Assessing Genetic Diversity and Population Differentiation of Colored Calla Lily (Zantedeschia Hybrid) for an Efficient Breeding Program
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. SSR Statistics and Distribution, and Functional Annotation of SSR-Containing Unigenes
2.3. EST-SSR Marker Cross-Species Transferability and Verification in Colored Calla Lily Accessions
2.4. Data Analysis
3. Results and Discussion
3.1. Features of SSRs from White Calla Lily EST-Unigenes
3.2. Functional Annotation of White Calla Lily ESTs Containing SSR Loci
3.3. Cross-Species Transferability and Genetic Diversity of EST-SSR Markers in Colored Calla Lily
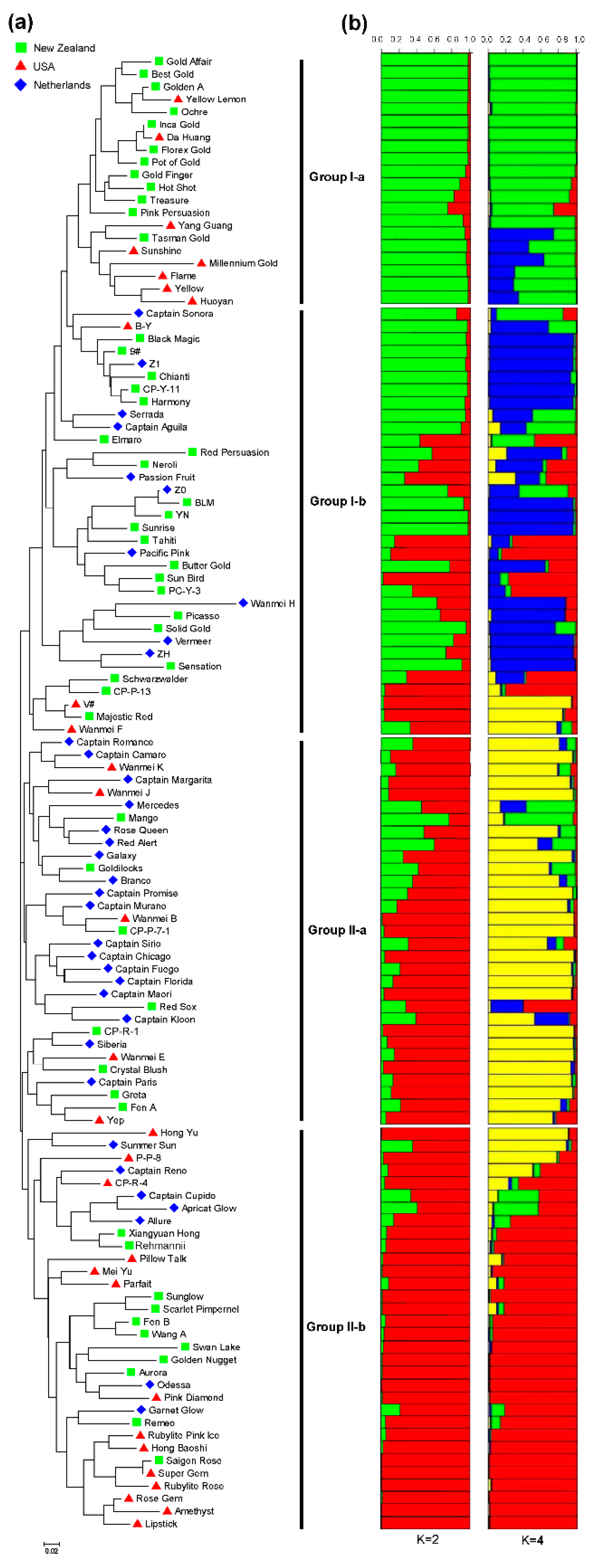
3.4. Genetic Relatedness and Population Structure among 117 Colored Calla Lily Accessions
3.5. Genetic Diversity and Population Differentiation within or among Countries and Inferred Groups or Sub-Groups of 117 Accessions of Colored Calla Lily
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Earl, G. Chromosome numbers and speciation in the genus Zantedeschia. Plant Life 1957, 13, 140–146. [Google Scholar]
- Letty, C. The genus Zantedeschia. Bothalia 1973, 11, 5–26. [Google Scholar] [CrossRef]
- Singh, Y.; Wyk, A.E.V.; Baijnath, H. Taxonomic notes on the genus Zantedeschia Spreng (Araceae) in Southern Africa. S. Afr. J. Bot. 1996, 66, 321–324. [Google Scholar] [CrossRef]
- Yao, J.L. Interspecific Hybridisation and Molecular Characterisation of Hybrids in the Genus Zantedeschia. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 1992. [Google Scholar]
- Funnell, K.A. Zantedeschia. In The Physiology of Flower Bulbs, 1st ed.; Hertogh, A.D., LeNard, M., Eds.; Elsevier: Amsterdam, The Netherlands, 1993; Volume 1, pp. 683–703. [Google Scholar]
- Aitken, A.G.; Hewett, E.W. Fresh Facts: New Zealand Horticulture. Available online: http//:www.hortresearch.co.nz/files/aboutus/factsandfigs/ff2011.pdf (accessed on 12 February 2012).
- Snijder, R.C. Genetics of Erwinia Resistance in Zantedeschia: Impact of Plastome-Genome Incompatibility. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2004. [Google Scholar]
- Snijder, R.C.; Brown, F.S.; Tuyl, J.M. The role of plastome-genome incompatibility and biparental plastid inheritance in interspecific hybridization in the genus Zantedeschia (Araceae). Floric. Ornam. Biotechnol. 2007, 1, 150–157. [Google Scholar]
- Friedt, W.R.; Snowdon, F.; Ordon, A.J. Plant breeding: Assessment of genetic diversity in crop plants and its exploitation in breeding. Prog. Bot. 2007, 68, 151–178. [Google Scholar]
- Hamada, K.; Hagimori, M. RAPD-based method for cultivar-identification of calla lily (Zantedeschia spp.). Sci. Hortic. 1996, 65, 215–218. [Google Scholar] [CrossRef]
- Lu, B.; Zheng, Y.H.; Peng, F.; Shu, X.C.; Chen, X.X. Optimization of RAPD reaction system by uniform design on Zantedeschica hybrid. North. Hortic. 2012, 11, 123–126. [Google Scholar]
- Zhang, Y.C.; Tang, G.G.; Chu, Y.X.; Wu, A.Z. Optimization of ISSR reaction system and preliminary study on Zantedeschia. Mol. Plant Breed. 2009, 7, 827–832. [Google Scholar]
- Chen, Z.; Xu, B.L.; Pu, C.J.; Xu, Q.; Tian, G. Physiological and biochemical and resistance changes and ISSR polymorphic analysis Exposed to 12C6+ heavy ion radiation on calla lily. J. Nucl. Agric. Sci. 2013, 27, 552–556. [Google Scholar]
- Zheng, Y.H.; Zhang, Z.; Lu, B.; Han, F.G.; Peng, F.; Hua, X.P. The influence of polyploidy induction and 60Co heavy ion radiation on calla lily cultivar ‘Parfait’ genetic variation. Jiangsu Agric. Sci. 2015, 43, 142–144. [Google Scholar]
- Li, X.L. Anther Culture of Zantedeschia aethiopica and the Characterization of Regenerated Plants. Master’s Thesis, Yunnan University, Kunming, China, 2011. [Google Scholar]
- Tan, C.C.; Wu, Y.Q.; Taliaferro, C.M.; Anderson, M.P. Development of simple sequence repeat markers for bermudagrass from its expressed sequence tag sequences and preexisting sorghum SSR markers. Mol. Breed. 2012, 29, 23–30. [Google Scholar] [CrossRef]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Squirrell, J.; Hollingsworth, P.M.; Woodhead, M.; Russell, J.; Lowe, A.J.; Gibby, M.; Powell, W. How much effort is required to isolate nuclear microsatellites from plants? Mol. Ecol. 2003, 12, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.Z.; Luo, L.B.; Zhang, H.L.; Xiong, M.; Wang, X.; Zhou, D. Identification and characterization of 43 novel polymorphic EST-SSR markers for arum lily, Zantedeschia aethiopica (Araceae). Am. J. Bot. 2012, 99, e493–e497. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.R.; Lüberstedt, T. Functional markers in plants. Trends Plant Sci. 2003, 8, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.Z.; Sun, Z.Z.; Cui, B.B.; Zhang, Q.X.; Xiong, M.; Wang, X.; Zhou, D. Transcriptome analysis of colored calla lily (Zantedeschia rehmannii Engl.) by Illumina sequencing: De novo assembly, annotation and EST-SSR marker development. PeerJ 2016, 4, e2378. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.Z.; Sun, Z.Z.; Xiong, M.; Wang, X.; Zhou, D. Characterization and cross-species transferability of EST-SSRs from the illumina-based de novo transcriptome sequencing of arum lily (Zantedeschia aethiopica). In Proceedings of the ISHS, XII International Symposium on Flower Bulbs and Herbaceous Perennials, Kunming, China, 28 June–2 July 2016; p. 51. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Cândido, E.D.S.; Fernandes, G.D.R.; Alencar, S.A.D.; Cardoso, M.H.E.S.; Lima, S.M.D.F.; Miranda, V.D.J.; Porto, W.F.; Nolasco, D.O.; Oliveira-Júnior, N.G.D.; Barbosa, A.E.A.D.; et al. Shedding some light over the floral metabolism by arum lily (Zantedeschia aethiopica) spathe de novo transcriptome assembly. PLoS ONE 2014, 9, e90487. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Xin, W.; Zhang, Y.X.; Fonceka, D.; Yang, W.J.; Diouf, D.; Liao, B.S.; Cissé, N.; Zhang, X.R. Analysis of genetic diversity and population structure of sesame accessions from Africa and Asia as major centers of its cultivation. Genes 2016, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Sun, Z.Z.; Zhou, D.; Xiong, M.; Wang, X.; Yang, J.M.; Wei, Z.Z. Development and characterization of novel EST-SSRs from Larix gmelinii and their cross-species transferability. Molecules 2015, 20, 12469–12480. [Google Scholar] [CrossRef] [PubMed]
- Batten, A. Flowers of Southern Africa, 1st ed.; Frandsen Publishers: Sandton, South Africa, 1986. [Google Scholar]
- Veatch-Blohm, M.E.; Morningstar, L. Calla lily growth and development under saline irrigation. Hortscience 2011, 46, 222–227. [Google Scholar]
- DuPlessis, N.M.; Duncan, G.; Bayer, B.; Bodley, E. Bulbous Plants of Southern Africa: A Guide to Their Cultivation and Propagation, 1st ed.; Tafelberg Publishers: Cape Town, South Africa, 1989. [Google Scholar]
- Aitken, A.G.; Kerr, J.P.; Hewett, E.W.; Hale, C.N.; Nixon, C. Calla lilies (Zantedeschia)—A novel export crop with a global impact. Grow. Futures Case Study Ser. 2005, 14, 1–6. [Google Scholar]
- Tjia, B. Hybrid calla lilies: A potential new crop for Florida. Proc. Fla. State Hort. Soc. 1985, 98, 127–130. [Google Scholar]
- Zhu, Z.; Zhang, L.Y.; Gao, L.X.; Tang, S.Q.; Zhao, Y.; Yang, J. Local habitat condition rather than geographic distance determines the genetic structure of Tamarix chinensis, populations in yellow river delta, china. Tree Genet. Genomes 2016, 12, 1–9. [Google Scholar] [CrossRef]
- Pérombelon, M.C.M.; Kelman, A. Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 1980, 18, 361–387. [Google Scholar] [CrossRef]
- Wright, P.J. A soft rot of calla (Zantedeschia spp.) caused by Erwinia carotovora subspecies carotovora. N. Z. J. Crop Hortic. Sci. 1998, 26, 331–334. [Google Scholar] [CrossRef]
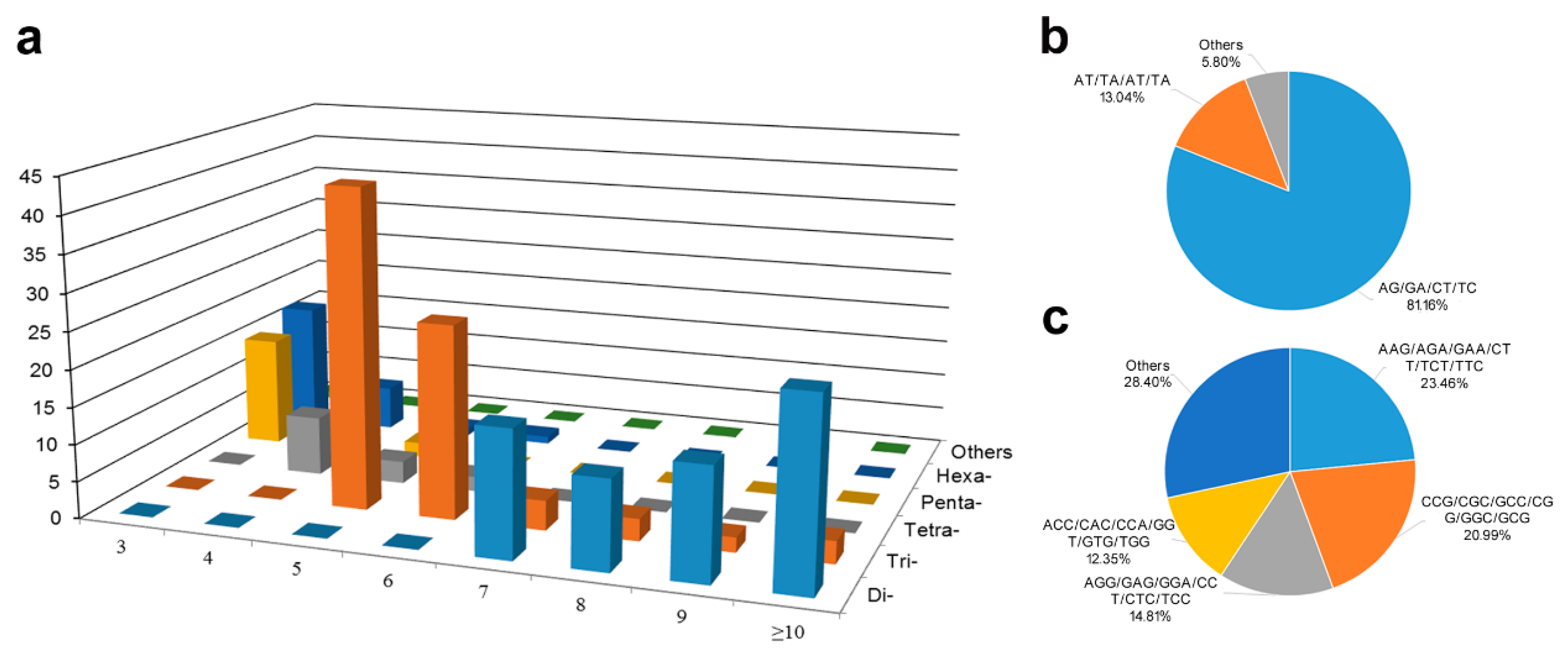

| Unigenes Features and SSR in Unigenes | Number |
|---|---|
| Total number of unigenes examined | 2175 |
| Number of contigs | 818 |
| Number of singletons | 1357 |
| Length range of contigs | 238–2073 bp |
| Length range of singletons | 53–1095 bp |
| Total size covered by examined unigenes | 1.52 Mb |
| Total number of SSRs identified | 209 |
| Number of unigenes containing one SSR | 160 |
| Number of unigenes containing more than one SSR | 22 |
| Frequency of SSR occurrence in total number of unigenes | 10.40 |
| Frequency of SSR occurrence in total size of unigenes | 7.2 kb |
| Loci | Na | Ne | Ho | He | PIC |
|---|---|---|---|---|---|
| ZW008 | 4 | 1.43 | 0.243 | 0.301 | 0.29 |
| ZW009 | 5 | 2.27 | 0.969 | 0.559 | 0.46 |
| ZW011 | 6 | 2.40 | 0.853 | 0.584 | 0.50 |
| ZW012 | 4 | 2.45 | 0.867 | 0.591 | 0.52 |
| ZW018 | 4 | 2.01 | 0.895 | 0.503 | 0.39 |
| ZW021 | 2 | 1.81 | 0.462 | 0.448 | 0.35 |
| ZW029 | 3 | 1.61 | 0.496 | 0.377 | 0.31 |
| ZW033 | 4 | 2.55 | 0.405 | 0.608 | 0.53 |
| ZW034 | 5 | 2.90 | 0.228 | 0.655 | 0.60 |
| ZW037 | 5 | 2.42 | 0.932 | 0.587 | 0.50 |
| ZW039 | 2 | 1.88 | 0.231 | 0.469 | 0.36 |
| ZW045 | 5 | 2.36 | 0.340 | 0.575 | 0.49 |
| ZW046 | 2 | 1.85 | 0.256 | 0.460 | 0.35 |
| ZW047 | 5 | 3.52 | 0.402 | 0.716 | 0.67 |
| ZW060 | 2 | 1.08 | 0.052 | 0.070 | 0.07 |
| ZW062 | 2 | 1.90 | 0.767 | 0.473 | 0.36 |
| ZW071 | 2 | 1.57 | 0.474 | 0.361 | 0.30 |
| ZW076 | 5 | 3.75 | 0.683 | 0.733 | 0.69 |
| ZW080 | 3 | 2.94 | 0.517 | 0.659 | 0.59 |
| ZW093 | 3 | 2.04 | 0.164 | 0.509 | 0.45 |
| ZW095 | 3 | 2.04 | 0.476 | 0.511 | 0.42 |
| ZW096 | 5 | 2.61 | 0.328 | 0.617 | 0.57 |
| ZW098 | 2 | 1.19 | 0.172 | 0.157 | 0.14 |
| ZW101 | 2 | 1.91 | 0.301 | 0.476 | 0.36 |
| ZW122 | 2 | 1.17 | 0.159 | 0.147 | 0.14 |
| ZW123 | 4 | 1.78 | 0.549 | 0.438 | 0.37 |
| ZW126 | 10 | 4.99 | 0.458 | 0.800 | 0.77 |
| ZW132 | 2 | 1.31 | 0.205 | 0.239 | 0.21 |
| ZW146 | 2 | 1.97 | 0.389 | 0.493 | 0.37 |
| ZW158 | 2 | 1.08 | 0.019 | 0.072 | 0.07 |
| ZW165 | 4 | 2.72 | 0.755 | 0.632 | 0.56 |
| Mean | 3.58 | 2.18 | 0.453 | 0.478 | 0.412 |
| Source | Population | N | Na | Ne | Ho | He | Private | PIC |
|---|---|---|---|---|---|---|---|---|
| Country | Netherlands | 34 | 3.194 | 2.104 | 0.478 | 0.459 | 6 | 0.396 |
| New Zealand | 52 | 3.226 | 2.157 | 0.470 | 0.471 | 4 | 0.404 | |
| USA | 31 | 3.161 | 2.042 | 0.411 | 0.463 | 4 | 0.394 | |
| Model-based population (K = 2) | G-I | 31 | 2.654 | 1.838 | 0.438 | 0.386 | 17 | 0.326 |
| G-II | 38 | 2.839 | 1.919 | 0.428 | 0.416 | 23 | 0.357 | |
| Model-based population (K = 4) | G-Ia | 12 | 2.000 | 1.606 | 0.394 | 0.292 | 1 | 0.241 |
| G-Ib | 12 | 2.323 | 1.799 | 0.508 | 0.382 | 8 | 0.319 | |
| G-IIa | 19 | 2.548 | 1.847 | 0.439 | 0.390 | 8 | 0.327 | |
| G-IIb | 19 | 2.323 | 1.820 | 0.437 | 0.381 | 4 | 0.321 |
| Source | d.f. (degree of freedom) | Sum of Squares | Mean of Squared Observations | Estimation Variance | Percentage of Variations % | Fixation Index | |
|---|---|---|---|---|---|---|---|
| Country | Among countries | 2 | 48.872 | 24.436 | 0.184 | 2% | Fst = 0.021 |
| Among individuals | 114 | 1195.107 | 10.483 | 2.022 | 23% | ||
| Within individuals | 117 | 753.500 | 6.440 | 6.440 | 74% | ||
| Total | 233 | 1997.479 | 8.646 | 100% | |||
| Model-based population (K = 2) | Among groups | 1 | 129.082 | 129.082 | 1.757 | 19% | Fst = 0.187 |
| Within populations | 67 | 610.041 | 9.105 | 1.491 | 16% | ||
| Among individuals | 69 | 422.500 | 6.123 | 6.123 | 65% | ||
| Total | 137 | 1161.623 | 9.371 | 100% | |||
| Model-based population (K = 4) | Among groups | 3 | 192.571 | 64.190 | 1.845 | 21% | Fst = 0.206 |
| Among individuals | 58 | 461.849 | 7.963 | 0.860 | 10% | ||
| Within individuals | 62 | 387.000 | 6.242 | 6.242 | 70% | ||
| Total | 123 | 1041.419 | 8.948 | 100% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Zhang, H.; Wang, Y.; Li, Y.; Xiong, M.; Wang, X.; Zhou, D. Assessing Genetic Diversity and Population Differentiation of Colored Calla Lily (Zantedeschia Hybrid) for an Efficient Breeding Program. Genes 2017, 8, 168. https://doi.org/10.3390/genes8060168
Wei Z, Zhang H, Wang Y, Li Y, Xiong M, Wang X, Zhou D. Assessing Genetic Diversity and Population Differentiation of Colored Calla Lily (Zantedeschia Hybrid) for an Efficient Breeding Program. Genes. 2017; 8(6):168. https://doi.org/10.3390/genes8060168
Chicago/Turabian StyleWei, Zunzheng, Huali Zhang, Yi Wang, Youli Li, Min Xiong, Xian Wang, and Di Zhou. 2017. "Assessing Genetic Diversity and Population Differentiation of Colored Calla Lily (Zantedeschia Hybrid) for an Efficient Breeding Program" Genes 8, no. 6: 168. https://doi.org/10.3390/genes8060168
APA StyleWei, Z., Zhang, H., Wang, Y., Li, Y., Xiong, M., Wang, X., & Zhou, D. (2017). Assessing Genetic Diversity and Population Differentiation of Colored Calla Lily (Zantedeschia Hybrid) for an Efficient Breeding Program. Genes, 8(6), 168. https://doi.org/10.3390/genes8060168





