Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of MYB Proteins in Sesame
2.2. Chromosomal Location and Gene Duplication of MYB Genes in Sesame
2.3. Phylogenetic Analysis and Functional Classification of MYB Proteins in Sesame
2.4. Analysis of the Expression Patterns of Sesame MYB Genes in Different Tissues, under Drought and Waterlogging Stresses Using RNA-seq Data
2.5. Plant Materials and Stress Treatments
2.6. RNA Isolation and Quantitative RT-PCR
3. Results
3.1. Identification of Sesame MYB Genes and Analysis of Their Sequence Features
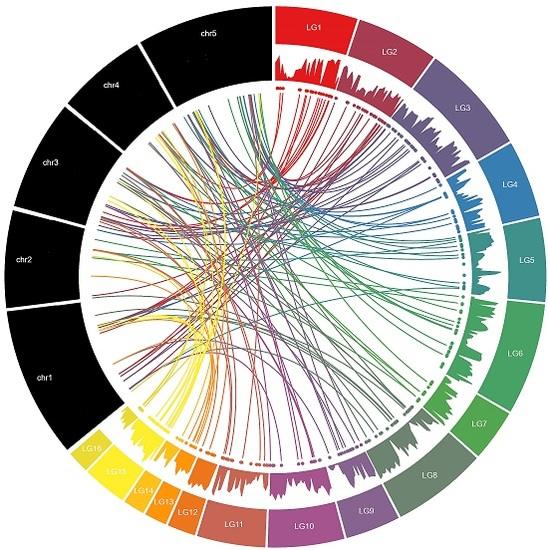
3.2. Chromosomal Distribution and Duplication Events among Sesame MYB Genes
3.3. Phylogenetic Relationships, Classification and Exon-Intron Structures of SIMYB Gene Family in Sesame
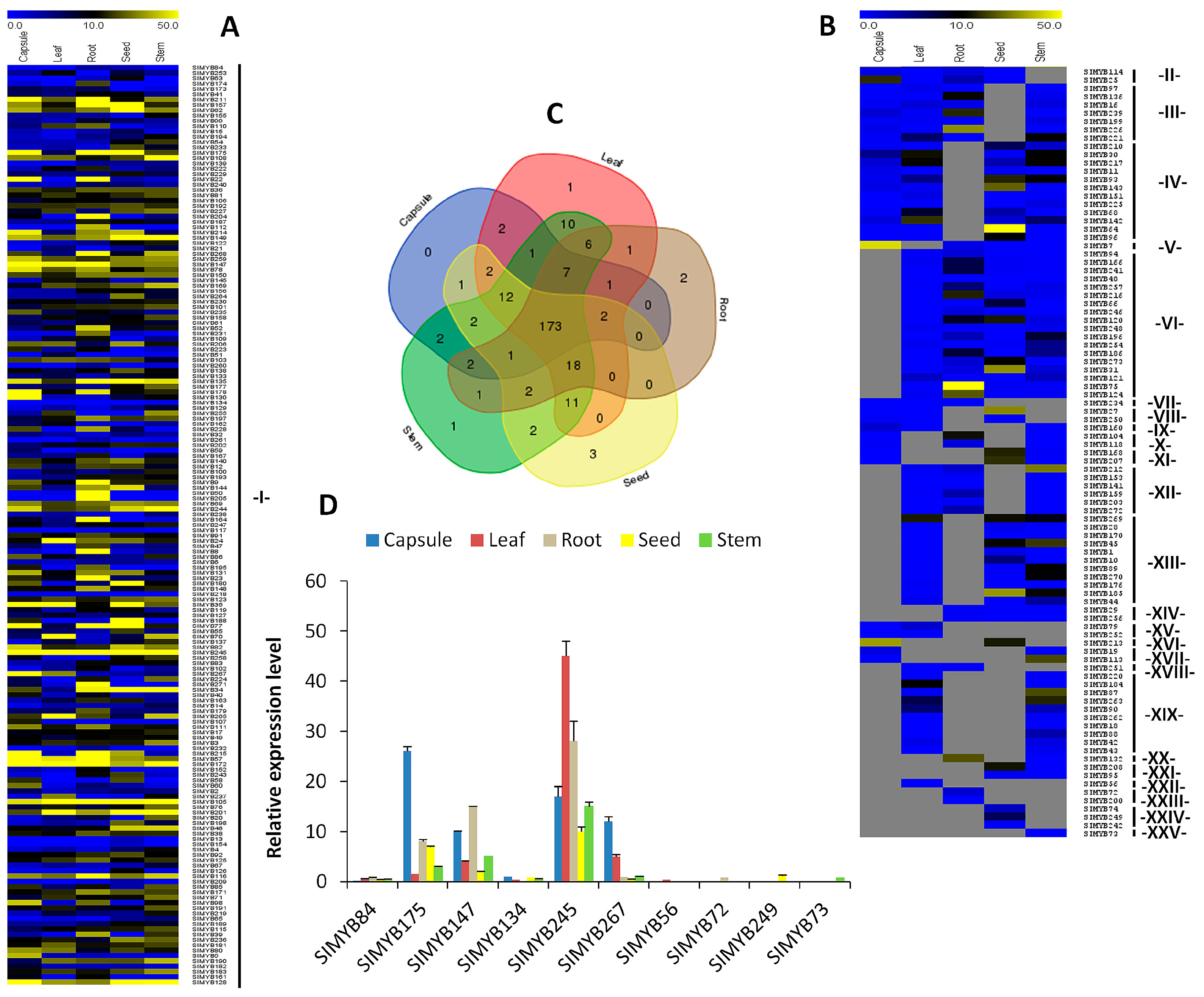
3.4. Expression Patterns of SIMYB Genes in Different Tissues of Sesame
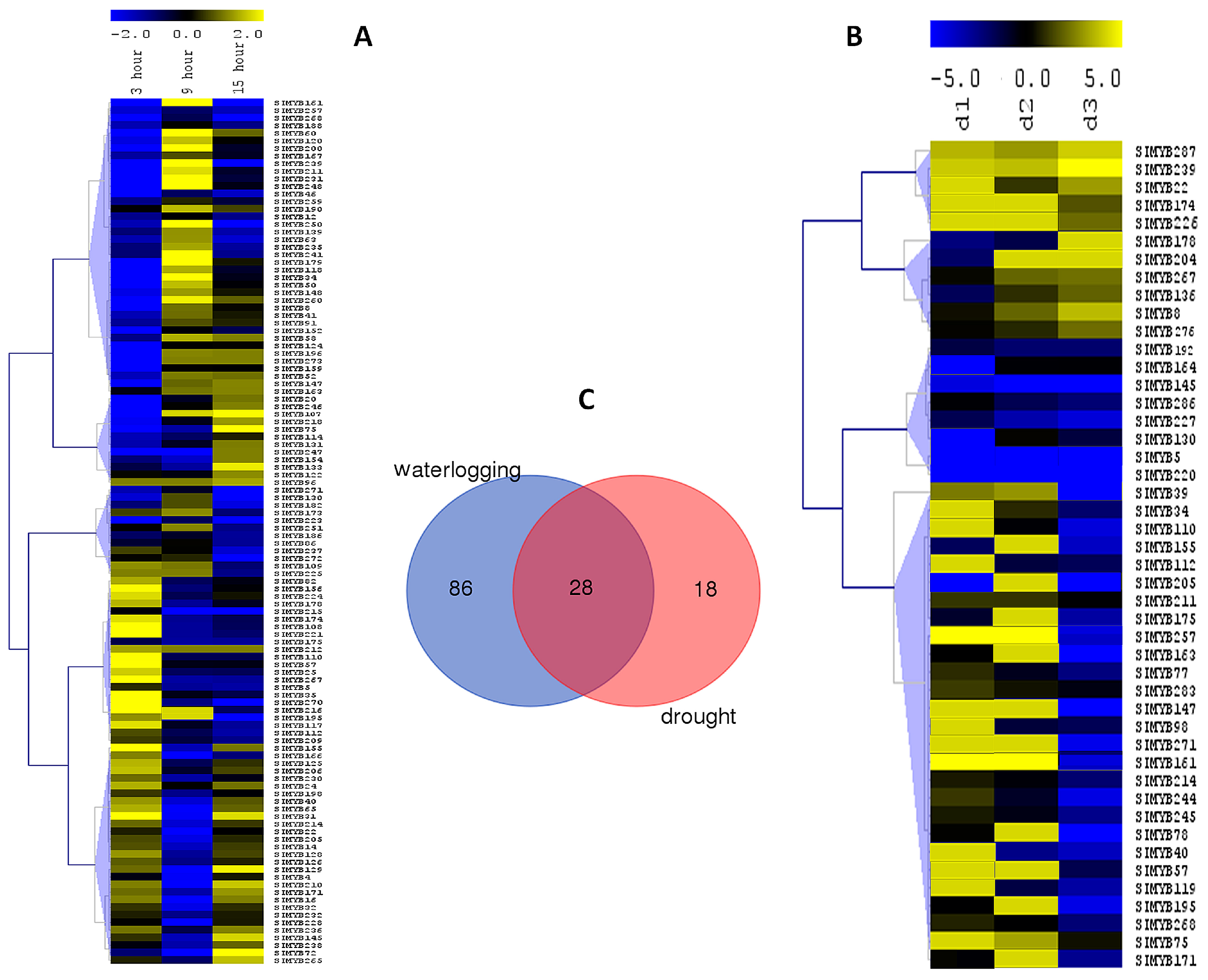
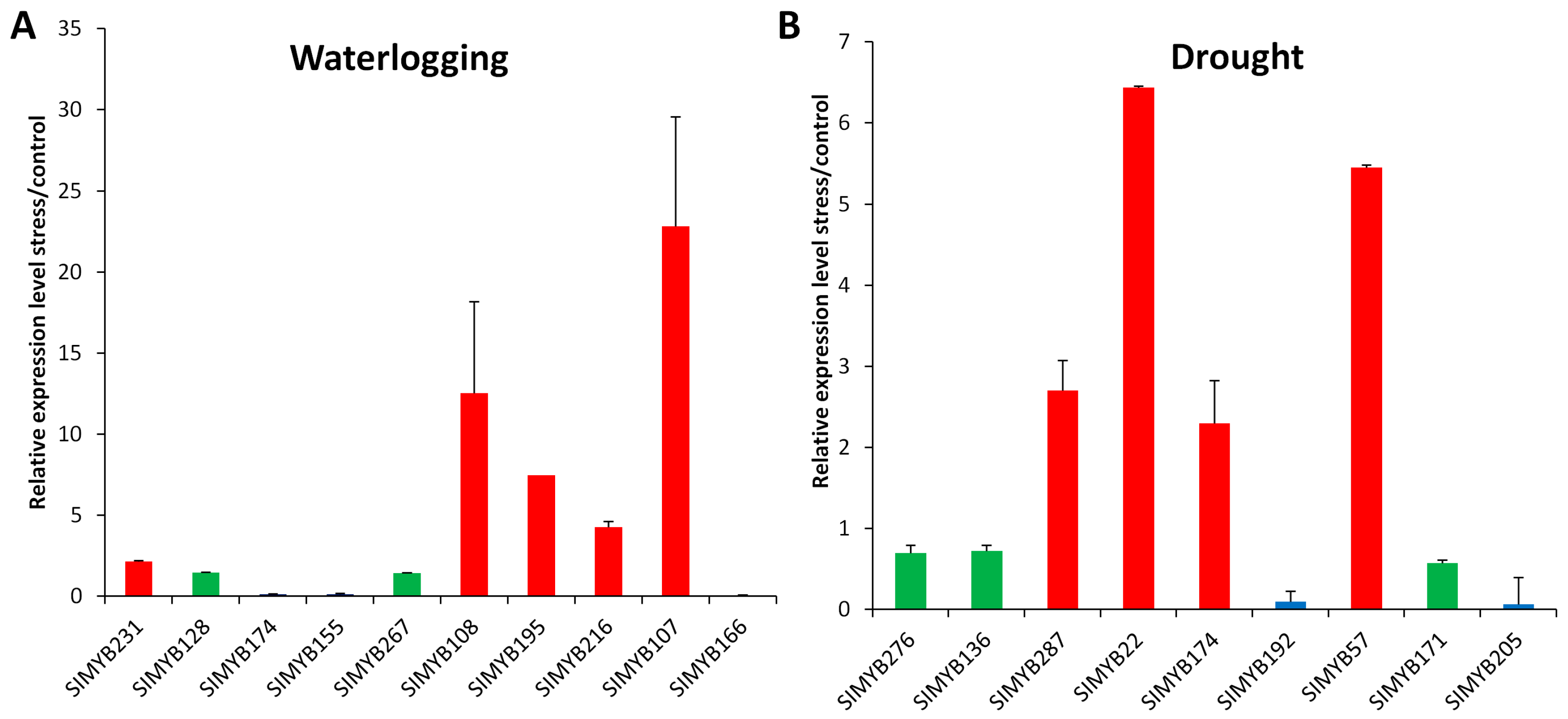
3.5. Expression Profiles of SIMYB Genes in Response to Waterlogging Stress
3.6. Expression Profiles of SIMYB Genes in Response to Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-wide identification, evolution and functional divergence of MYB transcription factors in Chinese white Pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Geethalakshmi, S.; Barathkumar, S.; Prabu, G. The MYB transcription factor family genes in sugarcane (Saccharum sp.). Plant Mol. Biol. Rep. 2015, 33, 512–531. [Google Scholar] [CrossRef]
- Lipsick, J.S. One billion years of Myb. Oncogene 1996, 13, 223–235. [Google Scholar] [PubMed]
- Hou, X.J.; Li, S.B.; Liu, S.R.; Hu, C.G.; Zhang, J.Z. Genome-wide classification and evolutionary and expression analyses of Citrus MYB transcription factor families in Sweet orange. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.I.; Ahmed, N.U.; Kayum, M.A.; Kang, K.K.; Nou, I.S. Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 2016, 104, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; Gong, W.; He, S.; Sun, G.; Sun, J.; Du, X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Hui, Z.; Li, F.; Zhao, M.-R.; Zhang, J.; Wang, W. Improvement of heat and drought photosynthetic tolerance in wheat by over accumulation of glycine betaine. Plant Biotechnol. Rep. 2010, 4, 213–222. [Google Scholar] [CrossRef]
- Liu, C.; Wang, X.; Xu, Y.; Deng, X.; Xu, Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol. Biol. Rep. 2014, 41, 6769–6785. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plant 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Yang, Q.; Pan, L.; Chi, X.; Chen, M.; Hu, D.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Genes 2014, 533, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luan, Y.; Yin, Y. SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene 2014, 547, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Smita, S.; Keshari, S.; Ravi, L.; Chinnusamy, R.V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J.; Su, Y.; Liu, H.; Chen, Y.; Luo, P.; Du, X.; Wang, D.; Zhang, H. TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Feng, K.; Que, F.; Wang, F.; Xiong, A.S. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xing, C.; Yao, Z.; Huang, X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017, 15, 1186–1203. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Cui, Y.; Wang, M.; Xia, X. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochem. Biophys. Res. Commun. 2017, 490, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.I.; Yang, Z.; Gong, Q.; Chen, E.; Wang, X.; Zhao, G.; Ge, X.; Zhang, X.; Li, F. GaMYB85 an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 2017, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Bedigian, D. Evolution of sesame revisited: Domestication, diversity and prospects. Genet. Resour. Crop Evol. 2003, 50, 779–787. [Google Scholar]
- Pathak, N.; Rai, A.K.; Kumari, R.; Thapa, A.; Bhat, K.V. Sesame crop: An underexploited oilseed holds tremendous potential for enhanced food value. Agric. Sci. 2014, 5, 519–529. [Google Scholar] [CrossRef]
- Sun, J.; Rao, Y.; Le, M.; Yan, T.; Yan, X.; Zhou, H. Effects of drought stress on sesame growth and yield characteristics and comprehensive evaluation of drought tolerance. Chin. J. Oil Crop Sci. 2010, 32, 525–533. [Google Scholar]
- Wang, L.; Zhang, Y.; Qi, X.; Li, D.; Wei, W.; Zhang, X. Global gene expression responses to waterlogging in roots of sesame (Sesamum indicum L.). Acta Physiol. Plant. 2012, 34, 2241–2249. [Google Scholar] [CrossRef]
- Wei, W.L.; Li, D.H.; Wang, L.H.; Ding, X.; Zhang, Y.X.; Gao, Y.; Zhang, X.R. Morpho-anatomical and physiological responses to waterlogging of sesame (Sesamum indicum L.). Plant Sci. 2013, 208, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Li, D.; Gao, Y.; Lu, H.; Zhang, X. Mapping of sesame waterlogging tolerance QTL and identification of excellent waterlogging tolerant germplasm. Sci. Agric. Sin. 2014, 47, 422–430. [Google Scholar]
- Wang, L.; Li, D.; Zhang, Y.; Gao, Y.; Yu, J.; Wei, X.; Zhang, X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, H.; Zehtab-Salmasi, S.; Ghassemi-Golezani, K.; Gharineh, M.H. Effects of water limitation on grain and oil yields of sesame cultivars. J. Food Agric. Environ. 2009, 7, 339–342. [Google Scholar]
- Anjum, A.H.; Xie, X.Y.; Wang, L.C.; Saleem, M.F.; Man, C.; Le, W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 2011, 6, 2026–2032. [Google Scholar]
- Kim, K.S.; Ryu, S.N.; Chung, H.G. Influence of drought stress on chemical composition of sesame seed. Korean J. Crop Sci. 2006, 51, 73–80. [Google Scholar]
- Ozkan, A.; Kulak, M. Effects of water stress on growth, oil yield, fatty acid composition and mineral content of Sesamum indicum. J. Anim. Plant Sci. 2013, 23, 1686–1690. [Google Scholar]
- Kadkhodaie, A.; Razmjoo, J.; Zahedi, M.; Pessarakli, M. Oil content and composition of sesame (Sesamum indicum L.) genotypes as affected by irrigation regimes. J. Am. Oil Chem. Soc. 2014, 91, 1737–1744. [Google Scholar] [CrossRef]
- Dossa, K.; Diouf, D.; Wang, L.; Wei, X.; Yu, J.; Niang, M.; Fonceka, D.; Yu, J.; Mmadi, M.A.; Yehouessi, L.W.; et al. The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 2017, 8, 1154. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, J.; Li, D.; Zhang, X. Sinbase: An integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2014, 56. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.-R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.H.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Diouf, D.; Cissé, N. Genome wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Gong, H.; Yu, J.; Liu, P.; Wang, L.; Zhang, Y.; Zhang, X. Sesame FG: An integrated database for the functional genomics of sesame. Sci. Rep. 2017, 7, 2342. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Yua, J.; Wei, X.; Fonceka, D.; Diouf, D.; Liao, B.; Cisse, N.; et al. Dynamic transcriptome landscape of Sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genom. Data 2017, 11, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Liu, A.; Yu, J.; Wei, X.; Zhou, R.; Fonceka, D.; Diouf, D.; et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.W.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Method Enzymol. 2006, 411, 134–193. [Google Scholar]
- Lalitha, S. Primer premier 5. Biotechnol. Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Liao, B.; Diouf, D.; Cissé, N.; et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, Y.; Wu, Z.; Lu, W.; Han, J.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the MYB gene family in physic nut (Jatropha curcas L.). Gene 2015, 572, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, L.; Guo, H.; Liao, B.; King, G.; Zhang, X. Genome evolutionary dynamics followed by diversifying selection explains the complexity of the Sesamum indicum genome. BMC Genom. 2017, 18, 257. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene conferring abiotic stress tolerance in plants. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, J.; Hu, R.; Wu, P.; Hou, X.L.; Song, X.M.; Xiong, A.S. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genom. 2015, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Hiratsuka, K.; Chua, N.H. Transcription factors in plant growth and development. Curr. Opin. Genet. Dev. 1994, 4, 642–646. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Gibbs, D.; Voss, U.; Harding, S.; Fannon, J.; Moody, L.; Yamada, E.; Swarup, K.; Nibau, C.; Bassel, G.; Choudhary, A.; et al. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol. 2014, 203, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jia, K.P.; Lian, H.L.; Yang, X.; Li, L.; Yang, H.Q. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 454, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Fu, J.; Zheng, J.; Zhou, X.; Huai, J.; Wang, J.; Wang, M.; Zhang, Y.; Chen, X.; Zhang, J. Annotation and expression profile analysis of full-length cDNAs from stress-induced maize (Zea mays L.) seedlings. Plant J. 2006, 48, 710–727. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Graham, N.S.; Vanholme, B.; Swennen, R.; May, S.T.; Keulemans, J. Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genom. 2009, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, M.; Matus, J.T.; Francia, P.; Rusconi, F.; Cañón, P.; Medina, C.; Conti, L.; Cominelli, E.; Tonelli, C.; Johnson, P.A. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 2011, 11, 142. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Luang, S.; Li, Y.; Bazanova, N.; Morran, S.; Song, Z.; Perera, M.A.; Hrmova, M.; Borisjuk, N.; Lopato, S. Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 2016, 67. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, W.; Qin, M.; Li, S.; Qiao, M.; Liu, Z.; Xiang, F. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017, 58, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Hoeren, F.U.; Dolferus, R.; Wu, Y.; Peacock, W.J.; Dennis, E.S. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase (ADH1) gene by low oxygen. Genetics 1998, 149, 479–490. [Google Scholar] [PubMed]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2007, 129, 375–385. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Mo, Z.H.; Xuan, J.P.; Jia, X.D.; Wang, G.; Guo, Z.R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015, 35, 208. [Google Scholar] [CrossRef]
- Peng, X.J.; Liu, H.; Wang, D.; Shen, S.H. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016, 17, 251. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; Yang, C. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Huang, P.; Yuan, X.; Chen, H.; Huang, J.; Zhang, H. CMYB1 encoding a MYB transcriptional activator is involved in abiotic stress and circadian rhythm in rice. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cao, G.; Qu, L.; Gu, H. Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 2009, 28, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.L.; Cao, Y.R.; Liu, Y.F.; Lei, G.; Zou, H.F.; Liao, Y.; Wang, H.W.; Zhang, W.K.; Ma, B.; Du, J.Z.; et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009, 19, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-Like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Scoville, R.; Barnett, L.L.; Roels, S.B.; Kelly, J.K.; Hileman, L.C. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 2011, 191, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Jang, I.C.; Henriques, R.; Chua, N.H. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 2009, 21, 1341–1359. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.J.; Lisa, J.; Perazza, D.E.; Juda, L.; Campbell, M.M. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004, 37, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V.; Lee, M.M.; Wester, K.; Herrmann, U.; Zheng, Z.; Oppenheimer, D.; Schiefelbein, J.; Hulskamp, M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 2005, 132, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [PubMed]
- Celenza, J.L.; Quiel, J.A.; Smolen, G.A.; Merrikh, H.; Silvestro, A.R.; Normanly, J.; Bender, J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005, 137, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhu, X.; Huang, X.; Liu, J.H. Overexpression of a stress responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol. Biochem. 2014, 78, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Andreasson, E.; Maslak, A.; Mock, P.H.; Mattsson, O.; Mundy, J. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004, 167, 1099–1107. [Google Scholar]
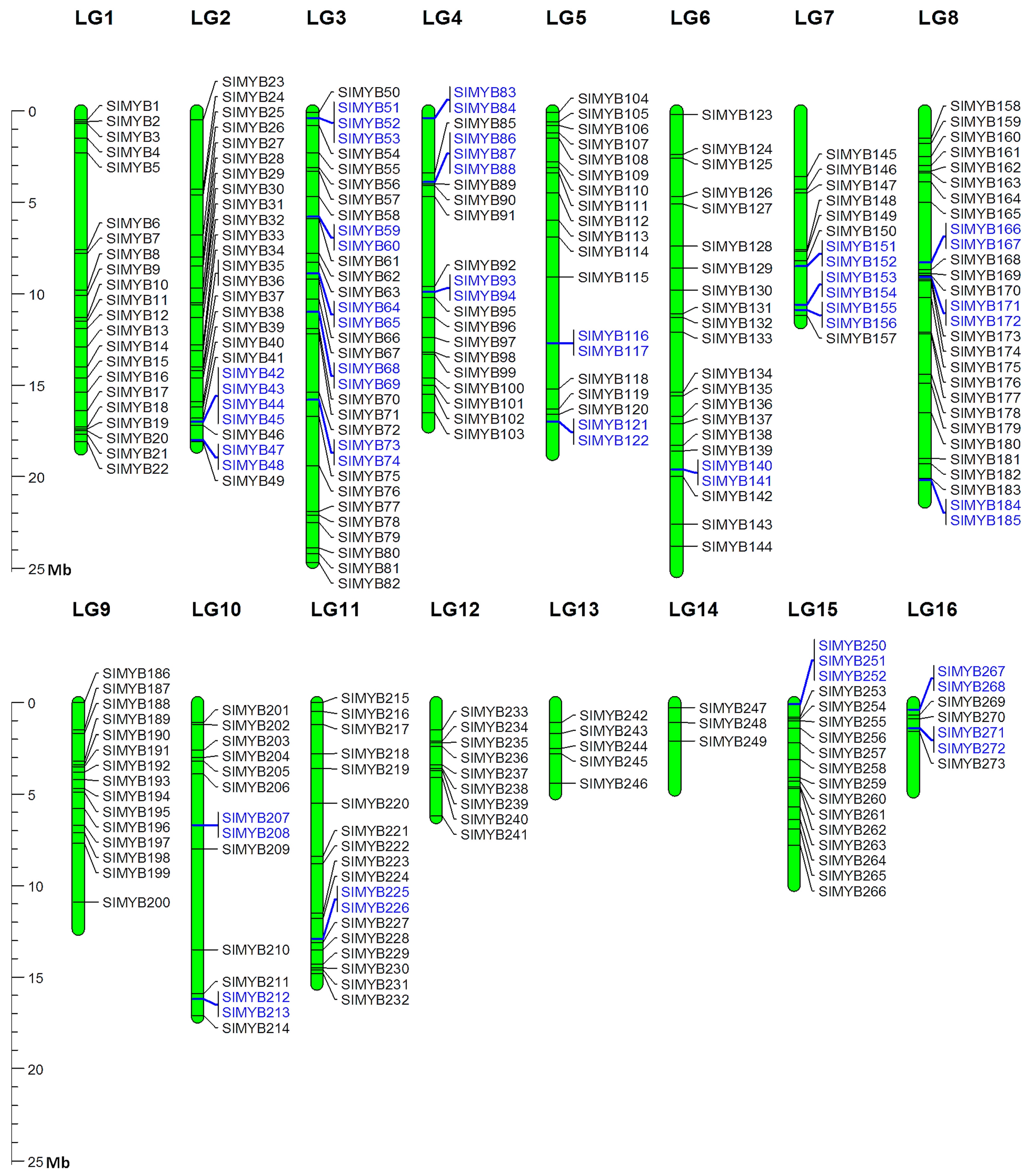
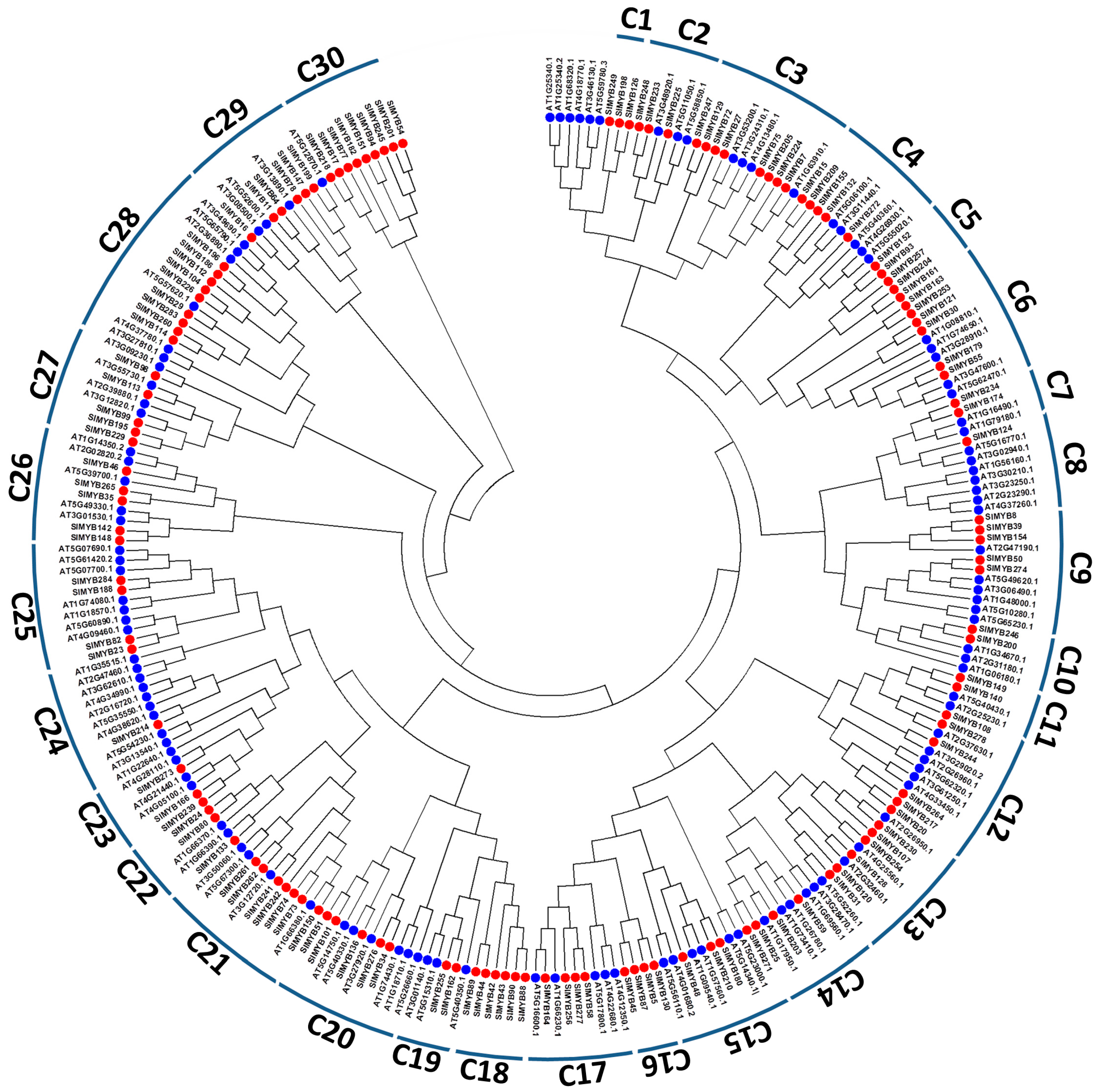
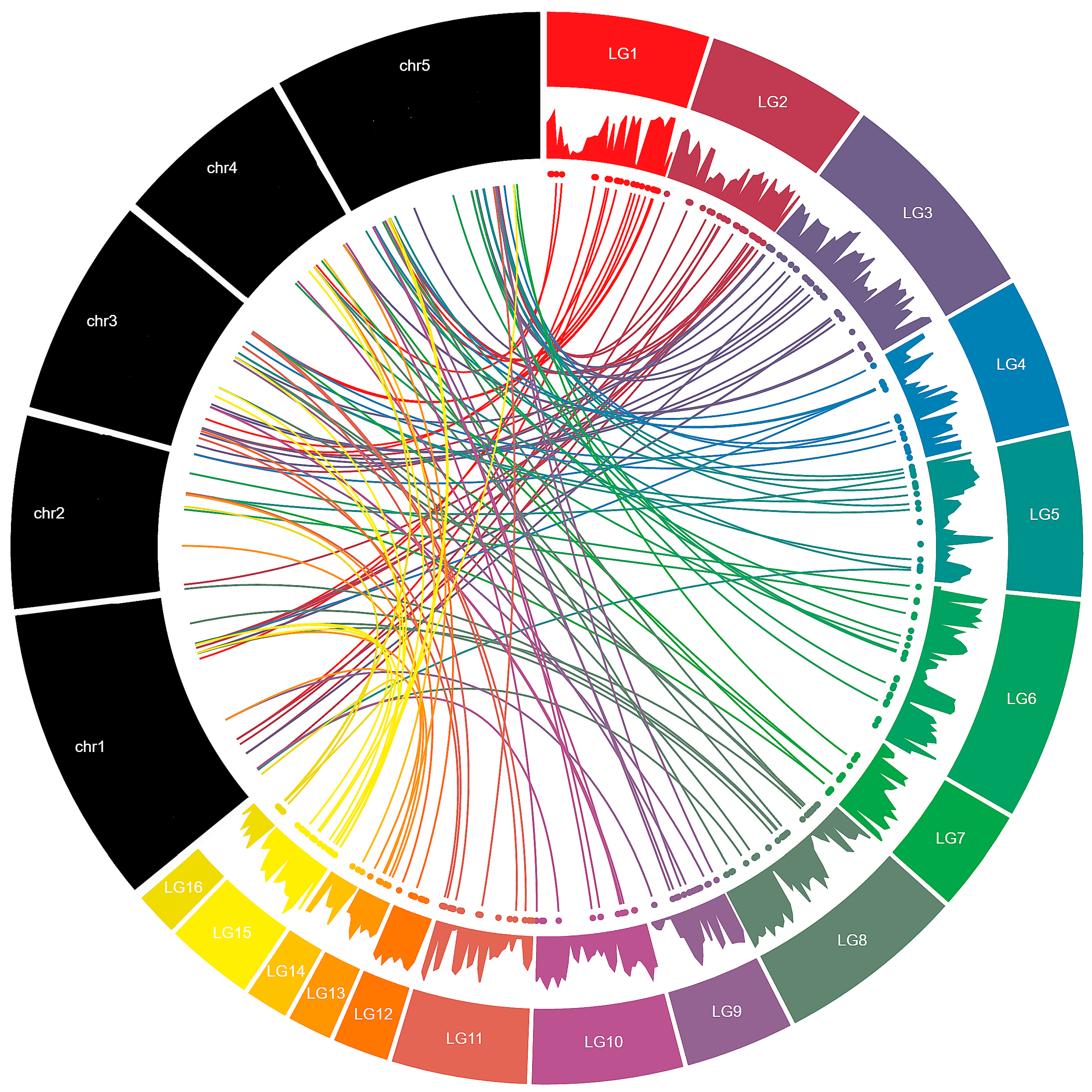
| Linkage Groups | R1-MYB | R1R2-MYB | R1R2R3-MYB | Atypical-MYB | Total |
|---|---|---|---|---|---|
| LG01 | 12 | 8 | 1 | 1 | 22 |
| LG02 | 11 | 16 | 27 | ||
| LG03 | 17 | 15 | 1 | 33 | |
| LG04 | 12 | 9 | 21 | ||
| LG05 | 10 | 8 | 1 | 19 | |
| LG06 | 11 | 10 | 1 | 22 | |
| LG07 | 5 | 8 | 13 | ||
| LG08 | 19 | 9 | 28 | ||
| LG09 | 8 | 7 | 15 | ||
| LG10 | 7 | 7 | 14 | ||
| LG11 | 10 | 7 | 17 | ||
| LG12 | 6 | 4 | 10 | ||
| LG13 | 1 | 4 | 5 | ||
| LG14 | 1 | 3 | 4 | ||
| LG15 | 7 | 10 | 17 | ||
| LG16 | 4 | 3 | 7 | ||
| Scaffold | 6 | 6 | 1 | 13 | |
| Total | 147 | 134 | 5 | 1 | 287 |
| Percentage (%) | 51.22 | 46.69 | 1.74 | 0.35 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mmadi, M.A.; Dossa, K.; Wang, L.; Zhou, R.; Wang, Y.; Cisse, N.; Sy, M.O.; Zhang, X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes 2017, 8, 362. https://doi.org/10.3390/genes8120362
Mmadi MA, Dossa K, Wang L, Zhou R, Wang Y, Cisse N, Sy MO, Zhang X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes. 2017; 8(12):362. https://doi.org/10.3390/genes8120362
Chicago/Turabian StyleMmadi, Marie Ali, Komivi Dossa, Linhai Wang, Rong Zhou, Yanyan Wang, Ndiaga Cisse, Mame Oureye Sy, and Xiurong Zhang. 2017. "Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame" Genes 8, no. 12: 362. https://doi.org/10.3390/genes8120362
APA StyleMmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., Cisse, N., Sy, M. O., & Zhang, X. (2017). Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes, 8(12), 362. https://doi.org/10.3390/genes8120362