SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Sequence and Phylogenetic Analysis
2.3. Abiotic Treatment Assays
2.4. RNA Isolation and Quantitative Real-Time PCR Analysis
2.5. Chlorophyll Content Measurement
2.6. SlbZIP38 Overexpression Vector Construction and Tomato Transformation
2.7. Measurement of Proline Content
2.8. Malondialdehyde Content Detection
2.9. Statistical Analysis
3. Results
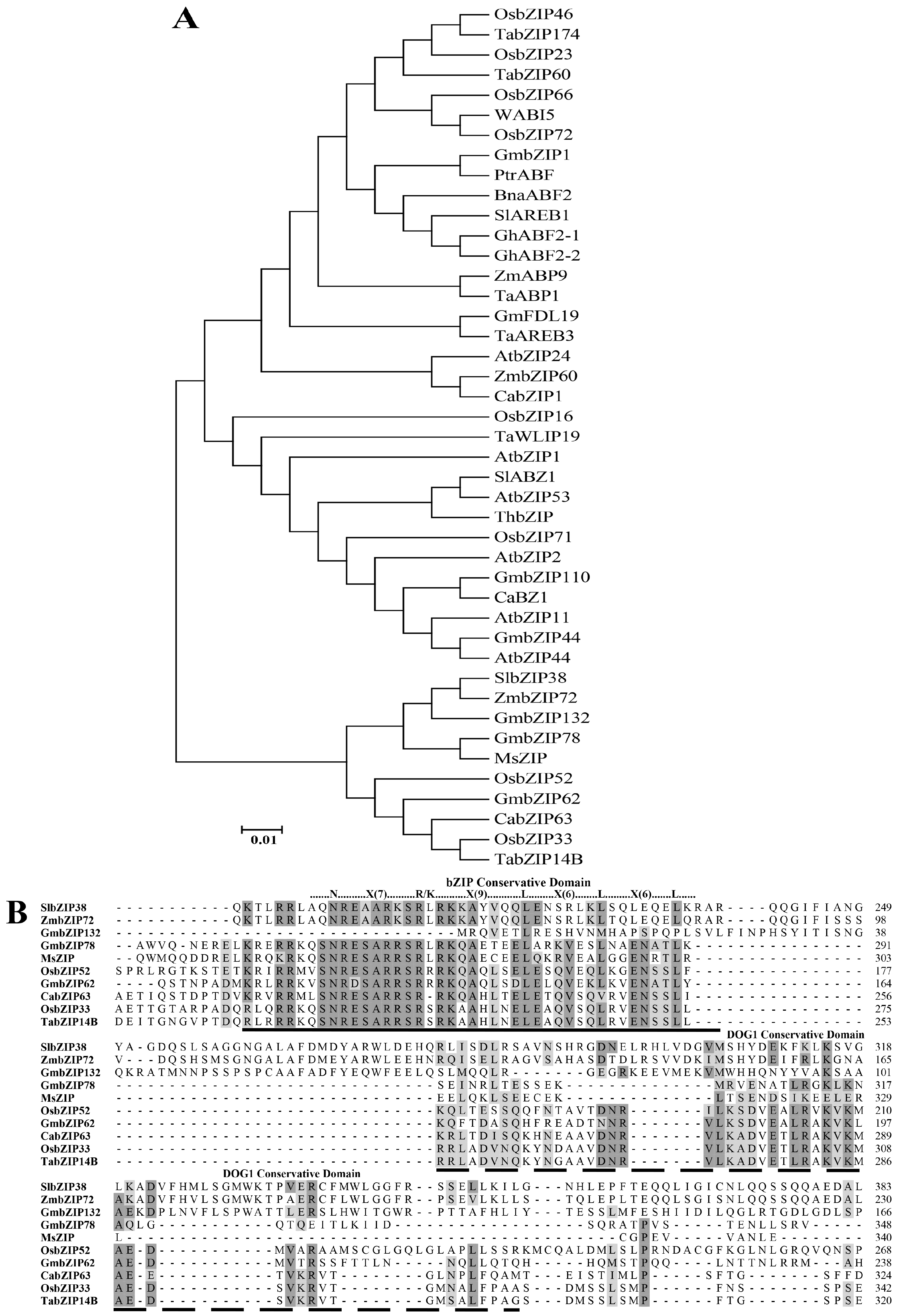
3.1. Structural and Phylogenetic Analysis of SlbZIP38 in Solanum lycopersicum
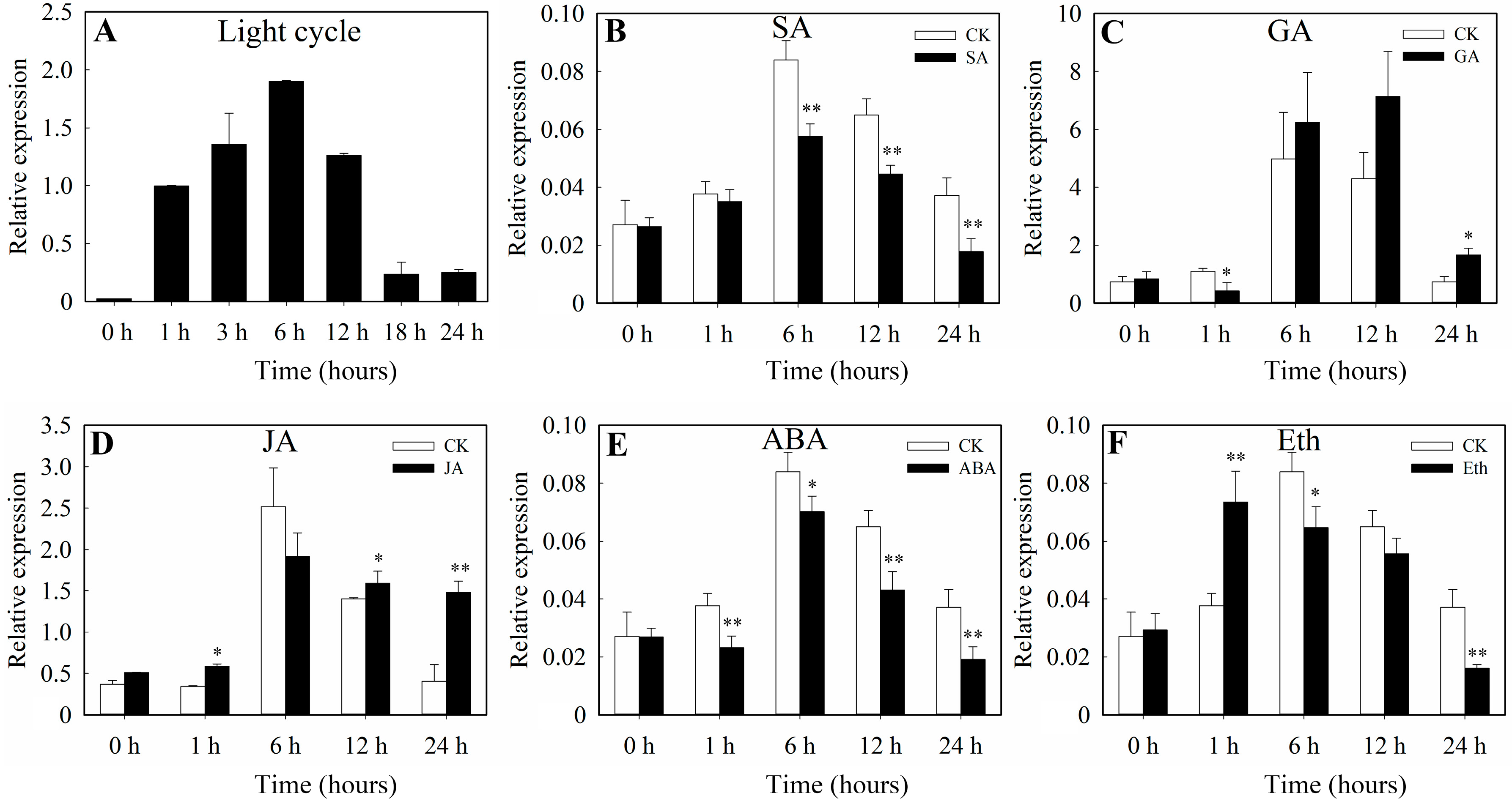
3.2. The Expression Profiles of SlbZIP38 under Abiotic Stress Treatments
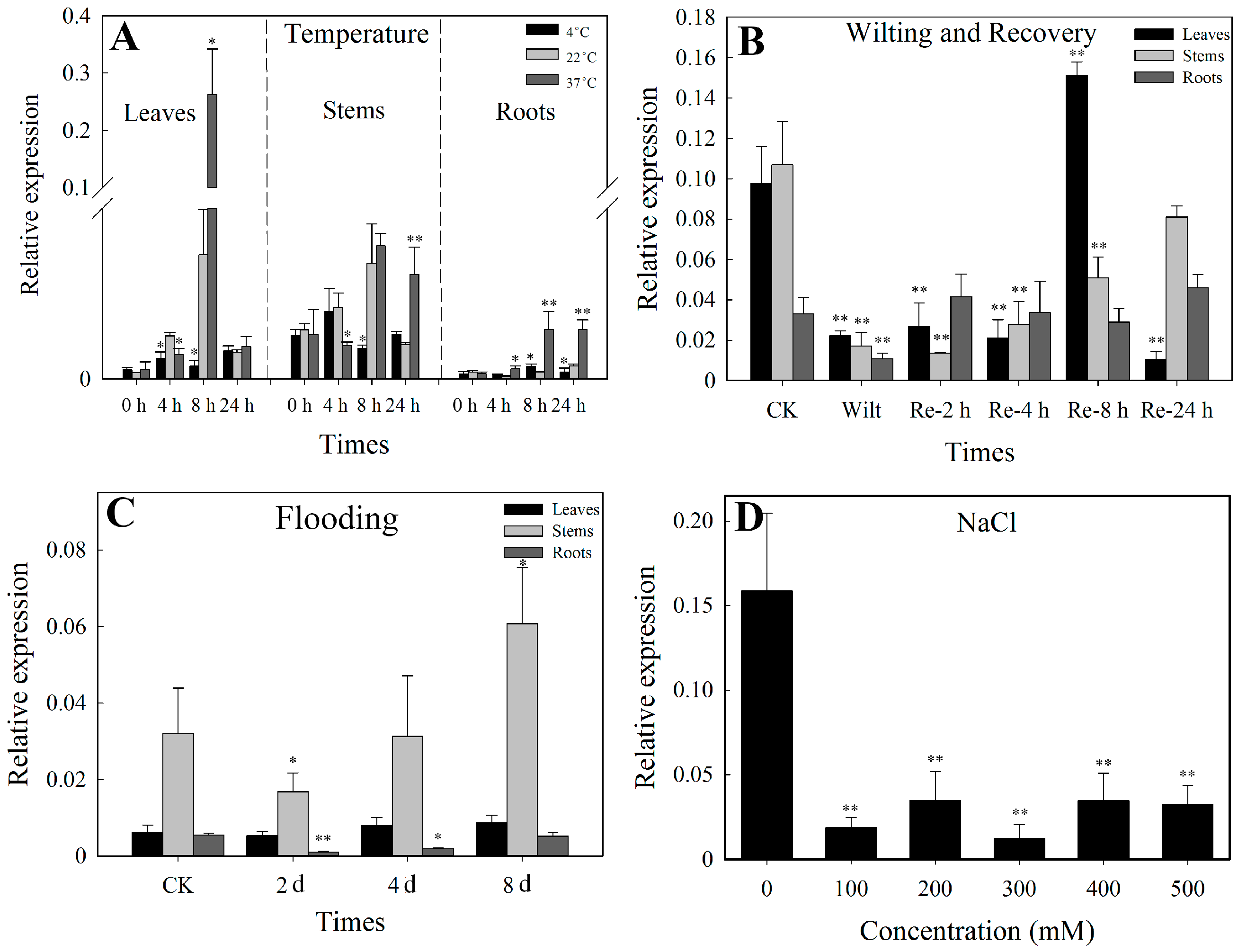
3.3. SlbZIP38 Expression Profiles under Various Abiotic Stresses
3.3.1. Temperature Treatment
3.3.2. Drought Stress
3.3.3. Flooding
3.3.4. NaCl Stress
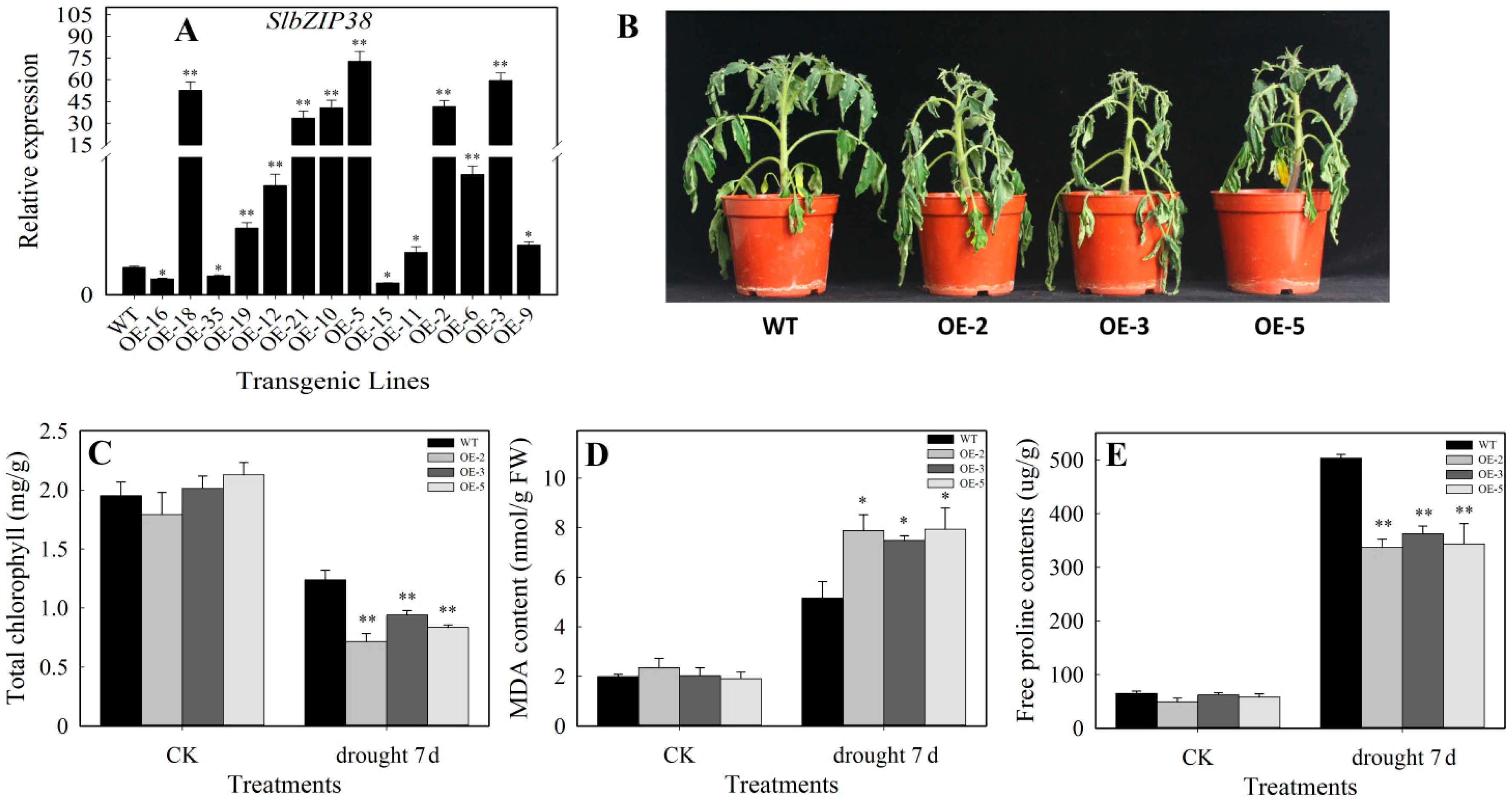
3.4. Performance Analysis of SlbZIP38-Overexpressing Plants under Drought Stress
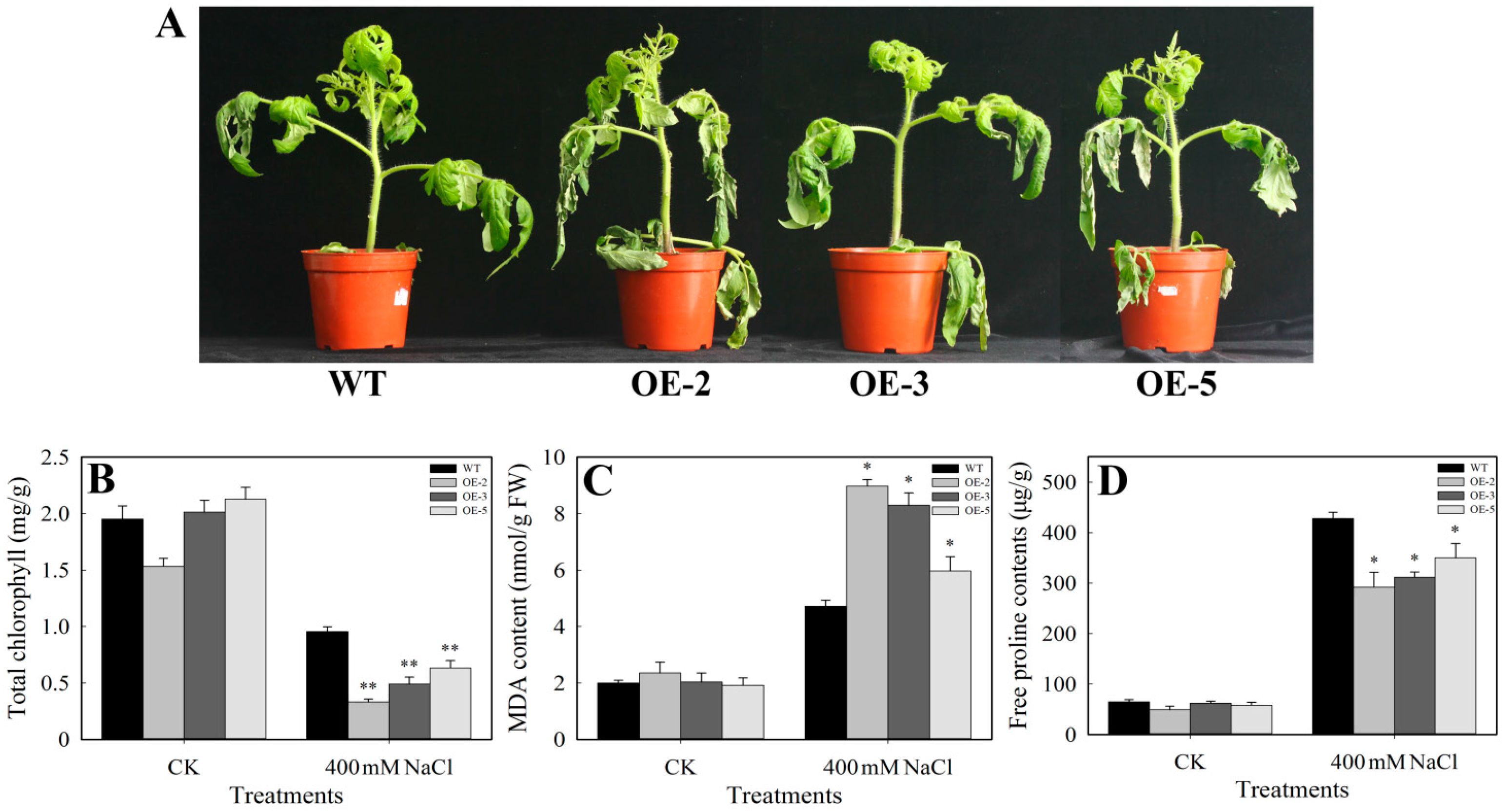
3.5. Performance Analysis of SlbZIP38-Overexpressing Plants under Salt Stress
3.6. Expression Analysis of the ABA Signaling Pathway Marker Genes under Drought and Salt Stress
4. Discussion
4.1. SlbZIP38 Is a Member of the Tomato bZIP Family
4.2. SlbZIP38-Overexpressing Plants Have a Reduced Tolerance to Drought and Salt Stress
4.3. SlbZIP38 Is a Negative Regulator of Drought and Salt Tolerance that Functions via the ABA Signaling Pathway
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Zeevaart, J.; Creelman, R. Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 439–473. [Google Scholar] [CrossRef]
- Leung, J.; Giraudat, J. Abscisic Acid Signal Transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Zhu, J.K. Before and beyond ABA: Upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem. Soc. Trans. 2005, 33, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Soon, F.F.; Ng, L.M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Hase, T.; Toyoda, T.; Shinozaki, K.; Okamoto, M. Origin and evolution of genes related to ABA metabolism and its signaling pathways. J. Plant Res. 2011, 124, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Ishitani, M.; Lee, H.; Zhu, J.K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 2001, 13, 2063–2084. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41. [Google Scholar] [CrossRef]
- Christmann, A.; Moes, D.; Himmelbach, A.; Yang, Y.; Tang, Y.; Grill, E. Integration of abscisic acid signalling into plant responses. Plant Biol. 2006, 8, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Drögelaser, W.; Vicentecarbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.-F.; Wei, W.; Hao, Y.-J.; Tian, A.G.; Huang, J.; Liu, Y.-F.; Zhang, J.-S.; Chen, S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Foster, R.; Chua, N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993, 230, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Lam, E.; Chua, N.H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 1989, 340, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Yoo, C.M.; Park, J.M.; Ryu, G.R.; Goekjian, V.H.; Nagao, R.T.; Key, J.L.; Cho, M.J.; Hong, J.C. STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J. 1998, 15, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Song, S.I.; Kim, Y.S.; Jang, H.J.; Kim, S.Y.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Kim, J.K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 Is a Transcription Activator of Novel ABRE-Dependent ABA Signaling That Enhances Drought Stress Tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchishinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Weltmeier, F.; Rahmani, F.; Ehlert, A.; Dietrich, K.; Schütze, K.; Wang, X.; Chaban, C.; Hanson, J.; Teige, M.; Harter, K.; et al. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: Availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 2009, 69, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Choi, H.W.; Hwang, I.S.; Du, S.C.; Hwang, B.K. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 2006, 224, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, W.; Zhou, J.; He, H.; Chen, L.; Deng, X.W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci. 2012, 193–194, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Avishek, D.; Kumar, S.M.; Srimonta, G.; Sen, S.K.; Maiti, M.K. Enhanced Gene Expression Rather than Natural Polymorphism in Coding Sequence of the OsbZIP23 Determines Drought Tolerance and Yield Improvement in Rice Genotypes. PLoS ONE 2016, 11, e0150763. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Jin, S.J.; Kang, H.L.; Kim, Y.S.; Yang, D.C.; Kim, J.K. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance. Plant Biotechnol. Rep. 2015, 9, 89–96. [Google Scholar] [CrossRef]
- Chen, H.; Dai, X.J.; Gu, Z.Y. OsbZIP33 is an ABA-Dependent Enhancer of Drought Tolerance in Rice. Crop Sci. 2015, 55, 1673. [Google Scholar] [CrossRef]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive Activation of Transcription Factor OsbZIP46 Improves Drought Tolerance in Rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, Y.; Wang, X. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta 2012, 235, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Nguyen, B.H.; Xie, Y.; Xiao, B.; Tang, N.; Zhu, W.; Mou, T.; Xiong, L. Co-overexpression of the Constitutively Active Form of OsbZIP46 and ABA-Activated Protein Kinase SAPK6 Improves Drought and Temperature Stress Resistance in Rice. Front. Plant Sci. 2017, 8, 1102. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.Y.; Chen, M.; Xu, Z.S.; Chen, Y.F.; Li, L.C.; Yu, Y.H.; Liu, Y.N.; Ma, Y.Z. Isolation and Functional Analysis of the bZIP Transcription Factor Gene TaABP1 from a Chinese Wheat Landrace. J. Integr. Agric. 2012, 11, 1580–1591. [Google Scholar] [CrossRef]
- Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant. 2015, 153, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qian, L.; Mao, X.; Li, A.; Jing, R. Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, F.; Maeta, E.; Terashima, A.; Takumi, S. Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol. Plant. 2008, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C.; Zhao, G.; Jia, J.; Kong, X. A novel wheat C-bZIP gene, TabZIP14-B, participates in salt and freezing tolerance in transgenic plants. Front. Plant Sci. 2017, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Lei, W.; Hui, M.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef]
- Wang, B.; Zheng, J.; Liu, Y.; Wang, J.; Wang, G. Cloning and characterization of the stress-induced bZIP gene ZmbZIP60 from maize. Mol. Biol. Rep. 2012, 39, 6319–6327. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ali, Z.; Xu, L.; He, X.; Huang, Y.; Yi, J.; Shao, H.; Ma, H.; Zhang, D. The nuclear protein GmbZIP110 has transcription activation activity and plays important roles in the response to salinity stress in soybean. Sci. Rep. 2016, 6, 20366. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Dong, C. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef] [PubMed]
- Orellana, S.; Yanez, M.; Espinoza, A.; Verdugo, I.; González, E.; Ruiz-Lara, S.; Casaretto, J. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, M.; Cáceres, S.; Orellana, S.; Bastías, A.; Verdugo, I.; Ruiz-Lara, S.; Casaretto, J.A. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep. 2009, 28, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Seong, E.S.; Kwon, S.S.; Ghimire, B.K.; Yu, C.Y.; Cho, D.H.; Lim, J.D.; Kim, K.S.; Heo, K.; Lim, E.S.; Chung, I.M. LebZIP2 induced by salt and drought stress and transient overexpression by Agrobacterium. BMB Rep. 2008, 41, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Sell, S.; Hehl, R. Functional dissection of a small anaerobically induced bZIP transcription factor from tomato. FEBS J. 2004, 271, 4534–4544. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/ SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Sun, Y.F.; Kai, W.B.; Liang, B.; Zhang, Y.S.; Zhai, X.W.; Jiang, L.; Du, Y.W.; Leng, P. Interactions of ABA signaling core components (SlPYLs, SlPP2Cs, and SlSnRK2s) in tomato (Solanum lycopersicon). J. Plant Physiol. 2016, 205, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Li, C. Closely Related NAC Transcription Factors of Tomato Differentially Regulate Stomatal Closure and Reopening during Pathogen Attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Muñozmayor, A.; Pineda, B.; Garciaabellán, J.O.; Antón, T.; Garciasogo, B.; Sanchezbel, P.; Flores, F.B.; Atarés, A.; Angosto, T.; Pintortoro, J.A. Overexpression of dehydrin tas14 gene improves the osmotic stress imposed by drought and salinity in tomato. J Plant Physiol. 2012, 169, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Guo, X.; Xiao, G.; Di, Z.; Wei, Z.; Chen, J.; Li, T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R. The Spectral Determination of Chlorophylls a and b, as well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- An, G. Binary ti vectors for plant transformation and promoter analysis. Methods Enzymol. 1987, 153, 292–305. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Bačkor, M.; Fahselt, D.; Wu, C.T. Free proline content is positively correlated with copper tolerance of the lichen photobiont Trebouxia erici (Chlorophyta). Plant Sci. 2004, 167, 151–157. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, J.S.; Chen, S.Y.; Zhang, W.K. Role of Soybean GmbZIP132 under Abscisic Acid and Salt Stresses. J. Integr. Plant Biol. 2008, 50, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, Y.; Kang, J.M.; Zhang, T.J.; Yang, Q.C.; Fang, F. Construction of RNAi Expression Vector of MsZIP Gene from Medicago sativa L. and Genetic Transformation in Alfalfa. Chin. J. Grassl. 2012, 3, 26–30. [Google Scholar]
- Yamauchi, Y.; Ai, F.; Seki, K.; Toyoda, Y.; Tanaka, K.; Sugimoto, Y. Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 2008, 46, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Zhu, S.; Tan, L.; Qi, W.; He, S.; Wang, G. Overexpression of OsLEA4 enhances drought, high salt and heavy metal stress tolerance in transgenic rice (Oryza sativa L.). Environ. Exp. Bot. 2016, 123, 68–77. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.K. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997, 114, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, P.; Thiery, L.; Buhot, N.; Grenier-De March, G.; Savouré, A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant. 2004, 120, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Nayyar, H.; Walia, D.P. Water stress induced proline accumulation in contrasting wheat genotypes as affected by calcium and abscisic acid. Biol. Plant. 2003, 46, 275–279. [Google Scholar] [CrossRef]
- Su, J.; Wu, R. Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci. 2004, 166, 941–948. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Lee, D.K.; Yu, I.J.; Kim, Y.S.; Yang, D.C.; Kim, J.K. Overexpression of the OsbZIP66 transcription factor enhances drought tolerance of rice plants. Plant Biotechnol. Rep. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Cai, H.; Bai, X.; Ji, W.; Ding, X.; Zhu, Y. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J. Plant Res. 2012, 125, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; Mccarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Luan, Y.S.; Liu, Z. SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissue Organ Cult. 2015, 123, 67–81. [Google Scholar] [CrossRef]
- Godoy, J.A.; Lunar, R.; Torresschumann, S.; Moreno, J.; Rodrigo, R.M.; Pintortoro, J.A. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol. Biol. 1994, 26, 1921–1934. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Primer Sequences (5′-3′) | |
|---|---|---|
| SlbZIP38 | CCATGCAAGCTTTCAAAGAAGCAGCTGT | GAGATGAATACGACGTACTAGAGTTGG |
| SlbZIP38-Q | GAGGTGTTTCATGTGGTTAGGTGGAT | CGGCTTGCTGAGAAGACTGTTGC |
| SlTAS14-Q | AGAAGGTGGGAGGAGAAAGAAG | ATGGAGATGAAAACAAAGGTGTT |
| SlNCED-Q | CCGGTGGTTTACGACAAGAA | TCCAGAGGTGGAAACAGAAAC |
| SlPP2C-Q | CAGTGATGGATTATGGGACGTGGTA | CCTAGCCAAGGCTAATTTCGTCAA |
| SlAREB1-Q | CAGGTGAGGGTGGAAGTGGTGGTGG | TGTTTGATTCTCCTCAGCATTCCAT |
| SlELF-α | ACCTTTGCTGAATACCCTCCATTG | CACACTTCACTTCCCCTTCTTCTG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Y.; Hu, X.; Li, C.; Xu, X.; Su, C.; Li, J.; Song, H.; Zhang, X.; Pan, Y. SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance. Genes 2017, 8, 402. https://doi.org/10.3390/genes8120402
Pan Y, Hu X, Li C, Xu X, Su C, Li J, Song H, Zhang X, Pan Y. SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance. Genes. 2017; 8(12):402. https://doi.org/10.3390/genes8120402
Chicago/Turabian StylePan, Yanglu, Xin Hu, Chunyan Li, Xing Xu, Chenggang Su, Jinhua Li, Hongyuan Song, Xingguo Zhang, and Yu Pan. 2017. "SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance" Genes 8, no. 12: 402. https://doi.org/10.3390/genes8120402
APA StylePan, Y., Hu, X., Li, C., Xu, X., Su, C., Li, J., Song, H., Zhang, X., & Pan, Y. (2017). SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance. Genes, 8(12), 402. https://doi.org/10.3390/genes8120402





