High Density Single Nucleotide Polymorphism (SNP) Mapping and Quantitative Trait Loci (QTL) Analysis in a Biparental Spring Triticale Population Localized Major and Minor Effect Fusarium Head Blight Resistance and Associated Traits QTL
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. FHB Inoculations and Phenotyping
2.3. Phenotyping of Other Traits
2.4. Phenotypic Data Evaluation
2.5. DNA Isolation and Genotyping
2.6. Construction of SNP Linkage Maps
2.7. Quantitative Trait Loci (QTL) Mapping
3. Results
3.1. FHB Development and DON Accumulation
3.2. Ergot Incidence, Test Weight, Grain Yield, Grain Protein Content, Plant Height and Lodging
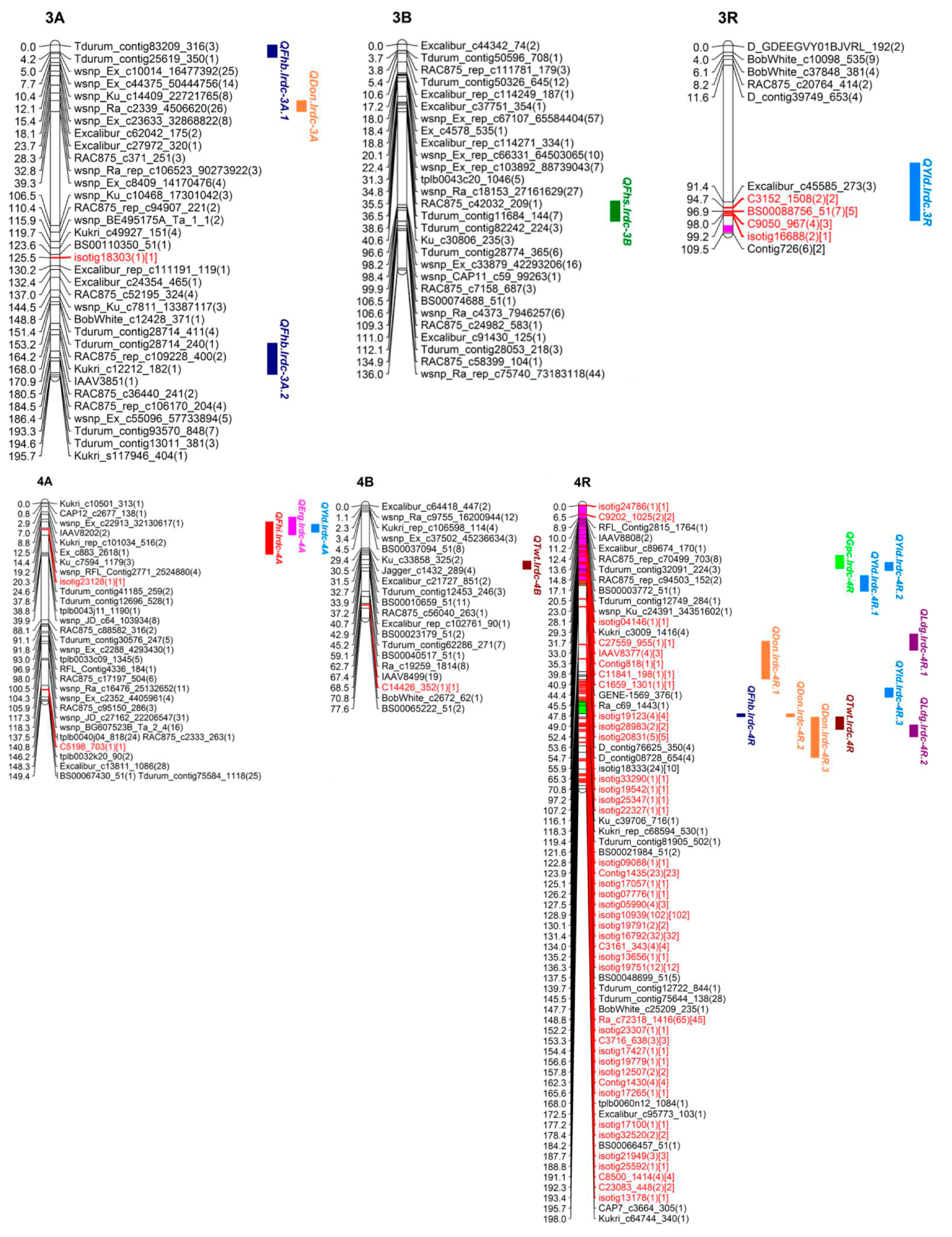
3.3. SNP Genetic Map
3.4. QTL Analysis
3.4.1. QTL for FHB Resistance and Low DON Content
3.4.2. QTL for Ergot Incidence
3.4.3. QTL for Grain Protein Content
3.4.4. QTL for Test Weight
3.4.5. QTL for Grain Yield
3.4.6. QTL for Plant Height and Lodging
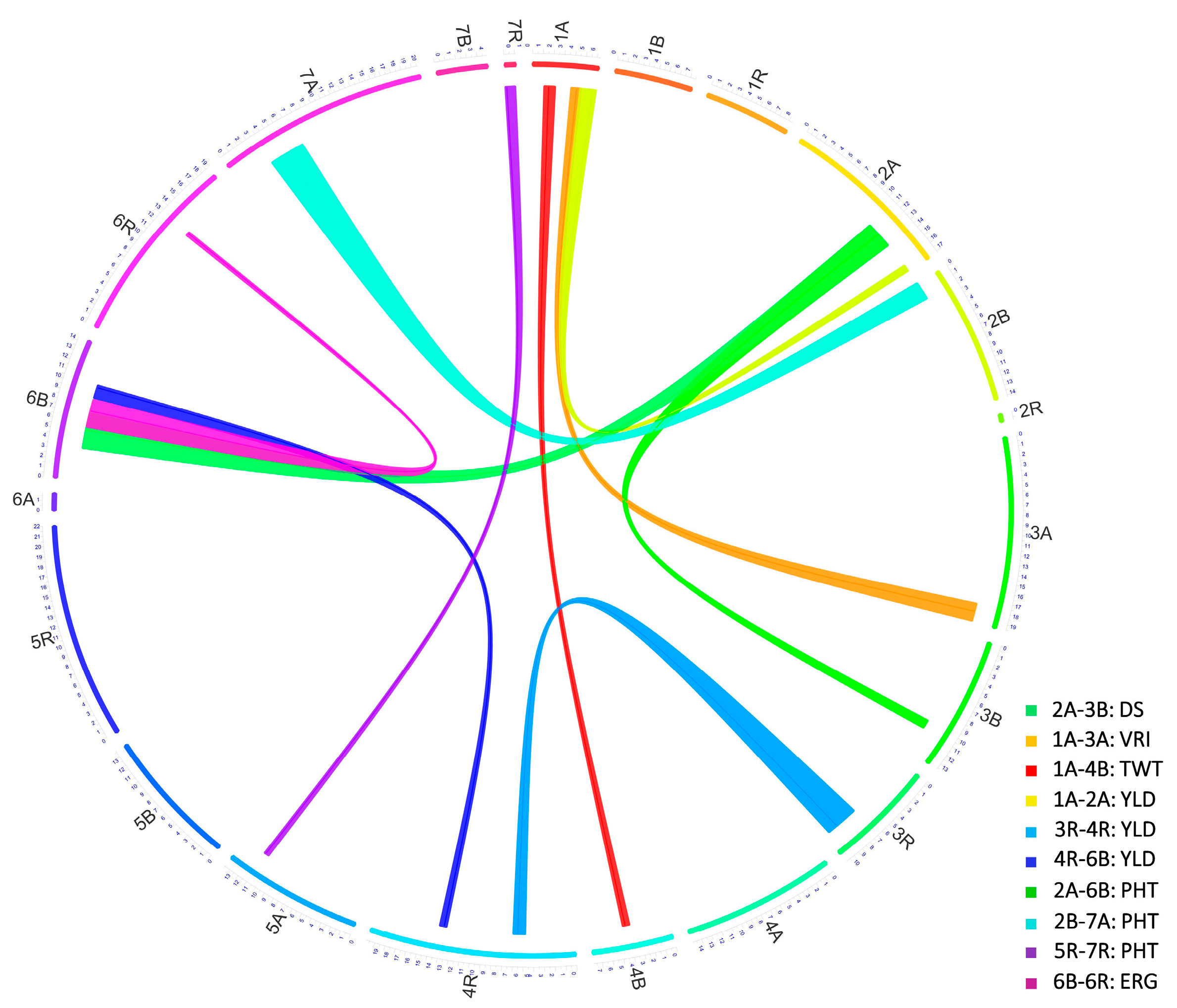
3.4.7. Co-Localized or Pleiotropic QTL
4. Discussion
4.1. Phenotypic Data
4.2. High Density Linkage Maps
4.3. Genetic Architecture of FHB Resistance in Triticale
4.4. Genetic Architecture of Other Studied Traits and Their Possible Relation with FHB Resistance
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Miedaner, T.; Schneider, B.; Oettler, G. Means and variances for Fusarium head blight resistance of F2-derived lines from winter triticale and winter wheat crosses. Euphytica 2006, 152, 405–411. [Google Scholar] [CrossRef]
- Miedaner, T. Breeding wheat and rye for resistance to Fusarium disease. Plant Breed. 1997, 116, 201–220. [Google Scholar] [CrossRef]
- Kalih, R.; Maurer, H.P.; Hackauf, B.; Miedaner, T. Effect of a rye dwarfing gene on plant height, heading stage and Fusarium head blight in triticale (×Triticosecale Wittmack). Theor. Appl. Genet. 2014, 127, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Kalih, R.; Maurer, H.P.; Miedaner, T. Genetic architecture of Fusarium head blight resistance in four winter triticale populations. Phytopathology 2015, 105, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Kalih, R.; Großmann, M.S.; Maurer, H.P. Correlation between Fusarium head blight severity and DON content in triticale as revealed by phenotypic and molecular data. Plant Breed. 2016, 135, 31–37. [Google Scholar] [CrossRef]
- Tamburic-Ilincic, L.; Falk, D.; Schaafsma, A. Fusarium ratings in Ontario winter wheat performance trial (OWWPT) using an index that combines Fusarium head blight symptoms an deoxynivalenol levels. Czech J. Genet. Plant Breed. 2011, 47, 115–122. [Google Scholar]
- Boeven, P.H.G.; Würschum, T.; Weissmann, S.; Miedaner, T.; Maurer, H.P. Prediction of hybrid performance for Fusarium head blight resistance in triticale (×Triticosecale Wittmack). Euphytica 2016, 207, 475–490. [Google Scholar] [CrossRef]
- Mesterhazy, A. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Mesterhazy, A.; Bartok, T.; Mirocha, C.G.; Komoroczy, R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999, 118, 97–110. [Google Scholar] [CrossRef]
- Oettler, G.; Heinrich, N.; Miedaner, T. Estimates of additive and dominance effects for Fusarium head blight resistance of winter triticale. Plant Breed. 2004, 123, 525–530. [Google Scholar] [CrossRef]
- Oettler, G.; Wahle, G. Genotypic and environmental variation of resistance to head blight in triticale inoculated with Fusarium culmorum. Plant Breed. 2001, 120, 297–300. [Google Scholar] [CrossRef]
- Miedaner, T.; Reinbrecht, C.; Lauber, U.; Schollenberger, M.; Geiger, H. Effects of genotype and genotype x environment interaction on deoxynivalenol accumulation and resistance to Fusarium head blight in rye, triticale and wheat. Plant Breed. 2001, 120, 97–105. [Google Scholar] [CrossRef]
- Randhawa, H.S.; Bona, L.; Graf, R.J. Triticale breeding—progress and prospect. In Triticale; Eudes, F., Ed.; Springer: New York, NY, USA, 2015; pp. 15–32. ISBN 978-3-319-22550-0. [Google Scholar] [CrossRef]
- Comeau, A.; Langevin, F.; Eudes, F. Mechanisms of resistance and tolerance to FHB. In Proceedings of the 3rd CWFHB meeting, Winnipeg, MB, Canada, 9–12 December 2003; pp. 88–106. [Google Scholar]
- Paul, P.A.; Lipps, P.E.; Madden, L.V. Relationship between visual estimates of Fusarium head blight intensity and deoxynivalenol accumulation in harvested wheat grain: A meta-analysis. Phytopathology 2005, 95, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Tamburic-Ilincic, L.; Schaafsma, A.; Falk, D.; Eudes, F. Breeding for Fusarium head blight resistance using conventional breeding and biotechnology. In Proceedings of the 9th European Fusarium Seminar, Wageningen, The Netherlands, 19–22 September 2006; p. 84. [Google Scholar]
- Nopsa, J.F.H. Fusarium head blight: Winter wheat cultivar responses and characterization of pathogen isolates. Ph.D. Thesis, University of Nebraska Lincoln, Lincoln, NE, USA, 23 November 2010. [Google Scholar]
- Voss, H.H.; Korzun, V.; Miedaner, T. Effect of the height-reducing allele Rht2 on Fusarium head blight in four segregating populations of winter wheat. In Proceedings of the 9th European Fusarium Seminar, Wageningen, The Netherlands, 19–22 September 2006; p. 118. [Google Scholar]
- Draeger, R.; Gosman, N.; Steed, A.; Chandler, E.; Thomsett, M.; Srinivasachary; Schondelmaier, J.; Buerstmayr, H.; Lemmens, M.; Schmolke, M.; et al. Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor. Appl. Genet. 2007, 115, 617–625. [Google Scholar] [CrossRef] [PubMed]
- McCartney, C.A.; Somers, D.J.; Fedak, G.; DePauw, R.M.; Thomas, J.; Fox, S.L.; Humphreys, D.G.; Lukow, O.; Savard, M.E.; McCallum, B.D.; et al. The evaluation of FHB resistance QTLs introgressed into elite Canadian spring wheat germplasm. Mol. Breed. 2007, 20, 209–221. [Google Scholar] [CrossRef]
- Miedaner, T.; Voss, H.H. Effect of dwarfing Rht genes on Fusarium head blight resistance in two sets of near-isogenic lines of wheat and check cultivars. Crop Sci. 2008, 48, 2115–2122. [Google Scholar] [CrossRef]
- Lu, Q.X.; Lillemo, M.; Skinnes, H.; He, X.Y.; Shi, J.R.; Ji, F.; Dong, Y.; Bjørnstad, Å. Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird’. Theor. Appl. Genet. 2013, 126, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Schmolke, M.; Zimmermann, G.; Buerstmayr, H.; Schweizer, G.; Miedaner, T.; Korzun, V.; Ebmeyer, E.; Hartl, L. Molecular mapping of Fusarium head blight resistance in the winter wheat Dream/Lynx. Theor. Appl. Genet. 2005, 111, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Häberle, J.; Schmolke, M.; Schweizer, G.; Korzun, V.; Ebmeyer, E.; Zimmermann, G.; Hartl, L. Effects of two major Fusarium head blight resistance QTL verified in a winter wheat backcross population. Crop Sci. 2007, 47, 1823–1831. [Google Scholar] [CrossRef]
- Zhang, X.; Fu, J.; Hiromasa, Y.; Pan, H.; Bai, G. Differentially expressed proteins associated with Fusarium head blight resistance in wheat. PLoS ONE 2013, 8, e82079. [Google Scholar] [CrossRef] [PubMed]
- Perlikowski, D.; Wiśniewska, H.; Góral, T.; Kwiatek, M.; Majka, M.; Kosmala, A. Identification of kernel proteins associated with the resistance to Fusarium head blight in winter wheat (Triticum aestivum L.). PLoS ONE 2014, 9, e110822. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Beaudoin, F.; Jenkins, J.; Griffiths-Jones, S.; Mills, E.N. Plant protein families and their relationships to food allergy. Biochem. Soc. Trans. 2002, 30, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.; Tang, H.; Belcram, H.; Paterson, A.; Gornicki, P.; Chalhoub, B. Sixty million years in evolution of soft grain trait in grasses: Emergence of the softness locus in the common ancestor of Pooideae and Ehrhartoideae, after their divergence from Panicoideae. Mol. Biol. Evol. 2009, 26, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Statistics Canada. Table 001-0017—Estimated areas, yield, production, average farm price and total farm value of principal field crops, in metric and imperial units, annual, CANSIM (Database). Available online: http://www5.statcan.gc.ca/cansim/a47#F1 (accessed on 18 December 2017).
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Loeffler, M.; Schon, C.C.; Miedaner, T. Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol. Breed. 2009, 23, 473–488. [Google Scholar] [CrossRef]
- McLeod, J.G.; Pfeiffer, W.F.; DePauw, R.M.; Clarke, J.M. AC Ultima spring triticale. Can. J. Plant Sci. 2000, 80, 831–833. [Google Scholar] [CrossRef]
- Eudes, F.; Amundsen, E. Isolated microspore culture of Canadian 6X triticale cultivars. Plant Cell Tissue Organ Cult. 2005, 82, 233–241. [Google Scholar] [CrossRef]
- Gilbert, J.; Morgan, K. Field–based rating of spring wheat infected with Fusarium graminearum, cause of Fusarium head blight. In Proceedings of the 6th European Seminar on Fusarium—Mycotoxins, Taxonomy and Pathogenicity, Berlin, Germany, 11–16 September 2000; p. 73. [Google Scholar]
- Sinha, R.C.; Savard, M.E. Comparison of immunoassay and gas chromatography methods for the detection of the mycotoxin deoxynivalenol in grain samples. Can. J. Plant. Pathol. 1996, 18, 233–236. [Google Scholar] [CrossRef]
- Griffin, G.M. Understanding and assessing lodging risk in winter wheat. Ph.D. Thesis, The University of Nottingham, Nottingham, UK, May 1998. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Wu, Y.; Bhat, P.R.; Close, T.J.; Lonardi, S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008, 4, e1000212. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distance from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Lorieux, M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012, 30, 1231–1235. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martis, M.M.; Zhou, R.; Haseneyer, G.; Schmutzer, T.; Vrána, J.; Kubaláková, M.; König, S.; Kugler, K.G.; Scholz, U.; Hackauf, B.; et al. Reticulate evolution of the rye genome. Plant Cell 2013, 25, 3685–3698. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenacc, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zeng, Z. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 1993, 90, 10972–10976. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [PubMed]
- Joehanes, R.; Nelson, J.C. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 2008, 24, 2788–2789. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, C.C.; Hu, H.; Yu, R.D.; Xia, Z.; Ye, X.Z.; Zhu, J. QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 2008, 24, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yan, C.; Hsu, C.H.; Chen, Q.R.; Niu, K.; Komatsoulis, G.A.; Meerzaman, D. OmicCircos: A simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 2014, 13, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Somers, D.J.; Thomas, J.; Depauw, R.; Fox, S.; Humphreys, G.; Fedak, G. Assembling complex genotypes to resist Fusarium in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2005, 111, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Fleury, D. FHB and Plant Breeding Advancements: What to Expect with increased disease resistant varieties. Top Crop Manager. 2014. Available online: http://www.topcropmanager.com/plant-breeding/fhb-and-plant-breeding-advancements-14757 (accessed on 2 November 2016).
- Ma, H.-X.; Bai, G.-H.; Zhang, X.; Lu, W.-Z. Main effects, epistasis and environmental interactions of quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population. Phytopathology 2006, 96, 534–541. [Google Scholar] [CrossRef] [PubMed]
- McCartney, C.A.; Brûlé-Babel, A.L.; Fedak, G.; Martin, R.A.; McCallum, B.D.; Gilbert, J.; Hiebert, C.W.; Pozniak, C.J. Fusarium Head blight resistance QTL in the spring wheat cross Kenyon/86ISMN 2137. Front. Microbiol. 2016, 7, 1542. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Prentice Hall: London, UK, 1996. [Google Scholar]
- Tyrka, M.; Tyrka, D.; Wędzony, M. Genetic map of triticale integrating microsatellite, DArT and SNP markers. PLoS ONE 2015, 10, e0145714. [Google Scholar] [CrossRef] [PubMed]
- Alheit, K.V.; Reif, J.C.; Maurer, H.P.; Hahn, V.; Weissmann, E.A.; Miedaner, T.; Würschum, T. Detection of segregation distortion loci in triticale (×Triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genom. 2011, 12, 380. [Google Scholar] [CrossRef] [PubMed]
- Giancaspro, A.; Giove, S.L.; Zito, D.; Blanco, A.; Gadaleta, A. Mapping QTLs for Fusarium head blight resistance in an interspecific wheat population. Front. Plant Sci. 2016, 7, 1381. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lillemo, M.; Shi, J.; Wu, J.; Bjørnstad, Å.; Belova, T.; Dreisigacker, S.; Duveiller, E.; Singh, P. QTL characterization of Fusarium head blight resistance in CIMMYT bread wheat line Soru#1. PLoS ONE 2016, 11, e0158052. [Google Scholar] [CrossRef]
- Wasternack, C. A plant’s balance of growth and defense—Revisited. New Phytol. 2017, 215, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
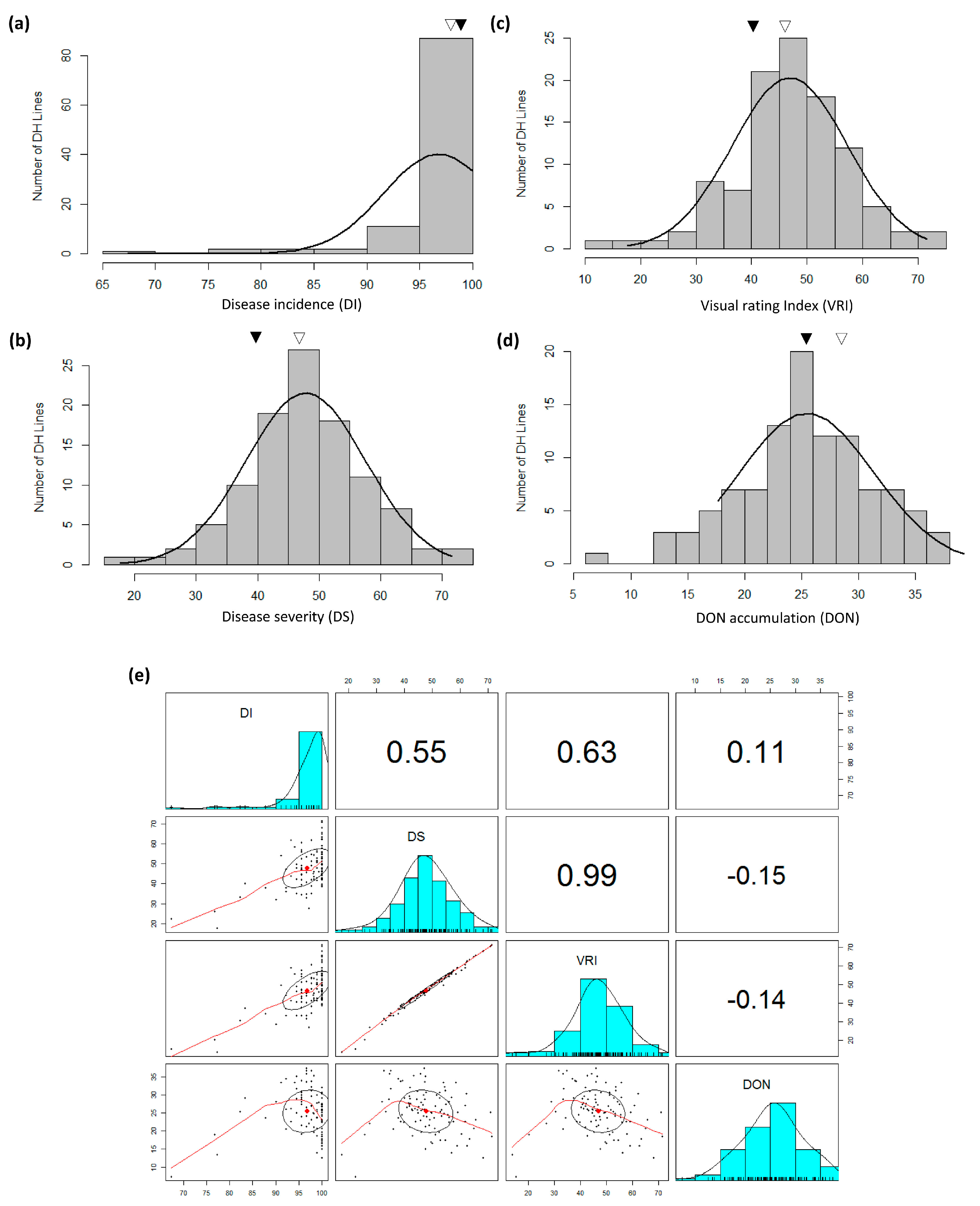
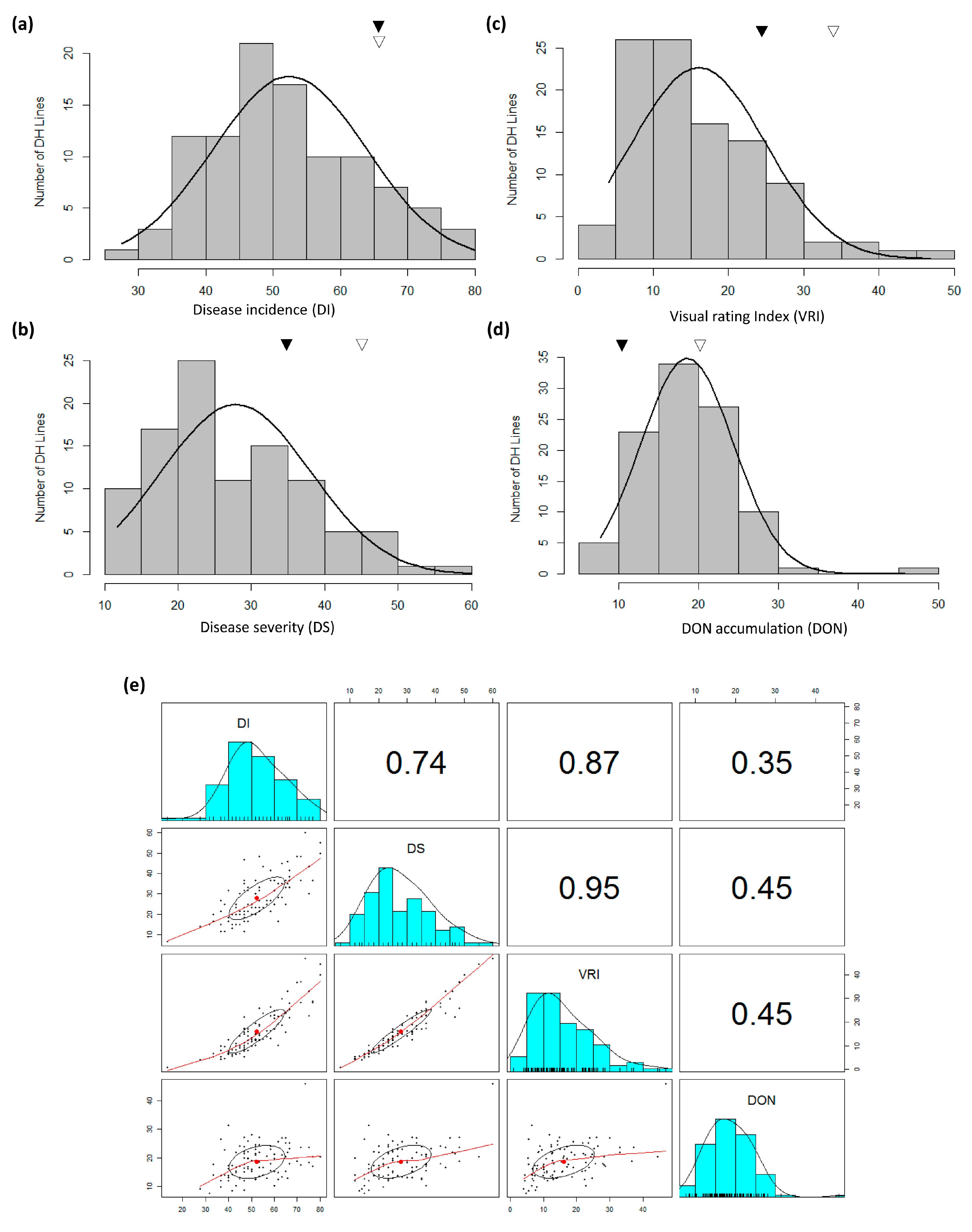
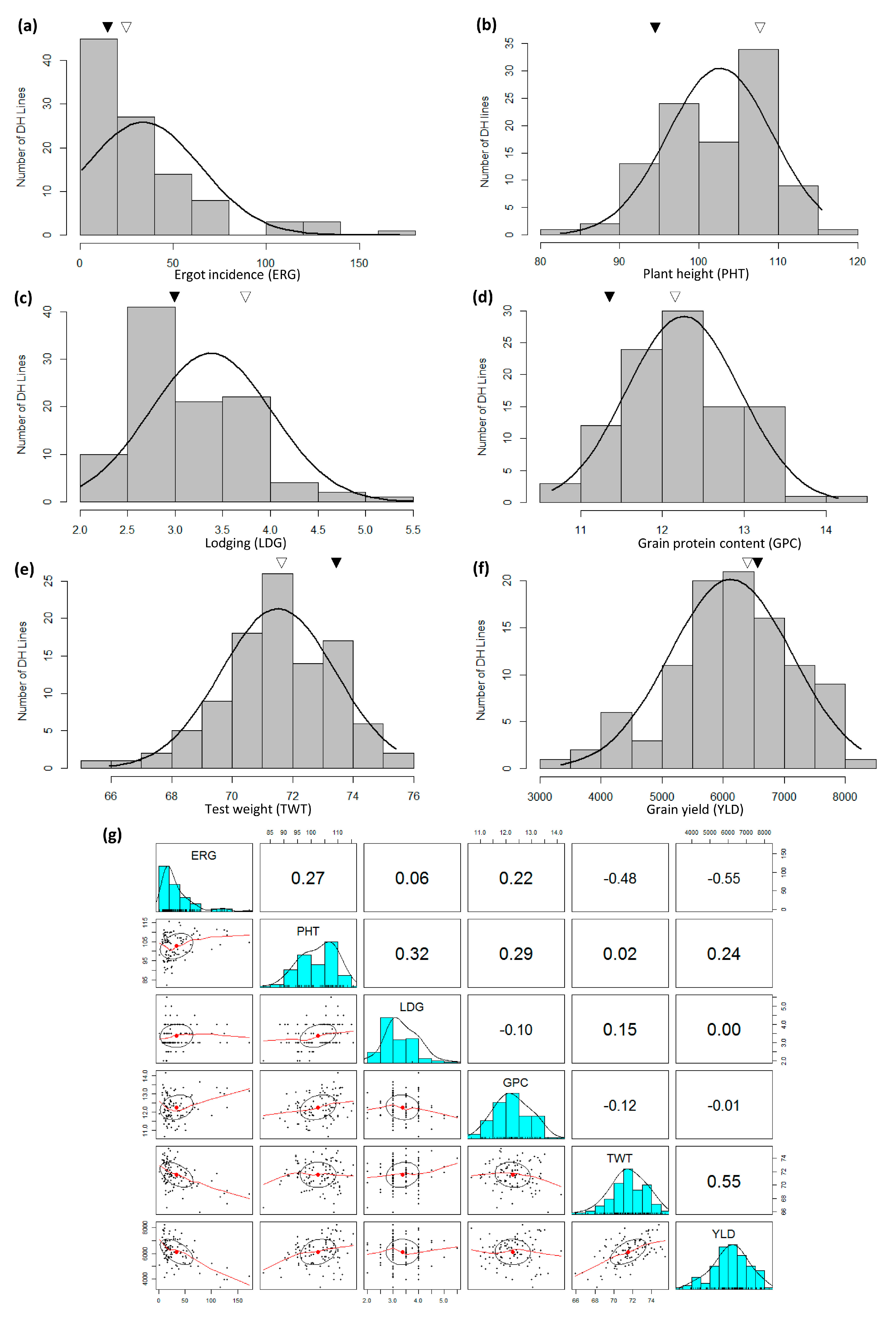


| Source | d.f. | DI | DS | VRI | DON |
|---|---|---|---|---|---|
| Environment (E) | 1 | 306,278.0 ** | 55,591.0 ** | 136,890.0 ** | 7495.1 ** |
| Block (within E) | 2 | 680.0 ** | 119.0 | 177.0 | 278.2 * |
| Treatment (T) | 104 | 439.0 ** | 496.0 ** | 474.0 ** | 148.7 ** |
| (E × T) | 100 | 212.0 ** | 205.0 | 201.0 | 70.9 ** |
| Error | 432 | 99.0 | 187.0 | 189.0 | 42.0 |
| CV, % | 13.5 | 36.7 | 44.7 | 29.5 |
| Source | d.f. | ERG | PHT | LDG | GPC | TWT | YLD |
|---|---|---|---|---|---|---|---|
| Block (B) | 1 | 290.9 | 103.9 | 0.8 | 0.4 | 1.4 | 2,624,730.0 |
| Genotype (G) | 100 | 1963.8 ** | 88.9 * | 0.8 * | 1.1 | 7.2 * | 2,066,536.0 |
| B × G | 100 | 370.1 | 26.5 | 0.3 | 0.2 | 0.3 | 680,597.0 |
| Error | 6 | 144.2 | 27.7 | 0.2 | 0.9 | 1.2 | 1,327,087.0 |
| CV, % | 36.0 | 5.1 | 14.8 | 7.7 | 1.5 | 18.7 |
| Genome and Chromosome | No. of SNPs (from R genome) | No. of Unique Loci (from R Genome) | Map Length (cM) | Marker Density | Map Density | ||
|---|---|---|---|---|---|---|---|
| SNPs/cM | Unique Loci/cM | cM/SNP | cM/Unique Locus | ||||
| A 1 | 238 (0) | 14 (0) | 63.09 | 3.77 | 0.22 | 0.27 | 4.51 |
| 2 | 362 (4) | 46 (4) | 172.09 | 2.10 | 0.26 | 0.48 | 3.74 |
| 3 | 152 (1) | 34 (1) | 195.65 | 0.78 | 0.17 | 1.29 | 5.75 |
| 4 | 195 (2) | 31 (2) | 149.44 | 1.30 | 0.21 | 0.77 | 4.82 |
| 5 | 272 (0) | 42 (0) | 136.00 | 2.00 | 0.31 | 0.50 | 3.24 |
| 6 | 10 (0) | 5 (0) | 17.76 | 0.56 | 0.28 | 1.78 | 3.55 |
| 7 | 792 (2) | 52 (2) | 206.14 | 3.84 | 0.25 | 0.26 | 3.96 |
| Total | 2021 (9) | 224 (9) | 940.17 | 2.15 | 0.24 | 0.47 | 4.20 |
| % | 38.30 (0.17) | 37.77 (1.52) | |||||
| B 1 | 955 (4) | 36 (4) | 75.40 | 12.67 | 0.48 | 0.08 | 2.09 |
| 2 | 274 (1) | 27 (1) | 141.59 | 1.94 | 0.19 | 0.52 | 5.24 |
| 3 | 225 (0) | 28 (0) | 135.99 | 1.65 | 0.20 | 0.60 | 4.86 |
| 4 | 94 (1) | 20 (1) | 77.58 | 1.21 | 0.26 | 0.83 | 3.88 |
| 5 | 479 (3) | 31 (2) | 137.16 | 3.49 | 0.23 | 0.29 | 4.42 |
| 6 | 97 (0) | 22 (0) | 143.39 | 0.68 | 0.15 | 1.48 | 6.52 |
| 7 | 125 (0) | 16 (0) | 49.36 | 2.53 | 0.32 | 0.39 | 3.09 |
| Total | 2249 (9) | 180 (8) | 760.48 | 2.96 | 0.24 | 0.34 | 4.22 |
| % | 42.60 (0.17) | 30.35 (1.35) | |||||
| R 1 | 33 (13) | 11 (6) | 83.62 | 0.39 | 0.13 | 2.53 | 7.60 |
| 2 | 21 (0) | 2 (0) | 7.59 | 2.77 | 0.26 | 0.36 | 3.80 |
| 3 | 45 (13) | 11 (5) | 109.46 | 0.41 | 0.10 | 2.43 | 9.95 |
| 4 | 405 (290) | 69 (42) | 197.97 | 2.05 | 0.34 | 0.49 | 2.87 |
| 5 | 215 (147) | 46 (37) | 219.79 | 0.98 | 0.21 | 1.02 | 4.78 |
| 6 | 260 (79) | 46 (31) | 192.04 | 1.35 | 0.24 | 0.74 | 4.17 |
| 7 | 25 (25) | 4 (4) | 10.99 | 2.27 | 0.36 | 0.44 | 2.75 |
| Total | 1004 (567) | 189 (125) | 821.47 | 1.22 | 0.23 | 0.82 | 4.35 |
| % | 19.04 (10.80) | 31.87 (20.08) | |||||
| A+B+R 1 | 1226 (17) | 61 (10) | 222.10 | 5.52 | 0.27 | 0.18 | 3.64 |
| 2 | 657 (5) | 75 (5) | 321.26 | 2.05 | 0.23 | 0.49 | 4.28 |
| 3 | 422 (14) | 73 (6) | 441.11 | 0.96 | 0.16 | 1.05 | 6.04 |
| 4 | 694 (293) | 120 (45) | 425.00 | 1.63 | 0.28 | 0.61 | 3.54 |
| 5 | 966 (150) | 119 (39) | 492.96 | 1.96 | 0.24 | 0.51 | 4.14 |
| 6 | 367 (79) | 73 (31) | 353.20 | 1.04 | 0.21 | 0.96 | 4.84 |
| 7 | 942 (27) | 72 (6) | 266.50 | 3.53 | 0.27 | 0.28 | 3.70 |
| Total | 5274 (585) | 593 (142) | 2522.13 | 2.09 | 0.23 | 0.48 | 4.25 |
| % | 100.00 (11.10) | 100.00 (23.95) | |||||
| Trait | QTL Name | Chr. | Flanking Markers | Interval | Position | LOD | %R2 | Add. Effect | A*E | D | Location | Other Associated Traits |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DI | QFhi.lrdc-2B | 2B | D_GDEEGVY01A58YG_63-RAC875_C6538_136 | 120.7–141.5 | 139.6 | 2.8 | 11.4 | 4.1 | 1.9 | U | B, O | DS,VRI,GPC |
| (Type-I | QFhi.lrdc-4A | 4A | TDURUM_CONTIG41185_259-TDURUM_CONTIG12696_528 | 14.4–38.8 | 33.6 | 3.1 | 14.1 | −3.4 | −1.5 | T | B, O | - |
| Resistance) | QFhi.lrdc-5R | 5R | TDURUM_CONTIG50875_569-GENE-3619_813 | 52.4–81.2 | 62.4 | 5.2 | 23.0 | −5.8 | −3.6 | T | B, O | DS, VRI, TWT |
| DS | QFhs.lrdc-1B | 1B | EXCALIBUR_C415_844-TDURUM_CONTIG100292_228 | 27.8–34.8 | 31.1 | 2.5 | 9.3 | −2.7 | −1.3 | T | B, O | - |
| (Type-II | QFhs.lrdc-2B | 2B | WSNP_EX_C1962_3696265-BOBWHITE_C2988_2161 | 117.2–128.8 | 123.4 | 2.7 | 11.4 | 3.5 | 1.3 | U | B, O | DI, VRI, GPC |
| Resistance) | QFhs.lrdc-5R.1 | 5R | TDURUM_CONTIG50875_569-GENE-3619_813 | 47.7–87.9 | 77.0 | 3.7 | 20.6 | −4.5 | - | T | B, O | DI, VRI, TWT |
| QFhs.lrdc-5R.2 | 5R | EXCALIBUR_C31769_793-BOBWHITE_C20682_107 | 113.5–116.8 | 114.7 | 3.2 | 15.1 | −3.9 | −1.6 | T | B, O | VRI, DON, PHT | |
| QFhs.lrdc-6B | 6B | TDURUM_CONTIG29162_378-CAP12_C2970_177 | 31.6–69.3 | 33.8 | 2.5 | 10.5 | −3.2 | −1.1 | T | B, O | ERG, PHT | |
| VRI | QFhb.lrdc-1A.1 | 1A | EXCALIBUR_REP_C113950_132-WSNP_KU_C3804_6986527 | 6.9–32.7 | 16.2 | 3.3 | 22.0 | −0.5 | - | T | B, O | DON, TWT |
| QFhb.lrdc-2B | 2B | D_GDEEGVY01A58YG_63-RAC875_C6538_136 | 117.7–141.5 | 127.8 | 7.7 | 32.0 | 3.8 | - | U | O | DI, DS, GPC | |
| QFhb.lrdc-3A.1 | 3A | TDURUM_CONTIG25619_350-CAP12_C1860_280 | 0.0–7.3 | 4.2 | 3.3 | 4.9 | 2.2 | - | U | B | - | |
| QFhb.lrdc-4R ❀ | 4R | IACX481-BOBWHITE_C25209_235 | 145.5–147.7 | 145.5 | 3.5 | 16.0 | −3.7 | −2.6 | T | B | DON, LDG, TWT | |
| QFhb.lrdc-5A | 5A | RAC875_C7132_134-EXCALIBUR_C92705_94 | 95.5–96.8 | 96.0 | 6.8 | 29.0 | 12.0 | - | U | B | TWT | |
| QFhb.lrdc-5R.1 | 5R | TDURUM_CONTIG50875_569-GENE-3619_813 | 58.4–86.2 | 77.2 | 3.4 | 17.5 | −3.5 | 1.7 | T | B, O | DI, DS, TWT | |
| QFhb.lrdc-5R.2 | 5R | ISOTIG29606-C23413_289 | 98.1–107.5 | 103.9 | 5.9 | 26.0 | −2.9 | - | T | B | DS, DON, PHT | |
| DON | QDon.lrdc-1A | 1A | EXCALIBUR_REP_C113950_132-WSNP_KU_C3804_6986527 | 6.9–32.7 | 16.2 | 8.3 | 34.1 | −3.0 | - | T | O | VRI, TWT |
| (Type-III | QDon.lrdc-3A | 3A | WSNP_CAP11_REP_C4157_1965583-WSNP_EX_C8409_14170476 | 32.8–39.3 | 34.1 | 5.9 | 26.0 | 2.4 | - | U | O | - |
| Resistance) | QDon.lrdc-4R.1 | 4R | ISOTIG22327-KU_C39706_716 | 94.8–121.4 | 114.2 | 3.3 | 15.0 | 1.8 | 1.3 | U | B, O | LDG |
| QDon.lrdc-4R.2 ❀ | 4R | IACX481-BOBWHITE_C25209_235 | 145.5–147.7 | 146.0 | 3.1 | 4.1 | −1.2 | - | T | B | VRI, LDG, TWT | |
| QDon.lrdc-4R.3 | 4R | ISOTIG23307-ISOTIG20098 | 147.9–176.2 | 152.3 | 3.0 | 2.9 | −1.2 | - | T | B | VRI, LDG, TWT | |
| QDon.lrdc-5A | 5A | TDURUM_CONTIG15970_217-EXCALIBUR_C32414_705 | 115.9–126.5 | 119.3 | 6.9 | 30.0 | 2.7 | - | U | B | PHT | |
| QDon.lrdc-5R | 5R | TPLB0059H22_1233-CONTIG2043 | 115.1–134.5 | 119.6 | 3.3 | 5.6 | −1.4 | - | T | O | DS, VRI, PHT |
| Trait | QTL Name | Chr. | Flanking Markers | Interval | Position | Add Effect | p Value | AA*E | p Value | D | Location | Other Associated Trait |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DS (Type-II | QFhs.lrdc-2A | 2A | BOBWHITE_C24021_254-BS00022456_51 | 120.1–124.3 | 122.3 | 0.9 | 0.000630 | 1.8 | 0.000054 | U | B, O | PHT |
| Resistance) | QFhs.lrdc-3B | 3B | RAC875_C7158_687-BS00074688_51 | 99.4–106.6 | 103.9 | - | ||||||
| VRI | QFhb.lrdc-1A.2 | 1A | WSNP_BF474340A_TA_2_1-EXCALIBUR_REP_C77271_230 | 38.8–49.5 | 44.2 | 1.8 | 0.000013 | −1.4 | 0.026233 | U | B, O | - |
| QFhb.lrdc-3A.2 | 3A | RAC875_REP_C106170_204-WSNP_EX_C55096_57733894 | 175.9–194.3 | 184.5 | - |
| Trait | QTL Name | Chr. | Flanking Markers | Interval | Position | LOD | %R2 | Add Effect | A*E | D | Other Associated Traits |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ergot | QErg.lrdc-4A | 4A | ISOTIG23128-TDURUM_CONTIG41185_259 | 10.8–24.3 | 20.3 | 2.5 | 11.0 | 8.9 | 6.2 | U | YLD |
| QErg.lrdc-5R | 5R | ISOTIG30930-ISOTIG29158 | 138.7–147.4 | 143.1 | 2.6 | 12.0 | −6.4 | −3.9 | T | GPC | |
| QErg.lrdc-7A | 7A | EXCALIBUR_C30713_591-WSNP_EX_C5177_9174930 | 132.5–142.5 | 136.3 | 8.1 | 34.0 | 34.0 | 5.6 | U | - | |
| GPC | QGpc.lrdc-2A | 2A | BS00022241_51-BS00081194_51 | 89.7–91.9 | 90.8 | 9.8 | 39.0 | −0.3 | - | U | - |
| QGpc.lrdc-2B | 2B | BOBWHITE_C2988_2161-BS00022572_51 | 106.5–138.6 | 127.8 | 2.5 | 6.1 | 0.1 | 0.1 | T | DI, DS, VRI | |
| QGpc.lrdc-4R ❀ | 4R | C11841_198-C1659_1301 | 35.0–43.9 | 39.8 | 8.9 | 36.0 | −0.7 | −0.3 | U | YLD | |
| QGpc.lrdc-5R | 5R | ISOTIG30930-ISOTIG29158 | 132.9–145.0 | 141.1 | 9.0 | 37.0 | −0.1 | −0.2 | U | ERG | |
| QGpc.lrdc-6R | 6R | TDURUM_CONTIG41906_1479-WSNP_EX_C7002_12063380 | 17.6–20.0 | 19.9 | 3.0 | 17.8 | −0.5 | - | U | PHT, LDG, YLD | |
| QGpc.lrdc-7B | 7B | EXCALIBUR_C3489_182-BOBWHITE_C20735_255 | 0.0–5.1 | 0.0 | 6.0 | 26.0 | −0.2 | −0.2 | U | - | |
| TWT | QTwt.lrdc.1R | 1R | RAC875_REP_C116934_270-ISOTIG11711 | 69.4–78.6 | 69.5 | 3.1 | 15.0 | 0.5 | - | T | - |
| QTwt.lrdc.4R ❀ | 4R | BOBWHITE_C25209_235-ISOTIG19779 | 147.7–156.6 | 152.1 | 4.5 | 21.0 | 0.6 | - | T | VRI, DON, LDG | |
| QTwt.lrdc-5A.1 | 5A | EXCALIBUR_C34451_370-KUKRI_C9358_269 | 2.0–7.4 | 3.4 | 4.7 | 21.5 | −0.6 | −0.9 | U | - | |
| QTwt.lrdc-5A.2 | 5A | RAC875_C60453_122-WSNP_EX_C31154_39982416 | 100.2–109.2 | 101.4 | 5.3 | 23.0 | 0.7 | - | T | VRI | |
| QTwt.lrdc-5R | 5R | TDURUM_CONTIG50875_569-GENE-3619_813 | 23.4–77.1 | 50.1 | 2.5 | 12.0 | 0.6 | - | T | DI, DS, VRI | |
| YLD | QYld.lrdc-4A | 4A | ISOTIG23128-TDURUM_CONTIG41185_259 | 16.4–22.3 | 20.3 | 2.9 | 14.1 | −342.0 | −128.1 | U | ERG |
| QYld.lrdc-4R.2 ❀ | 4R | C1659_1301-GENE-1569_376 | 39.8–45.4 | 41.9 | 3.1 | 15.0 | −97.6 | −203.3 | U | GPC | |
| QYld.lrdc-6A | 6A | BS00110902_51-KUKRI_C20401_505 | 7.2–17.3 | 11.8 | 2.7 | 13.0 | −358.4 | - | U | - | |
| QYld.lrdc-6R | 6R | TDURUM_CONTIG41906_1479-WSNP_EX_C7002_12063380 | 16.2–19.9 | 17.6 | 2.5 | 12.0 | −371.0 | −251.1 | U | GPC, PHT, LDG | |
| PHT | QPht.lrdc-5A | 5A | CAP8_C317_307-GENE-2725_244 | 109.3–112.7 | 112.4 | 2.7 | 13.0 | −1.9 | 2.2 | T | DON |
| QPht.lrdc-5R | 5R | TPLB0059H22_1233-CONTIG2043 | 108.7–117.9 | 116.9 | 2.7 | 13.0 | −2.4 | −2.3 | T | DS, VRI, DON | |
| QPht.lrdc-6R | 6R | KUKRI_C9940_659-TDURUM_CONTIG41906_1479 | 2.1–27.1 | 16.3 | 9.4 | 38.0 | −3.2 | - | T | GPC, LDG, YLD | |
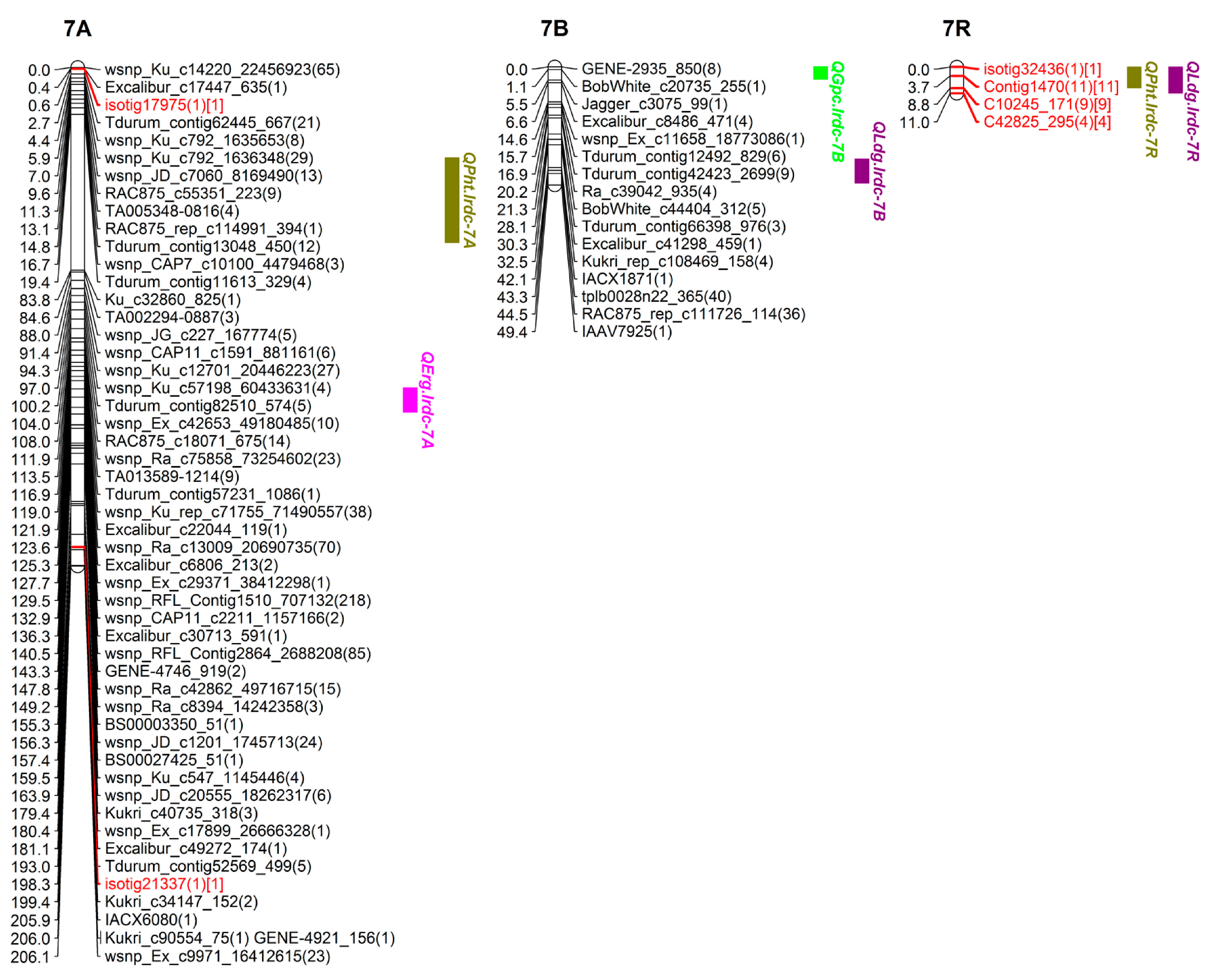
| QPht.lrdc-7R | 7R | ISOTIG32436-ISOTIG07139 | 0.0–8.7 | 1.0 | 2.5 | 11.0 | −1.0 | −1.7 | T | LDG | |
| LDG | QLdg.lrdc-2R | 2R | EXCALIBUR_C46590_290-BS00061187_51 | 1.0–7.0 | 7.0 | 2.5 | 11.7 | −0.1 | −0.1 | T | - |
| QLdg.lrdc-4R.1 | 4R | ISOTIG25347-ISOTIG22327 | 89.8–101.2 | 97.2 | 6.0 | 26.0 | 0.3 | 0.1 | U | DON | |
| QLdg.lrdc-4R.2 | 4R | ISOTIG17427-ISOTIG19779 | 153.3–161.7 | 156.4 | 5.9 | 26.0 | 0.1 | 0.1 | U | VRI, DON, TWT | |
| QLdg.lrdc-6R ❀ | 6R | TDURUM_CONTIG8849_343-ISOTIG24496 | 8.2–9.9 | 9.8 | 2.9 | 14.1 | −0.14 | −0.1 | T | GPC, PHT, YLD | |
| QLdg.lrdc-7B | 7B | WSNP_KU_C10355_17149304-IBV7925 | 38.5–48.5 | 44.5 | 2.5 | 6.0 | −0.1 | −0.1 | T | - | |
| QLdg.lrdc-7R | 7R | C10245_171-C6434_652 | 0.0–10.8 | 8.8 | 3.0 | 14.1 | −0.2 | - | T | PHT |
| Trait | QTL Name | Chr. | Flanking Markers | Interval | Position | Add Effect | p Value | AA*E | p Value | D | Other Associated Traits |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERG | QErg.lrdc-6B | 6B | CAP12_C2970_177-CAP7_C3697_87 | 57.3–86.1 | 74.3 | 3.3 | 0.002321 | 6.0 | 0.000859 | U | DS, PHT |
| QErg.lrdc-6R | 6R | ISOTIG07765-ISOTIG30810 | 128.6–132.2 | 131.2 | - | ||||||
| TWT | QTwt.lrdc-1A | 1A | RAC875_C89908_105-WSNP_KU_C3804_6986527 | 11.6–22.3 | 16.3 | 0.6 | 0.015108 | 0.9 | 0.029488 | T | DON, VRI |
| QTwt.lrdc-4B | 4B | EXCALIBUR_REP_C102761_90-BS00023179_51 | 38.2–43.9 | 41.7 | - | ||||||
| YLD | QYld.lrdc.1A | 1A | WSNP_EX_C18499_27344859-BS00070695_51 | 48.5–63.0 | 60.2 | 62.2 | 0.043176 | 102.1 | 0.026707 | T | - |
| QYld.lrdc.2A | 2A | ISOTIG09181-EXCALIBUR_REP_C102984_157 | 165.1–171.8 | 170.8 | - | ||||||
| QYld.lrdc.3R | 3R | IACX7129-C3152_1508 | 68.6–102.2 | 92.4 | −108.9 | 0.000227 | −219.0 | 0.000010 | U | - | |
| QYld.lrdc.4R.1 ❀ | 4R | TDURUM_CONTIG42642_656-ISOTIG33290 | 49.0–59.9 | 55.9 | - | ||||||
| QYld.lrdc-4R.3 | 4R | ISOTIG29781-ISOTIG09995 | 127.6–134.0 | 129.9 | 85.1 | 0.005414 | 159.4 | 0.001273 | T | - | |
| QYld.lrdc-6B | 6B | RFL_CONTIG2206_1694-IACX2322 | 89.5–102.4 | 100.4 | - | ||||||
| PHT | QPht.lrdc-2A | 2A | EXCALIBUR_REP_C111191_119-EXCALIBUR_C24354_465 | 110.4–137.0 | 131.2 | 1.5 | 0.001168 | 3.0 | 0.000160 | U | DS |
| QPht.lrdc-6B | 6B | CAP12_C2970_177-CAP7_C3697_87 | 33.8–71.3 | 53.3 | ERG, DS | ||||||
| QPht.lrdc-2B | 2B | EXCALIBUR_REP_C114249_187-EXCALIBUR_C37751_354 | 0.0–18.4 | 14.6 | 2.2 | 0.001360 | 4.1 | 0.000319 | U | - | |
| QPht.lrdc-7A | 7A | CAP11_C3214_133-KU_C32860_825 | 37.4–72.4 | 53.4 | - | ||||||
| QPht.lrdc-5A | 5A | CAP8_C317_307-GENE-2725_244 | 109.3–112.7 | 112.4 | 0.9 | 0.013789 | 1.4 | 0.012962 | T | DON | |
| QPht.lrdc-7R | 7R | ISOTIG32436-ISOTIG07139 | 0.0–8.7 | 1.0 | LDG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhariwal, R.; Fedak, G.; Dion, Y.; Pozniak, C.; Laroche, A.; Eudes, F.; Randhawa, H.S. High Density Single Nucleotide Polymorphism (SNP) Mapping and Quantitative Trait Loci (QTL) Analysis in a Biparental Spring Triticale Population Localized Major and Minor Effect Fusarium Head Blight Resistance and Associated Traits QTL. Genes 2018, 9, 19. https://doi.org/10.3390/genes9010019
Dhariwal R, Fedak G, Dion Y, Pozniak C, Laroche A, Eudes F, Randhawa HS. High Density Single Nucleotide Polymorphism (SNP) Mapping and Quantitative Trait Loci (QTL) Analysis in a Biparental Spring Triticale Population Localized Major and Minor Effect Fusarium Head Blight Resistance and Associated Traits QTL. Genes. 2018; 9(1):19. https://doi.org/10.3390/genes9010019
Chicago/Turabian StyleDhariwal, Raman, George Fedak, Yves Dion, Curtis Pozniak, André Laroche, François Eudes, and Harpinder Singh Randhawa. 2018. "High Density Single Nucleotide Polymorphism (SNP) Mapping and Quantitative Trait Loci (QTL) Analysis in a Biparental Spring Triticale Population Localized Major and Minor Effect Fusarium Head Blight Resistance and Associated Traits QTL" Genes 9, no. 1: 19. https://doi.org/10.3390/genes9010019
APA StyleDhariwal, R., Fedak, G., Dion, Y., Pozniak, C., Laroche, A., Eudes, F., & Randhawa, H. S. (2018). High Density Single Nucleotide Polymorphism (SNP) Mapping and Quantitative Trait Loci (QTL) Analysis in a Biparental Spring Triticale Population Localized Major and Minor Effect Fusarium Head Blight Resistance and Associated Traits QTL. Genes, 9(1), 19. https://doi.org/10.3390/genes9010019





