Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Chicken Primary Myoblast Isolation and Culture
2.3. Immunofluorescence
2.4. RNA Isolation, Library Preparation, and Sequencing
2.5. Quality Control
2.6. Sequencing Data Analysis and Transcriptome Assembly
2.7. Identification of lncRNAs
2.8. lncRNAs Targets Prediction and Annotation
2.9. Screening and Clustering Analysis of Differentially Expressed lncRNAs and mRNAs
2.10. Gene Ontology and KEGG Enrichment Analysis
2.11. Validation of Gene Expression by Quantitative Real-Time Polymerase Chain Reaction Analysis
3. Results
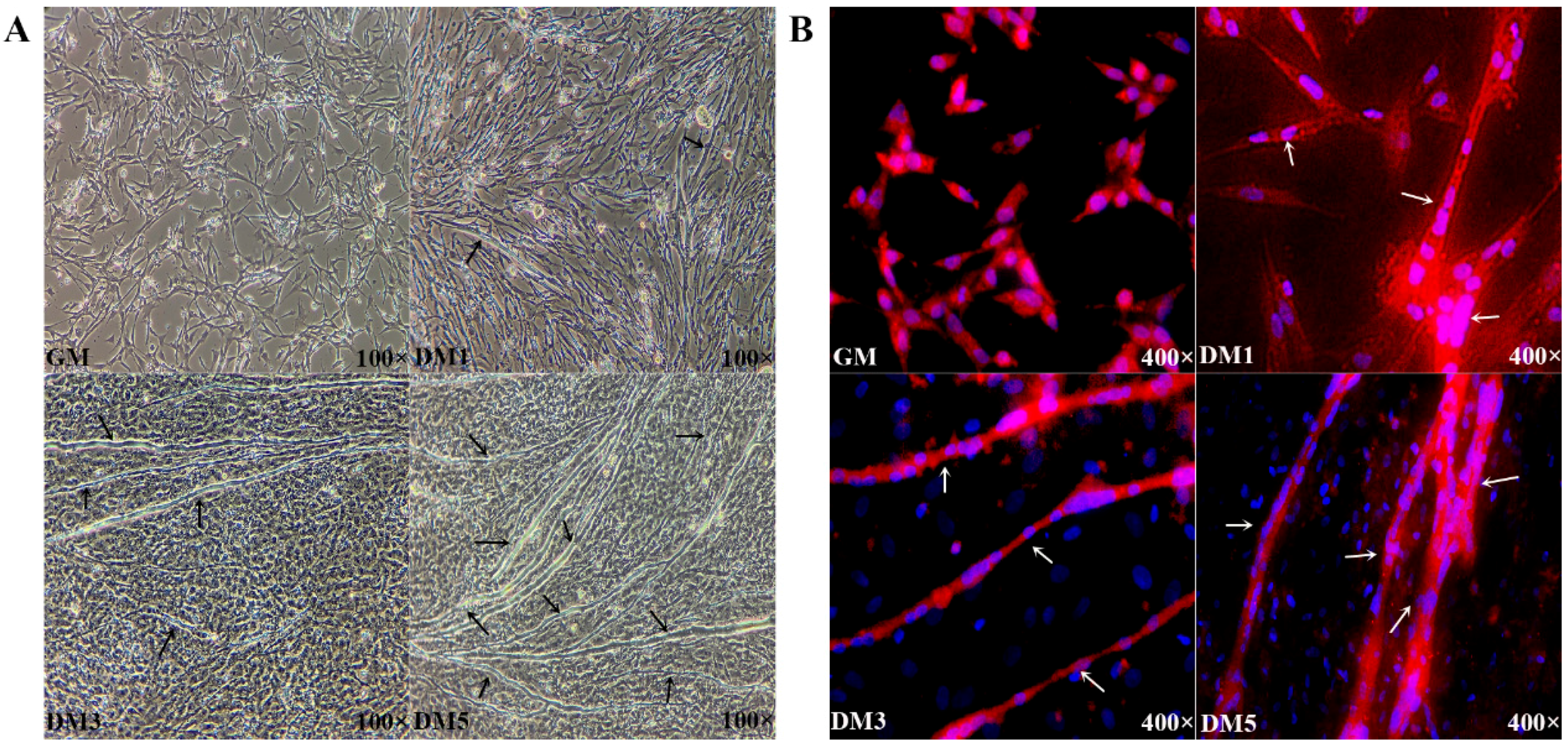
3.1. Proliferation and Differentiation of Embryonic Primary Myoblasts
3.2. Overview of RNA-Sequencing
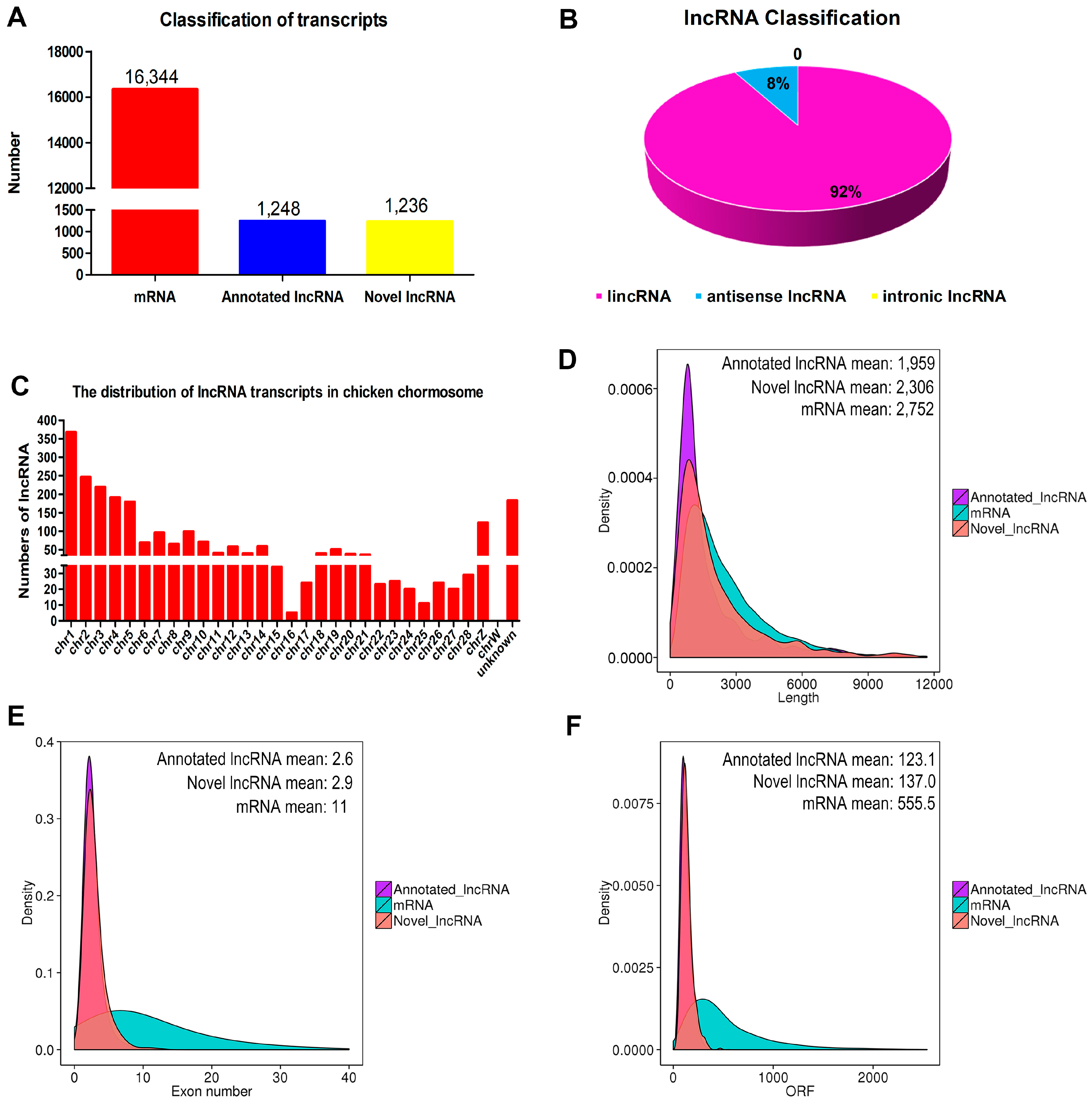
3.3. Identification and Characterization of lncRNAs in Chicken Myoblasts
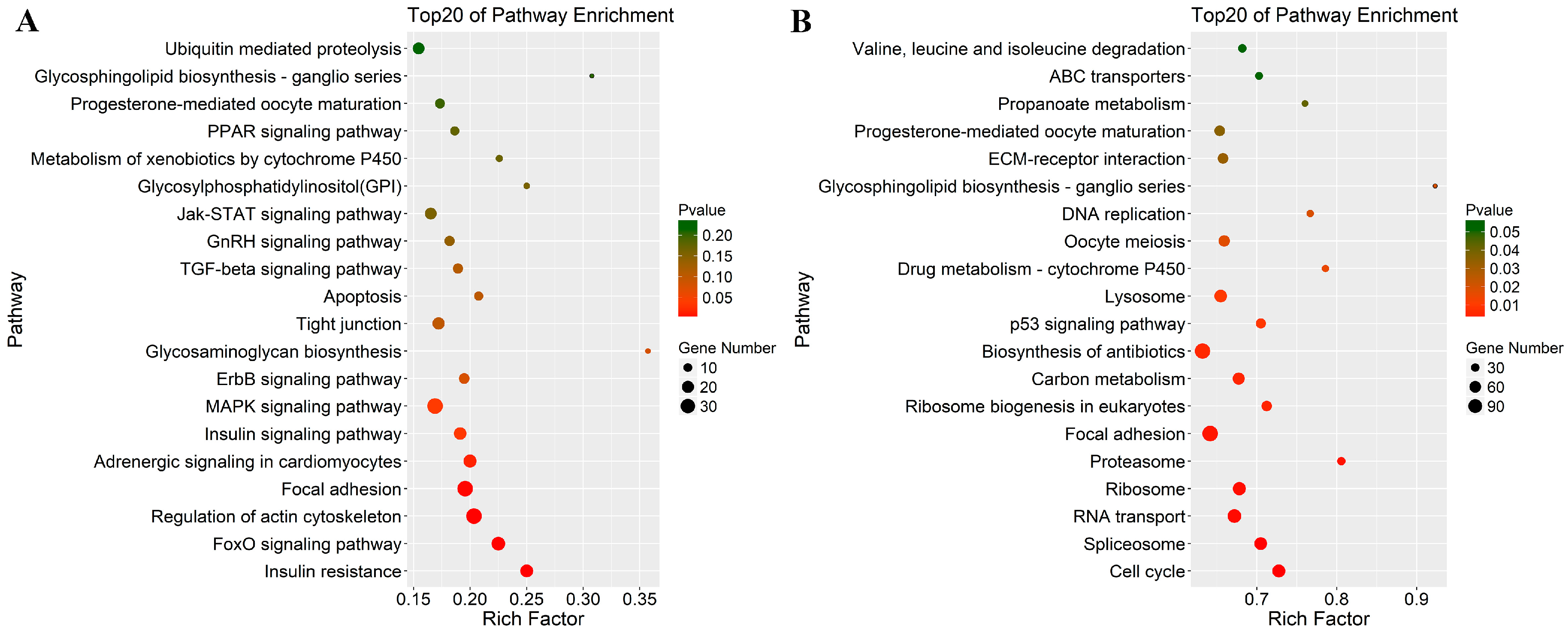
3.4. Functional Enrichment Analysis: GO and KEGG
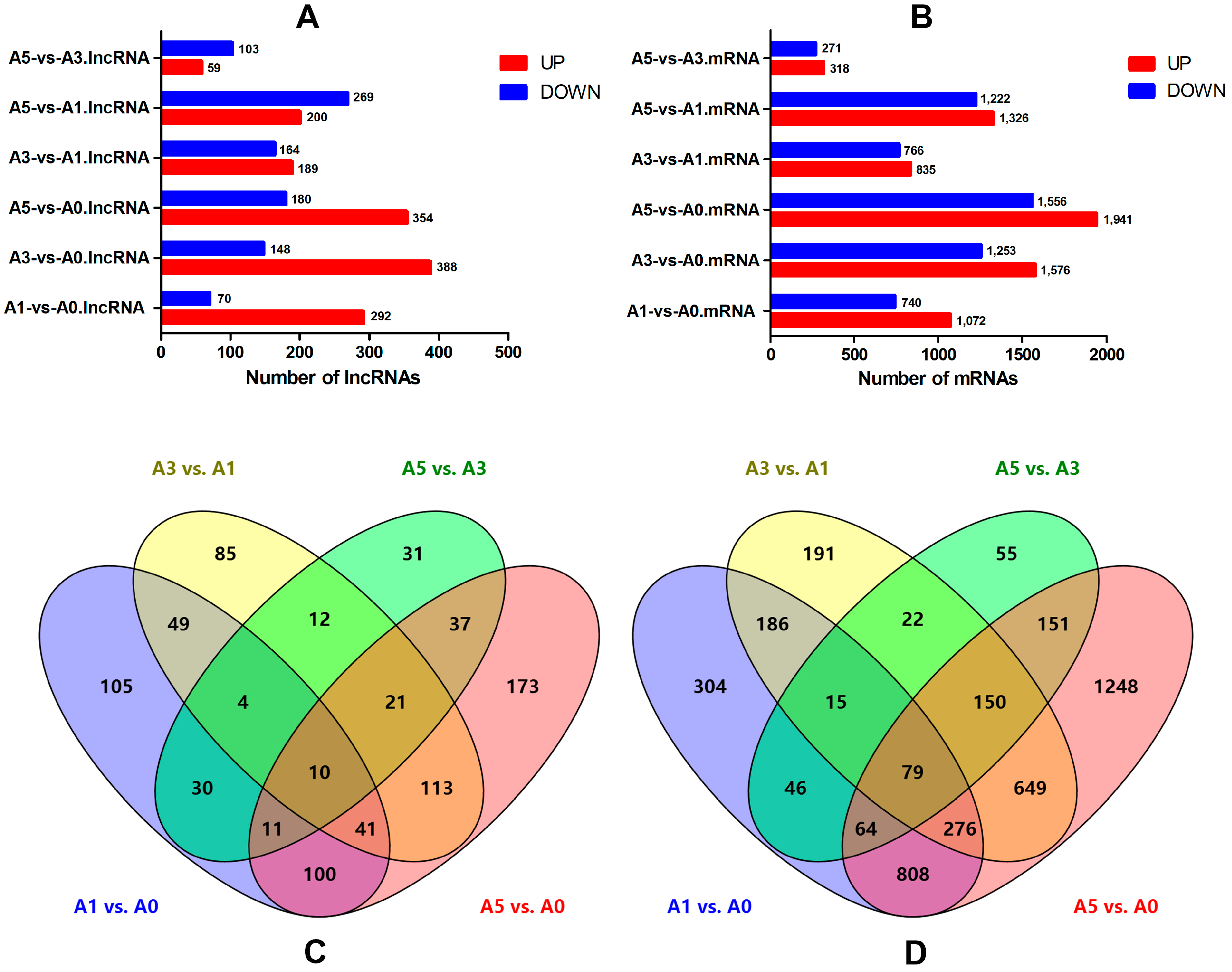
3.5. Differentially-Expressed lncRNAs and mRNAs during Primary Myoblast Differentiation
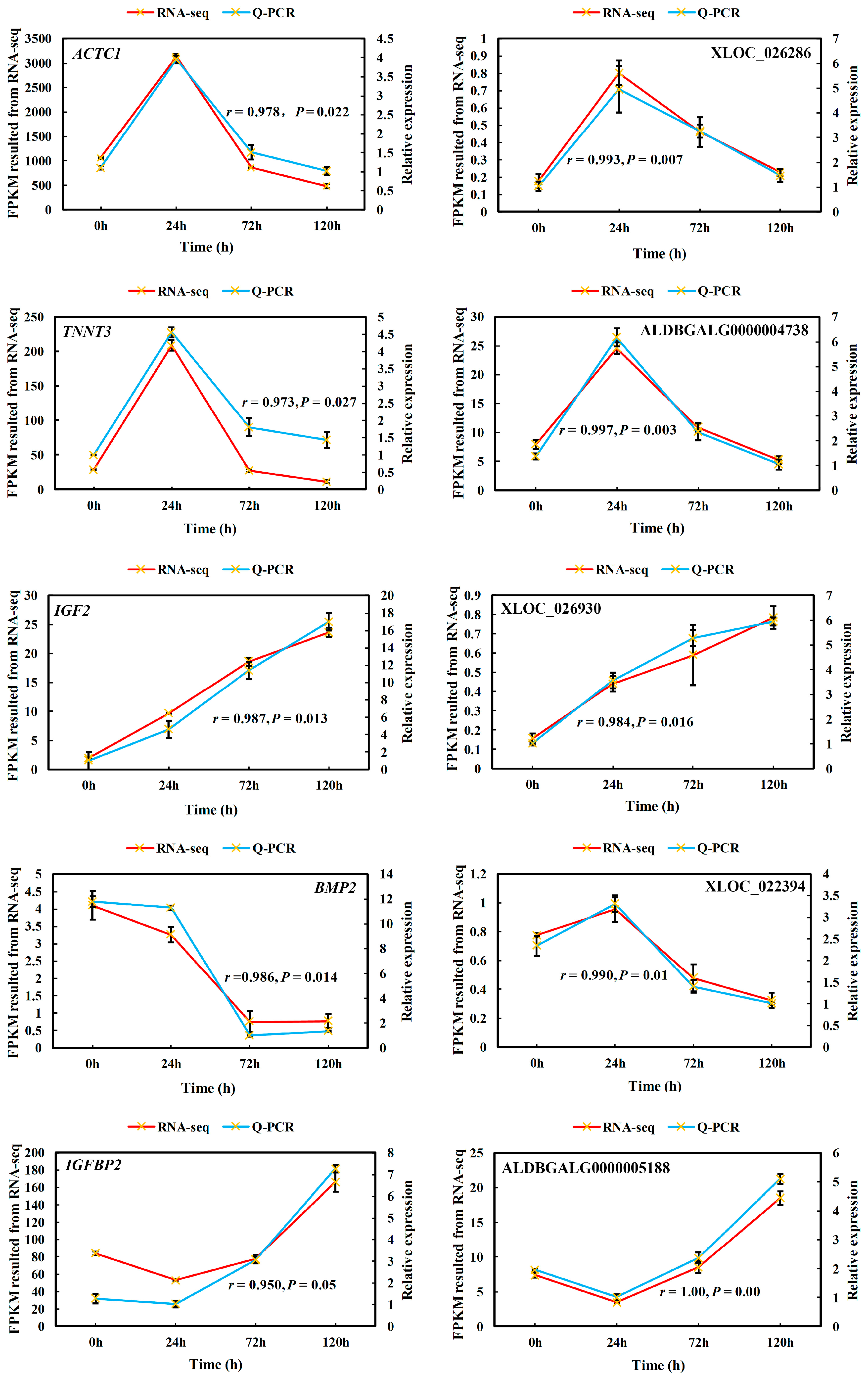
3.6. Verification of Gene Expression Profiles Using qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Coelen, R.J.; Wiggers, J.K.; Nio, C.Y.; Besselink, M.G.; Busch, O.R.; Gouma, D.J.; van Gulik, T.M. Preoperative computed tomography assessment of skeletal muscle mass is valuable in predicting outcomes following hepatectomy for perihilar cholangiocarcinoma. HPB 2015, 17, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Szulc, P.; Munoz, F.; Marchand, F.; Chapurlat, R.; Delmas, P.D. Rapid loss of appendicular skeletal muscle mass is associated with higher all-cause mortality in older men: The prospective MINOS study. Am. J. Clin. Nutr. 2010, 91, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Koomkrong, N.; Theerawatanasirikul, S.; Boonkaewwan, C.; Jaturasitha, S.; Kayan, A. Breed-related number and size of muscle fibres and their response to carcass quality in chickens. Ital. J. Anim. Sci. 2015, 14, 638–642. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Fiedler, I. Number and size of muscle fibres in relation to meat production. In Muscle Development of Livestock Animals: Physiology, Genetic, and Meat Quality; te Pas, M.F.W., Everts, M.E., Haagaman, H.P., Eds.; CABI Publishing: Wallingford, UK, 2004; pp. 1–38. ISBN 0851998119. [Google Scholar]
- Sato, F.; Kurokawa, M.; Yamauchi, N.; Hattori, M.A. Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. Am. J. Physiol.-Cell Physiol. 2006, 291, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Tao, Y.; Lin, X.; Dai, Y.; Yang, T.; Yue, X.; Jiang, X.; Li, X.; Jiang, D.S.; Andrade, K.C.; et al. Histone methyltransferase Setd2 is critical for the proliferation and differentiation of myoblasts. Biochim. Biophys. Acta 2017, 1864, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Billerey, C.; Boussaha, M.; Esquerré, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.F.; Guo, X.H.; Du, M.; Cao, G.Q.; Yang, Q.C.; Pu, Z.D.; Wang, Z.Y.; Zhang, Q.; Li, M.; Jin, Y.S.; et al. LncRNA profiling of skeletal muscles in Large White pigs and Mashen pigs during development. J. Anim. Sci. 2017, 95, 4239–4250. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Deng, M.; Fan, Y.; Yang, H.; Zhang, G.; Feng, X.; Li, F.; Wang, D.; Wang, F.; Zhang, Y. Genome-wide analysis reveals extensive changes in lncRNAs during skeletal muscle development in Hu sheep. Genes 2017, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated analysis of long non-coding RNAs (lncRNAs) and mrna expression profiles reveals the potential role of lncRNAs in skeletal muscle development of the chicken. Front. Physiol. 2016, 7, 687. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, Y.; Li, Y.; Bai, H.; Xue, F.; Xu, S.; Xu, H.; Shi, L.; Yang, N.; Chen, J. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 2017, 7, 9055. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 encodes a micropeptide to activate Six1 in cis and is involved in cell proliferation and muscle growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Cong, F.J.; Yan, L.; Xiang, B.D.; Xin, L.; Lin, L.Z.; Xin, F.L.; Hong, G. Lnc133b, a novel, long non-coding RNA, regulates bovine skeletal muscle satellite cell proliferation and differentiation by mediating miR-133b. Gene 2017, 630, 35–43. [Google Scholar]
- Sun, K.; Zhou, L.; Zhao, Y.; Wang, H.; Sun, H. Genome-wide RNA-seq and ChiP-seq reveal Linc-YY1 function in regulating YY1/PRC2 activity during skeletal myogenesis. Genom. Data 2016, 7, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ji, S.; Li, W.; Yi, B.; Li, H.; Zhang, H.; Ma, W. LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/Foxo1. Cell. Mol. Biol. Lett. 2017, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell diffrentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwiller, L.; Eddy, S.R.; Griffithsjones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Irwin, J.; Manolis, K. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Lin, C.; Ouyang, M.; Guo, Y.K. Biocloud: Cloud computing for biological, genomics, and drug design. Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Syatematic and intergrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Ginestet, C. Ggplot2: Elegant graphics for data analysis by H. Wickham. J. R. Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; Fitzhugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Nie, Q.; Zhang, X. MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genom. 2013, 40, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Deng, Z.L.; Liu, J.; Wang, D.Z. Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. BioMed Res. Int. 2015, 2015, 676575. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q. Integration analysis of lncRNA regulatory network involving miRNA and mRNA in chicken breast muscle. In Proceedings of the Plant and Animal Genome Asia, Singapore, 13–15 July 2015. [Google Scholar]
- Xue, Q.; Zhang, G.; Li, T.; Ling, J.; Zhang, X.; Wang, J. Transcriptomic profile of leg muscle during early growth in chicken. PLoS ONE 2017, 12, e0173824. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Mao, H.; Huang, M.; Chen, L.; Chen, J.; Cai, Z.; Ying, W.; Xu, N. Long noncoding RNA and mRNA expression profiles in the thyroid gland of two phenotypically extreme pig breeds using Ribo-Zero RNA sequencing. Genes 2016, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Lyer, M.K.; Stuart, P.E.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Satkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 30, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, X.; Mu, M.; Wang, J.; Wang, X.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Guo, L.; et al. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Dong, Y.; Zhao, W.; Guo, J.; Zhong, T.; Wang, L.; Li, L.; Zhang, H. Genome-wide identification and characterization of long non-coding RNAs in developmental skeletal muscle of fetal goat. BMC Genom. 2016, 17, 666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Qin, C.; Cao, G.; Xin, W.; Feng, C.; Zhang, W. Systematic analysis of long noncoding RNAs in the senescence-accelerated mouse prone 8 brain using RNA sequencing. Mol. Ther. Nucleic Acids 2016, 5, e343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xiong, Q.; Chen, H.; Yang, C.; Fan, Y. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front. Mol. Neurosci. 2017, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Ilkovski, B.; Clement, S.; Sewry, C.; North, K.N.; Cooper, S.T. Defining α-skeletal and α-cardiac actin expression in human heart and skeletal muscle explains the absence of cardiac involvement in ACTA1 nemaline myopathy. Neuromuscul. Disord. 2005, 15, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, J.; Bugaisky, G.; Buckingham, M. Simultaneous expression of skeletal muscle and heart actin proteins in various striated muscle tissues and cells. A quantitative determination of the two actin isoforms. J. Biol. Chem. 1986, 261, 1838–1843. [Google Scholar] [PubMed]
- Nowak, K.J.; Ravenscroft, G.; Jackaman, C.; Filipovska, A.; Davies, S.M.; Lim, E.M.; Squire, S.E.; Potter, A.C.; Baker, E.; Clément, S.; et al. Rescue of skeletal muscle α-actin–null mice by cardiac (fetal) α-actin. J. Cell Biol. 2009, 185, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Ravenscroft, G.; Mcnamara, E.; Griffiths, L.M.; Papadimitriou, J.M.; Hardeman, E.C.; Bakker, A.J.; Davies, K.E.; Laing, N.G.; Nowak, K.J. Cardiac α-actin over-expression therapy in dominant ACTC1 disease. Hum. Mol. Genet. 2013, 22, 3987–3997. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.J.; Ravenscroft, G.; Laing, N.G. Skeletal muscle α-actin diseases (actinopathies): Pathology and mechanisms. Acta Neuropathol. 2013, 125, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.S.; Brassington, A.M.; Krakowiak, P.A.; Carey, J.C.; Jorde, L.B.; Bamshad, M. Mutations in TNNT3 cause multiple congenital contractures: A second locus for distal arthrogryposis type 2B. Am. J. Hum. Genet. 2003, 73, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Florini, J.R.; Magri, K.A.; Ewton, D.Z.; James, P.L.; Grindstaff, K.; Rotwein, P.S. ‘Spontaneous’ differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 1991, 266, 15917–15923. [Google Scholar] [PubMed]
- Ge, Y.; Sun, Y.; Chen, J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011, 192, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, K.; Yamane, A.; Suga, T.; Suzuki, E.; Fukui, T.; Nakamura, Y. Bone morphogenetic protein-2 functions as a negative regulator in the differentiation of myoblasts, but not as an inducer for the formations of cartilage and bone in mouse embryonic tongue. BMC Dev. Biol. 2011, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Millay, D.P.; O’Rourke, J.R.; Sutherland, L.B.; Bezprozvannaya, S.; Shelton, J.M.; Basselduby, R.; Olson, E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 2013, 499, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Millay, D.P.; Sutherland, L.B.; Bassel-Duby, R.; Olson, E.N. Myomaker is essential for muscle regeneration. Genes Dev. 2014, 28, 1641–1646. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.L.; Guan, J.L. Differential regulation of cell migration and cell cycle progression by FAK complexes with Src, Pi3k, Grb7 and Grb2 in focal contacts. FEBS Lett. 2001, 499, 176–181. [Google Scholar] [CrossRef]
- Liu, L. Tyrosine kinases Abl and Fak inhibit muscle adhesion and migration in dissociated Drosophila embryonic cultures. Ph.D. Thesis, Southeast University, Nanjing, China, 8 June 2014. [Google Scholar]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Ramoni, M.F.; Sebastiani, P.; Kohane, I.S. Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. USA 2002, 99, 9121–9126. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, C.; Su, Y.; Zhang, K.; Zhang, Y.; Chen, M.; Ge, M.; Gu, L.; Lu, T.; Li, N.; et al. MiRNA-34c inhibits myoblasts proliferation by targeting YY1. Cell Cycle 2017, 16, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.P.; Liu, H.H.; Wang, H.H.; Wang, Y.; Han, C.C.; Li, L.; He, H.; Xu, H.Y.; Xu, F.; Wang, J.W. Silencing Pax3 by shRNA inhibits the proliferation and differentiation of duck (Anas platyrhynchos) myoblasts. Mol. Cell. Biochem. 2014, 386, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Wu, G.; Xiang, A.; Zhang, Q.; Li, W.; Yang, G.; Shi, X.; Sun, S.; Li, X. Over-expression of miR-125a-5p inhibits proliferation in C2C12 myoblasts by targeting E2F3. Chin. J. Biochem. Mol. Biol. 2015, 47, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jint, H.; Liu, H.; Sun, L.; Li, X.; Yang, C.; Zhang, R.; Li, L.; Wang, J. Molecular cloning and expression pattern of duck Six1 and its preliminary functional analysis in myoblasts transfected with eukaryotic expression vector. Indian J. Biochem. Biophys. 2014, 51, 271–281. [Google Scholar] [PubMed]
- Panda, A.C.; Kotb, A.; Martindale, J.L.; Clara, D.G.; Yang, X.; Ioannis, G.; Heon, N.J.; Zhang, Y.; Elin, L.; Dudekula, D.B. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016, 44, 2393–2408. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.A.; Kalter, V.; Strowitzki, M.; Schneider, M.; Lichter, P. IGF2 knockdown in two colorectal cancer cell lines decreases survival, adhesion and modulates survival-associated genes. Tumour Biol. 2016, 37, 12485–12495. [Google Scholar] [CrossRef] [PubMed]
- Girona, J.; Rosales, R.; Plana, N.; Saavedra, P.; Masana, L.; Vallvé, J.C. FABP4 induces vascular smooth muscle cell proliferation and migration through a MAPK-dependent pathway. PLoS ONE 2013, 8, e81914. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Su, Y.; Song, Q.; Tung, B.; Oyinlade, O.; Liu, S.; Ying, M.; Ming, G.L.; Song, H.; Qian, J.; et al. Methylatedcis-regulatory elements mediate KLF4-dependent gene transactivation and cell migration. eLIFE 2017, 6, e20068. [Google Scholar] [CrossRef] [PubMed]
- Clemente, C.F.M.Z.; Corat, M.A.F.; Saad, S.T.O.; Franchini, K.G. Differentiation of C2C12 myoblasts is critically regulated by fak signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 289, R862–R870. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.C.; Kapur, R.; King, A.J.; Tao, W.; Kim, C.; Borneo, J.; Breese, R.; Marshall, M.; Dinauer, M.C.; Williams, D.A. Rac2 stimulates Akt activation affecting BAD/Bcl-XL expression while mediating survival and actin function in primary mast cells. Immunity 2000, 12, 557–568. [Google Scholar] [CrossRef]
- Millay, D.P.; Gamage, D.G.; Quinn, M.E.; Min, Y.L.; Mitani, Y.; Basselduby, R.; Olson, E.N. Structure-function analysis of myomaker domains required for myoblast fusion. Proc. Natl. Acad. Sci. USA 2016, 113, 2116–2121. [Google Scholar] [CrossRef] [PubMed]
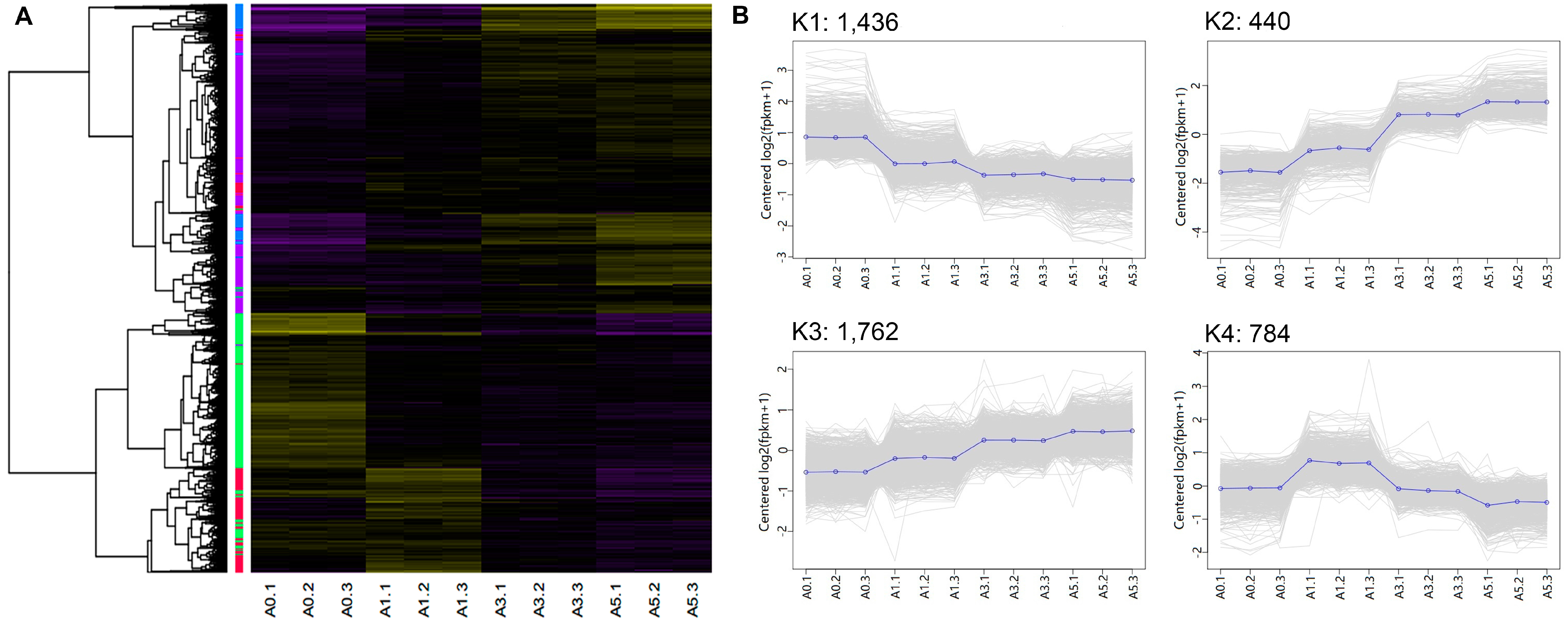
| Cluster (# of genes) | Example genes | Enriched GO terms | # of gens | p-value |
|---|---|---|---|---|
| K1 (1,436) | YY1, TGFβ1, SOX8, SIX1, SIX2, PAX3, PAX7, MYF5, MYF6, ID1, ID2, E2F7, E2F3, E2F1, E2F2, CDC7, CDC6, CDC45, CDC20, CDK1, CDK2, CDK6, CDK7 | Cell cycle process | 138 | 3.81 × 10−22 |
| DNA metabolic process | 123 | 5.17 × 10−22 | ||
| Cell cycle | 156 | 9.67 × 10−22 | ||
| Chromosome organization | 132 | 4.51 × 10−21 | ||
| DNA replication | 59 | 2.94 × 10−20 | ||
| Mitotic cell cycle process | 92 | 1.86 × 10−17 | ||
| Nuclear division | 73 | 5.56 × 10−17 | ||
| DNA repair | 76 | 8.60 × 10−17 | ||
| Mitotic cell cycle | 98 | 3.19 × 10−16 | ||
| Chromosome segregation | 54 | 2.18 × 10−15 | ||
| K2 (440) | MYOC, IGF2, MYO1D, FGF7, FGF18, FABP4, ECM2, DCX, FAP, FAT4, KLF15, LBP, SHOX, TF | Extracellular structure organization | 18 | 5.03 × 10−7 |
| Extracellular matrix organization | 18 | 5.03 × 10−7 | ||
| Chemotaxis | 24 | 1.0 6 ×10−5 | ||
| Taxis | 24 | 1.13 × 10−5 | ||
| Cellular response to chemical stimulus | 59 | 2.49 × 10−5 | ||
| Response to external stimulus | 52 | 3.50 × 10−5 | ||
| Regulation of cell migration | 27 | 6.87 × 10−5 | ||
| Cellular response to organic substance | 49 | 7.19 × 10−5 | ||
| Cell adhesion | 41 | 9.90 × 10−5 | ||
| Biological adhesion | 41 | 1.09 × 10−4 | ||
| K3 (1,762) | FLI1, KLF4, WNT9A, CTR, WNT5A, EGFR, BMP5, FAK, FGF14, FGF9, RAC2, MYH6, MYH10, MYCN, MXD4, IGFBP2, IGFBP4, IGFBP5 | Locomotion | 139 | 6.05 × 10−8 |
| Movement of cell or subcellular component | 148 | 1.79 × 10−6 | ||
| Enzyme linked receptor protein signaling pathway | 86 | 3.16 × 10−6 | ||
| Transmembrane receptor protein tyrosine kinase signal | 61 | 7.29 × 10−6 | ||
| Organ morphogenesis | 93 | 7.34 × 10−6 | ||
| Biological adhesion | 118 | 9.30 × 10−6 | ||
| Skeletal system development | 59 | 1.04 × 10−5 | ||
| Cell adhesion | 117 | 1.25 × 10−5 | ||
| Skeletal system morphogenesis | 36 | 1.42 × 10−5 | ||
| Neurogenesis | 127 | 1.59 × 10−5 | ||
| K4 (784) | TNNT3, SRF, RGS2, Myosin, MYOG, TMEM8C, DLL1 NOTCH1, MYOD, MEF2C, GDF8, MEF2D, MAPK13, DIK2, BMP4, β-FGF, ACTC1, MTOR, SMYD1 | Muscle structure development | 61 | 3.24 × 10−20 |
| Muscle system process | 41 | 6.97 × 10−18 | ||
| Muscle contraction | 37 | 3.07 × 10−17 | ||
| Striated muscle cell differentiation | 35 | 2.35 × 10−15 | ||
| System process | 87 | 5.03 × 10−14 | ||
| Muscle cell differentiation | 41 | 1.18 × 10−13 | ||
| Muscle tissue development | 41 | 2.72 × 10−13 | ||
| Muscle organ development | 36 | 1.40 × 10−12 | ||
| Striated muscle tissue development | 38 | 1.51 × 10−12 | ||
| Muscle cell development | 26 | 3.83 × 10−12 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Zhang, G.; Wu, P.; Duan, L.; Li, G.; Liu, Q.; Wang, J. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes 2018, 9, 34. https://doi.org/10.3390/genes9010034
Li T, Zhang G, Wu P, Duan L, Li G, Liu Q, Wang J. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes. 2018; 9(1):34. https://doi.org/10.3390/genes9010034
Chicago/Turabian StyleLi, Tingting, Genxi Zhang, Pengfei Wu, Lian Duan, Guohui Li, Qiuhong Liu, and Jinyu Wang. 2018. "Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis" Genes 9, no. 1: 34. https://doi.org/10.3390/genes9010034
APA StyleLi, T., Zhang, G., Wu, P., Duan, L., Li, G., Liu, Q., & Wang, J. (2018). Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes, 9(1), 34. https://doi.org/10.3390/genes9010034





