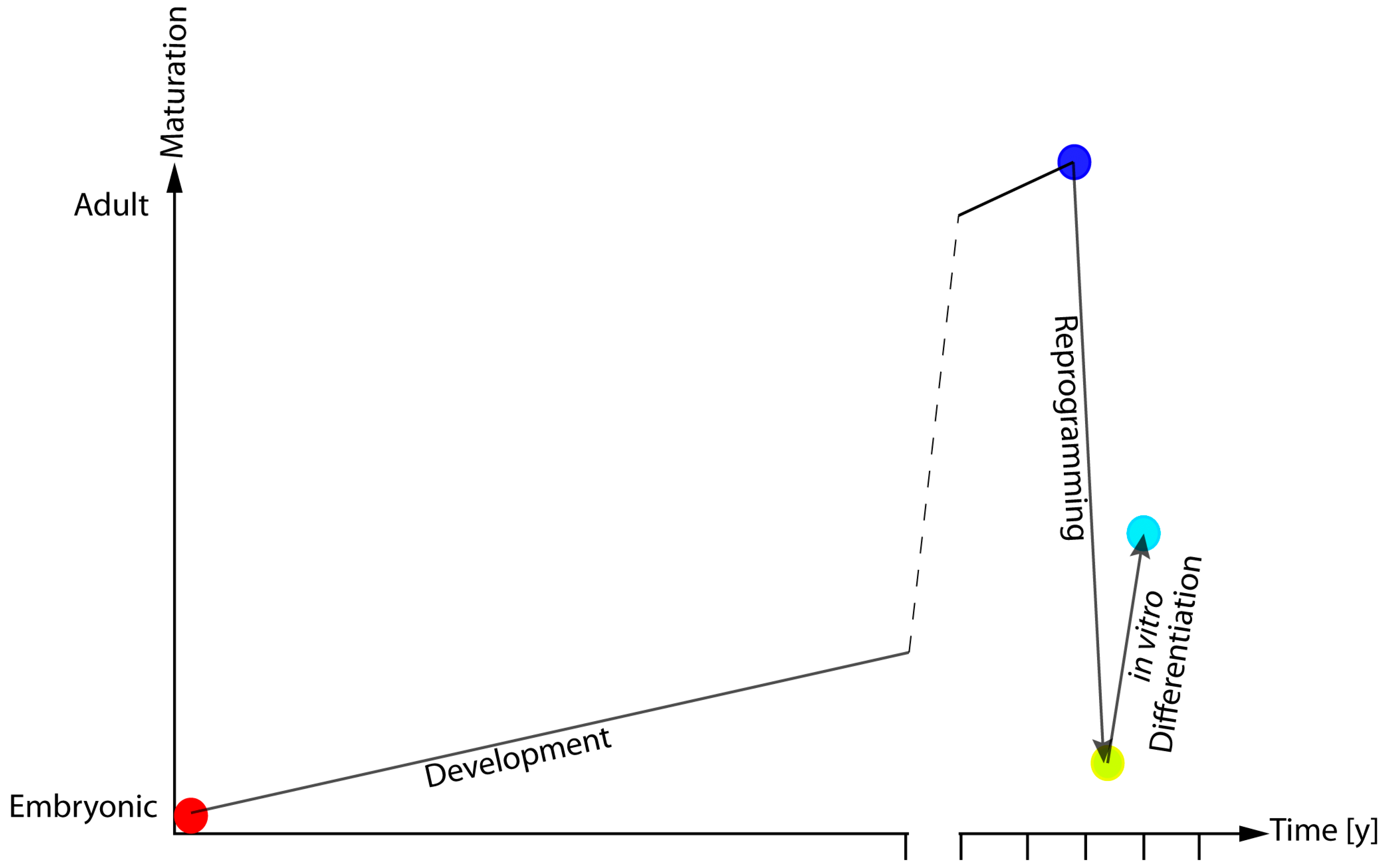
| Reference | Species | Age of Donor | Tissue | Vector | Factors | Efficiency |
|---|---|---|---|---|---|---|
| Kim et al. [15] | Mouse | Juvenile and 12 m | DF; BM | Retroviral | OSKM | 5 times higher in juvenile than 12 m old |
| Cheng et al. [17] | Mouse | 2 m and 23 m | BM | Retroviral | OSKM | 5 times higher and 2 times faster in 2 m than 23 m old |
| Wang et al. [16] | Mouse | 1.5 m; 6 m and 14 m | DF | Retroviral | OSKM | 6 times higher in 1.5 m and 6 m than 14 m old |
| Miller et al. [21] | Human | 11 y.o.; 31–55 y.o.; 71–96 y.o. | DF | Sendai | OSKM | Not assessed |
| Lapasset et al. [22] | Human | 92 y.o.; 94 y.o.; 96 y.o.; 101 y.o. | DF | Lentiviral | OSKMNL | Not assessed |
| Yagi et al. [23] | Human | 106 y.o. | DF | Retroviral | OSKM | Not assessed |
| Wen et al. [24] | Human | 47 y.o. and 78 y.o. | DF | Lentiviral | OSKM | 1.3 times more fully reprogrammed lines from 47 y.o. compared to 78 y.o. |
| Boulting et al. [20] | Human | 29–82 y.o. | DF | Retroviral | OSK | Not assessed |
| Frobel et al. [25] | Human | 56 y.o.; 63 y.o.; 74 y.o. | MSC | Retroviral | OSKM | Not assessed |
| Ohmine et al. [26] | Human | 56–78 y.o. | Keratinocytes | Lentiviral | OSKM | Not assessed |
| Trokovic et al. [18] | Human | 0-83 y.o. | DF | Retroviral | OSKM | Negative correlation between donor’s age and reprogramming efficiency (r = −0.89; p value = 0.0002) |
| Lo Sardo et al. [14] | Human | 20–100 y.o. | PBMCs | Plasmid + Electroporation | OSKL | No differences in reprogramming efficiency were noticed with increasing age |
| Reference | Species | Tissues | Transfection Vector | Reprogramming Factors | Methylation Analysis Technique |
|---|---|---|---|---|---|
| Ma et al. [40] | Human | FF | Sendai virus | OSKM | Infinium HumanMethylation450 Illumina |
| Lister et al. [39] | Human | ADS | Retrovirus | OSKM | Methyl-C Seq |
| Nishino et al. [42] | Human | FLF; AM; E; PDE; MB | Retrovirus | OSKM | Infinium HumanMethylation27 Illumina |
| Planello et al. [41] | Human | FF | Retrovirus | OSKM/OSKL | Infinium HumanMethylation450 Illumina |
| He et al. [38] | Human | FF; AF | Lentivirus | OSKM | Infinium HumanMethylation450 Illumina |
| Episomal | OSKMNL | ||||
| Frobel et al. [25] | HUMAN | BM-MSC | Retrovirus | OSKM | Infinium HumanMethylation450 Illumina |
| Shao et al. [43] | Human | MSC | Retrovirus | OSKM | Infinium HumanMethylation450 Illumina |
| Lo Sardo et al. [14] | HUMAN | PBMCs | eD_Plasmid | OSKL | Infinium HumanMethylation450 Illumina |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravaioli, F.; Bacalini, M.G.; Franceschi, C.; Garagnani, P. Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly. Genes 2018, 9, 39. https://doi.org/10.3390/genes9010039
Ravaioli F, Bacalini MG, Franceschi C, Garagnani P. Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly. Genes. 2018; 9(1):39. https://doi.org/10.3390/genes9010039
Chicago/Turabian StyleRavaioli, Francesco, Maria G. Bacalini, Claudio Franceschi, and Paolo Garagnani. 2018. "Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly" Genes 9, no. 1: 39. https://doi.org/10.3390/genes9010039
APA StyleRavaioli, F., Bacalini, M. G., Franceschi, C., & Garagnani, P. (2018). Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly. Genes, 9(1), 39. https://doi.org/10.3390/genes9010039





