Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants and Growing Conditions
2.2. Cloning and Sequence Analysis
2.3. Subcellular Localization
2.4. Quantitative Real Time-PCR
2.5. Plasmid Constructs and Plant Transformation
2.6. Yeast Two-Hybrid Assay
2.7. BIFC Assay
3. Results
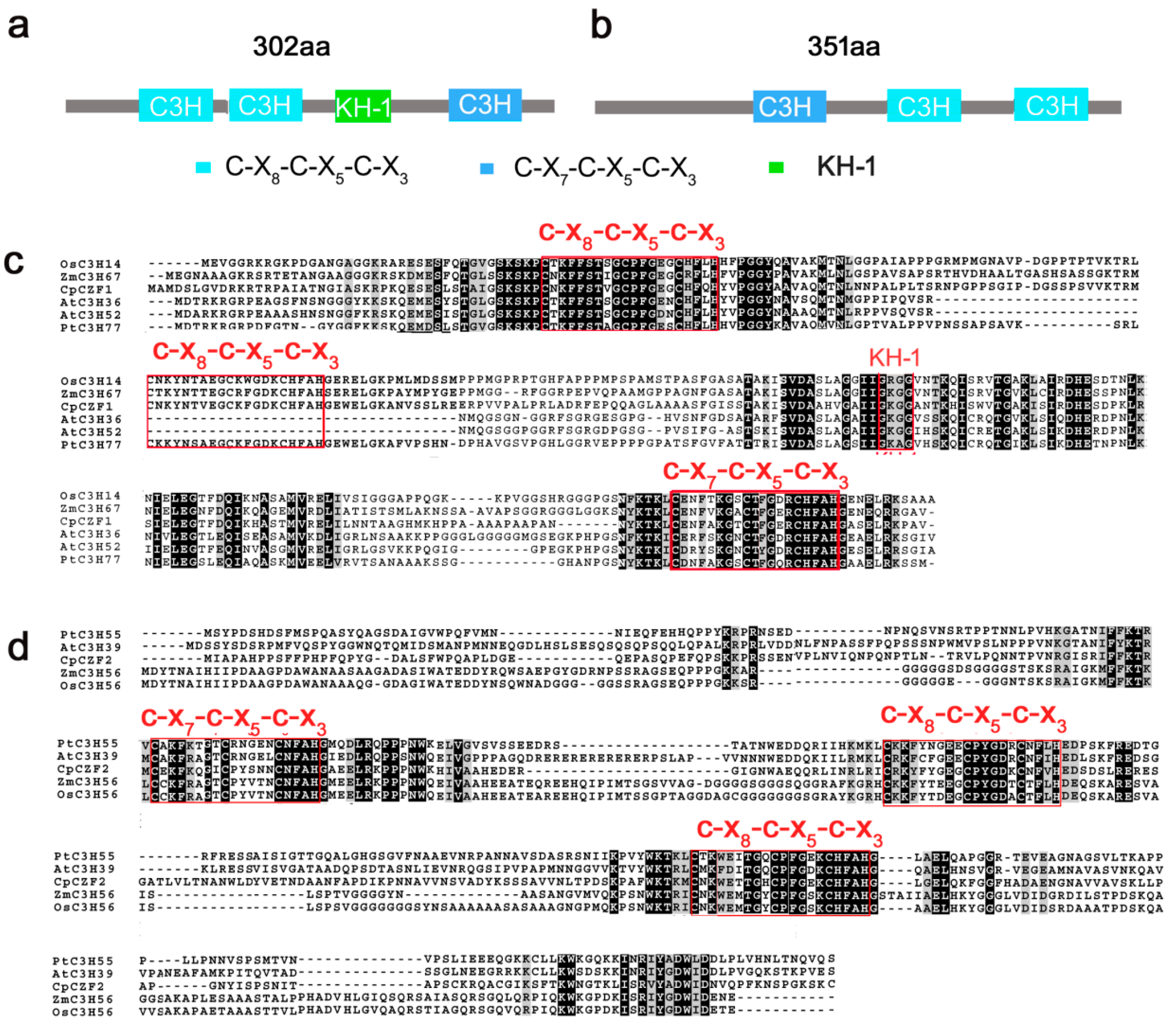
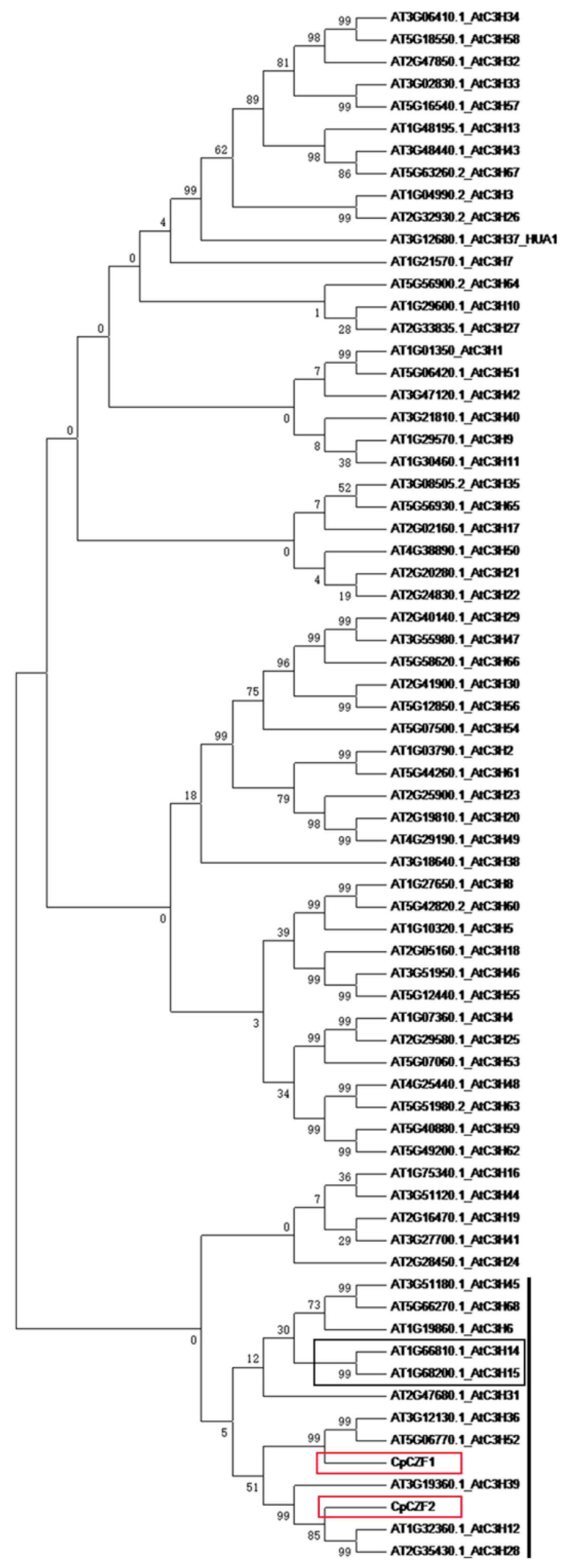
3.1. Isolation and Characterization of CpCZF1 and CpCZF2
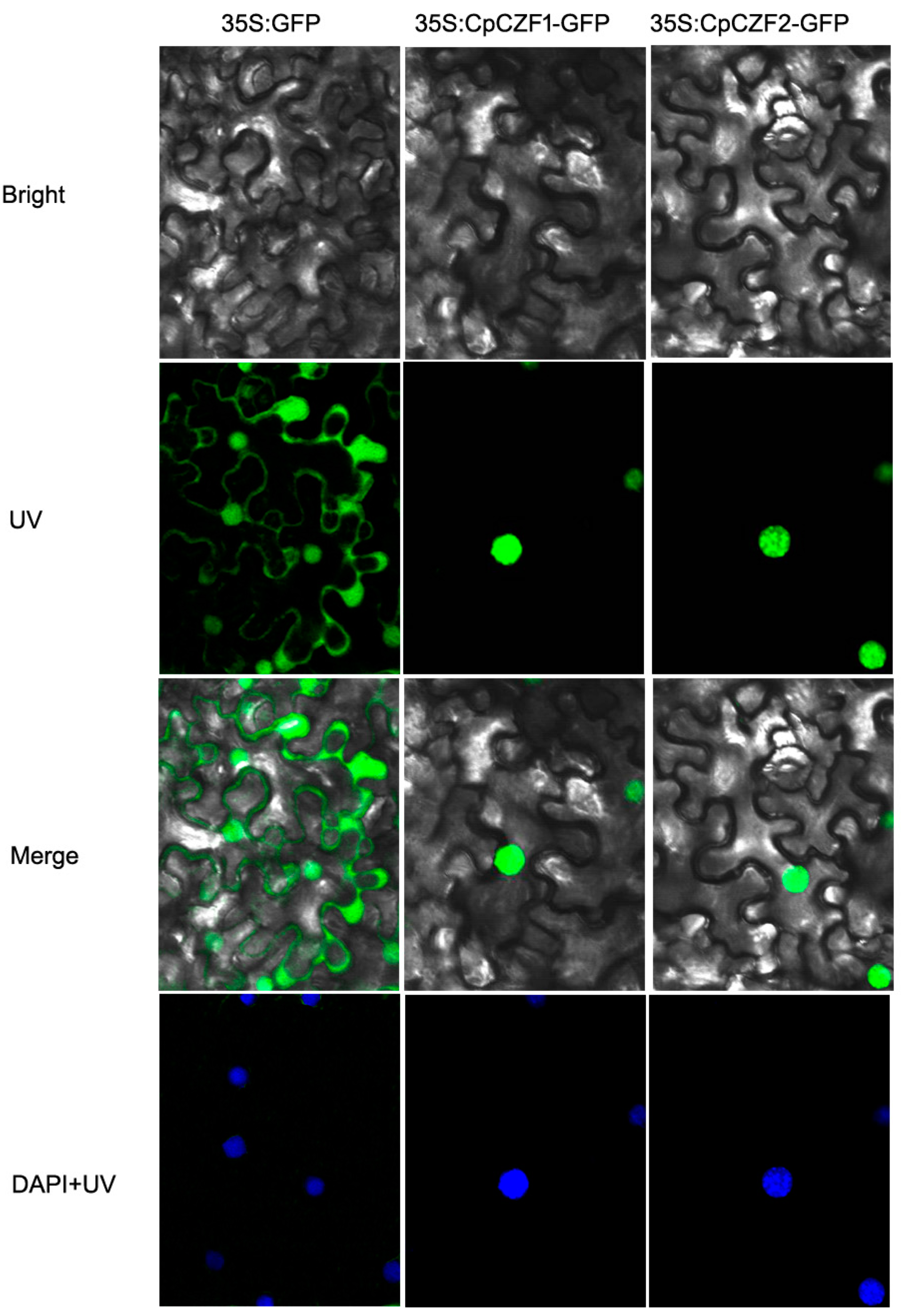
3.2. CpCZF1 and CpCZF2 Located to Nucleus
3.3. The Spatiotemporal Expression Patterns of CpCZF1 and CpCZF2
3.4. Effects of CpCZF1 and CpCZF2 Genes on Flower Organ Identities in Arabidopsis
3.5. Expression of the Stamen Identity-Related Genes
3.6. Protein Interaction of CpCZF1 with CpCZF2
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| C3H | Cysteine3Histidine |
| DDO/X | SD/-Trp/-Leu/X-α-Gal |
| QDO/X/A | SD/-Trp/-Leu/-His/-Ade/X-α-Gal/Aureobasidin |
| qRT-PCR | Quantitative reverse transcriptase–polymerase chain reaction |
| Y2H | Yeast two hybrid |
| WT | Wild type |
| OE | Overexpression |
References
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201. [Google Scholar] [CrossRef]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Jack, T.; Fox, G.L.; Meyerowitz, E.M. Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 1994, 76, 703–716. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Irish, F.V.; Sussex, I.M. Function of the APETALA-1 gene during Arabidopsis floral developmen. Plant Cell 1990, 2, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; de Boer, B.G.W.; MontaguIb, M.V.; Okamuroa, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Alvarez-Buylla, E.R.; Benítez, M.; Corvera-Poiré, A.; Chaos Cador, A.; de Folter, S.; Gamboa de Buen, A.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V.; et al. Flower development. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2010; Volume 8, pp. 1–57. [Google Scholar]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E.; et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.B.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [PubMed]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [PubMed]
- Li, J.; Jia, D.; Chen, X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Meyerowitz, E.M. HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol. Cell 1999, 3, 349–360. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, J.; Kato, N.; Chen, X.; Wang, W. Two RNA binding proteins, HEN4 and HUA1, act in the processing of agamous Pre-mRNA in Arabidopsis thaliana. Dev. Cell 2003, 4, 53–66. [Google Scholar] [CrossRef]
- Li, W.; He, M.; Wang, J.; Wang, Y. Zinc finger proteins(ZFP) in plants—A review. Plant Omics J. 2013, 6, 474–480. [Google Scholar]
- Berg, J.M.; Shi, Y.G. The galvanization of biology: A growing appreciation for the roles of zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Bogamuwa, S.P.; Jang, J.C. Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 2014, 55, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhao, Y.; Cao, J.; Zhang, W.; Jiang, H.; Li, X.; Ma, Q.; Zhu, S.; Cheng, B. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.H.; Kim, S.K.; Xu, Z.; Chong, K. OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Tazuke, A.; Asayama, M. Expression of CsSEF1 gene encoding putative CCCH zinc finger protein is induced by defoliation and prolonged darkness in cucumber fruit. Planta 2013, 237, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Bogamuwa, S.; Jang, J.C. The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ. 2013, 36, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jiang, H.; Xu, Y.; Li, H.; Wu, X.; Xie, Q.; Li, C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007, 48, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Kong, Y.; Zhu, M.; Yu, L.; Qi, G.; Tang, X.; Wang, Z.; Cao, Y.; Yu, C.; Zhou, G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J. Exp. Bot. 2015, 66, 2595–2609. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.C.; Kim, J.Y.; Ko, J.H.; Kang, H.; Kim, J.; Han, K.H. AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mrnas in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2014, 80, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.H.; Yu, Y.P.; Wang, D.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zheng, C.C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21a and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Zhang, T.; Yang, Q.; Kang, J.; Sun, Y.; Gruberc, M.Y.; Qin, Z. Expression of the alfalfa CCCH-type zinc finger protein gene MsZFN delays flowering time in transgenic Arabidopsis thaliana. Plant Sci. Int. J. Exp. Plant Biol. 2014, 215–216, 92–99. [Google Scholar]
- Sui, S.; Luo, J.; Ma, J.; Zhu, Q.; Lei, X.; Li, M. Generation and analysis of expressed sequence tags from Chimonanthus praecox (wintersweet) flowers for discovering stress-responsive and floral development-related genes. Comp. Funct. Genom. 2012. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Sui, S.; Ma, J.; Li, Z.; Guo, Y.; Luo, D.; Yang, J.; Li, M. Transcriptomic analysis of flower development in wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-H.; Jia, B.; Zhuo, R.-Y.; Liu, J.-L.; Pan, H.-Y.; Baldwin, T.C.; Zhang, S.-H. An Acyl-Acyl carrier protein thioesterase gene isolated from wintersweet (Chimonanthus praecox), CpFATB, enhances drought tolerance in transgenic tobacco (Nicotiana tobaccum). Plant Mol. Biol. Rep. 2011, 30, 433–442. [Google Scholar] [CrossRef]
- Cao, L.; Wu, B.; Wu, W.; Chen, L.; Hu, X.; Xu, Z.; Zheng, Y. Analysis of essential oil from pericarps of Chimonanthus praecox. Asian J. Chem. 2013, 25, 671–674. [Google Scholar]
- Wang, B.-G.; Duan, K.; Zhang, Q.; Pan, A.-H.; Sui, S.-Z.; Li, M.-Y.; Wang, L.-G.; Xue-Min, T. The AGL6-like gene CpAGL6, a potential regulator of floral time and organ identity in wintersweet (Chimonanthus praecox). Plant Growth Regul. 2011, 30, 343–352. [Google Scholar] [CrossRef]
- Wu, C.; Hu, N. Studies on the flower form and blooming characteristics of the wintersweet. Acta Hortic. Sin. 1995, 22, 277–282. (In Chinese) [Google Scholar]
- Ma, J.; Li, Z.; Wang, B.; Sui, S.Z.; Li, M.Y. Cloning of an expansin gene from chimonanthus praecox flowers and its expression in flowers treated with ethephon or 1-methylcyclopropene. Hortscience 2012, 47, 1472–1477. [Google Scholar]
- Weinthal, D.; Tzfir, T. Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 2009, 14, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Sanchez, I.E.; Maruri-Lopez, I.; Ferrando, A.; Carbonell, J.; Graether, S.P.; Jimenez-Bremont, J.F. Nuclear localization of the dehydrin OPSDHN1 is determined by histidine-rich motif. Front. Plant Sci. 2015, 6, 702. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The jasmonate-zim domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Tonaco, I.; Boutilier, K.; Immink, R. A cautionary note on the use of split-YFP/BIFC in plant protein-protein interaction studies. Int. J. Mol. Sci. 2014, 15, 9628–9643. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiang, H.; Xiang, Y.; Zhang, H.; Zhu, S.; Zhao, Y.; Cheng, B. Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 2013, 32, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Hu, R.; Zhang, D.; Qi, G.; Zuo, R.; Cao, Y.; Chen, P.; Kong, Y.; Zhou, G. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genom. 2012, 13, 253. [Google Scholar] [CrossRef] [PubMed]
- Xu, R. Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Thomas, T.L. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 1998, 10, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Qi, G.; Cao, Y.; Wang, Z.; Yu, L.; Tang, X.; Yu, Y.; Wang, D.; Kong, Y.; Zhou, G. Poplar PdC3H17 and pdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol. 2014, 203, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Saedler, H. Floral quartets. Nature 2001, 409, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Melzer, R.; Rumpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Lewis, M.W.; Fletcher, J.C. RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J. Cell Mol. Biol. 2006, 45, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Matsumoto, N.; Okada, K. RABBIT EARS, encoding a Superman-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 2011, 138, 3591. [Google Scholar] [CrossRef]
- Ingram, G.C.; Goodrich, J.; Wilkinson, M.D.; Simon, R.; Haughn, G.W.; Coen, E.S. Parallels between unusual floral organs and fimbriata, genes controlling flower development in Arabidopsis and Antirrhinu. Plant Cell 1995, 7, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Li, W.; Dong, X.; Guo, W.; Shu, H. Ectopic expression of a hyacinth AGL6 homolog caused earlier flowering and homeotic conversion in Arabidopsis. Sci. China C Life Sci. 2007, 50, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qi, T.; Ma, J.; Ma, T.; Ma, L.; Lin, X. Ectopic expression of a SOC1 homolog from phyllostachys violascens alters flowering time and identity of floral organs in Arabidopsis thaliana. Trees 2016, 30, 2203–2215. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis Wrky18, Wrky40, and Wrky60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [PubMed]


| Genotype | No. of Transgenic Lines | Phenotype |
|---|---|---|
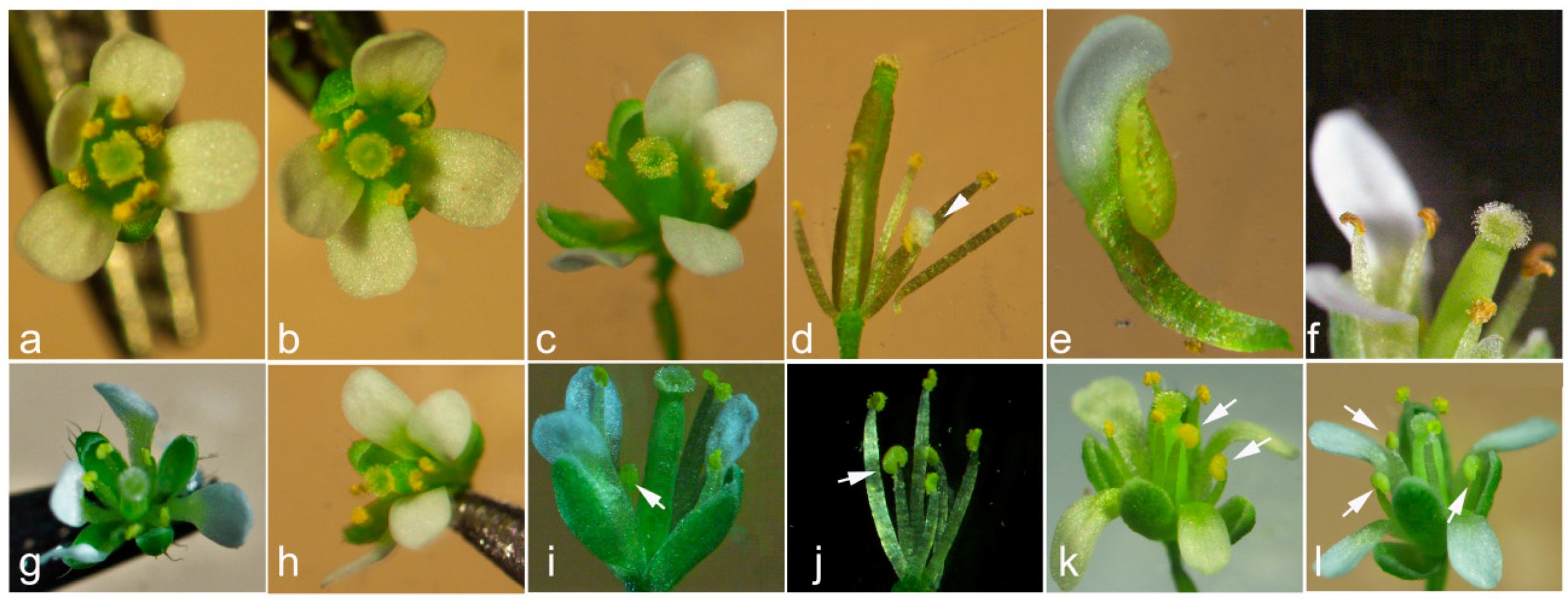
| 35S:CpCZF1 | 6 | stamen number reduced and/or anthers withered before the pistil could be fertilized (type 1: Figure 5b,c,f) |
| 12 | stamen number reduced and /or partial petaliod lateral stamen (type 2: Figure 5b–e) | |
| 35S:CpCZF2 | 9 | stamen number reduced (type 3: Figure 5g,h) |
| 21 | stamen number reduced and /or stamen(s) changed to staminode (s) (type 4: Figure 5g–i) |
| Genotype | Type | 1–30 a/90 b | |||||
|---|---|---|---|---|---|---|---|
| Lost 1 Stamen | Lost 2 Stamens | Petaliod | Withered Anther | Staminode(s) | Ratio | ||
| WT | 4 | 0 | 0 | 0 | 0 | 0.04 | |
| 35S:CpCZF1 | 1 | 32 | 14 | 0 | 12 | 0 | 0.64 |
| 2 | 30 | 12 | 11 | 0 | 0 | 0.59 | |
| 35S:CpCZF2 | 3 | 33 | 12 | 0 | 0 | 0 | 0.5 |
| 4 | 33 | 16 | 0 | 0 | 13 | 0.69 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Huang, R.; Ma, J.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes 2017, 8, 199. https://doi.org/10.3390/genes8080199
Liu H, Huang R, Ma J, Sui S, Guo Y, Liu D, Li Z, Lin Y, Li M. Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes. 2017; 8(8):199. https://doi.org/10.3390/genes8080199
Chicago/Turabian StyleLiu, Huamin, Renwei Huang, Jing Ma, Shunzhao Sui, Yulong Guo, Daofeng Liu, Zhineng Li, Yechun Lin, and Mingyang Li. 2017. "Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis" Genes 8, no. 8: 199. https://doi.org/10.3390/genes8080199
APA StyleLiu, H., Huang, R., Ma, J., Sui, S., Guo, Y., Liu, D., Li, Z., Lin, Y., & Li, M. (2017). Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes, 8(8), 199. https://doi.org/10.3390/genes8080199






