Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and RNA Extraction
2.2. Illumina Sequencing
2.3. Raw Sequence Processing and Read Mapping
2.4. Identification of Differential Expression Genes
2.5. Clustering Analyses
2.6. GO and KEGG Enrichment Analyses
2.7. DEG Network Analysis
2.8. Quantitative Real-Time PCR
3. Results and Discussion
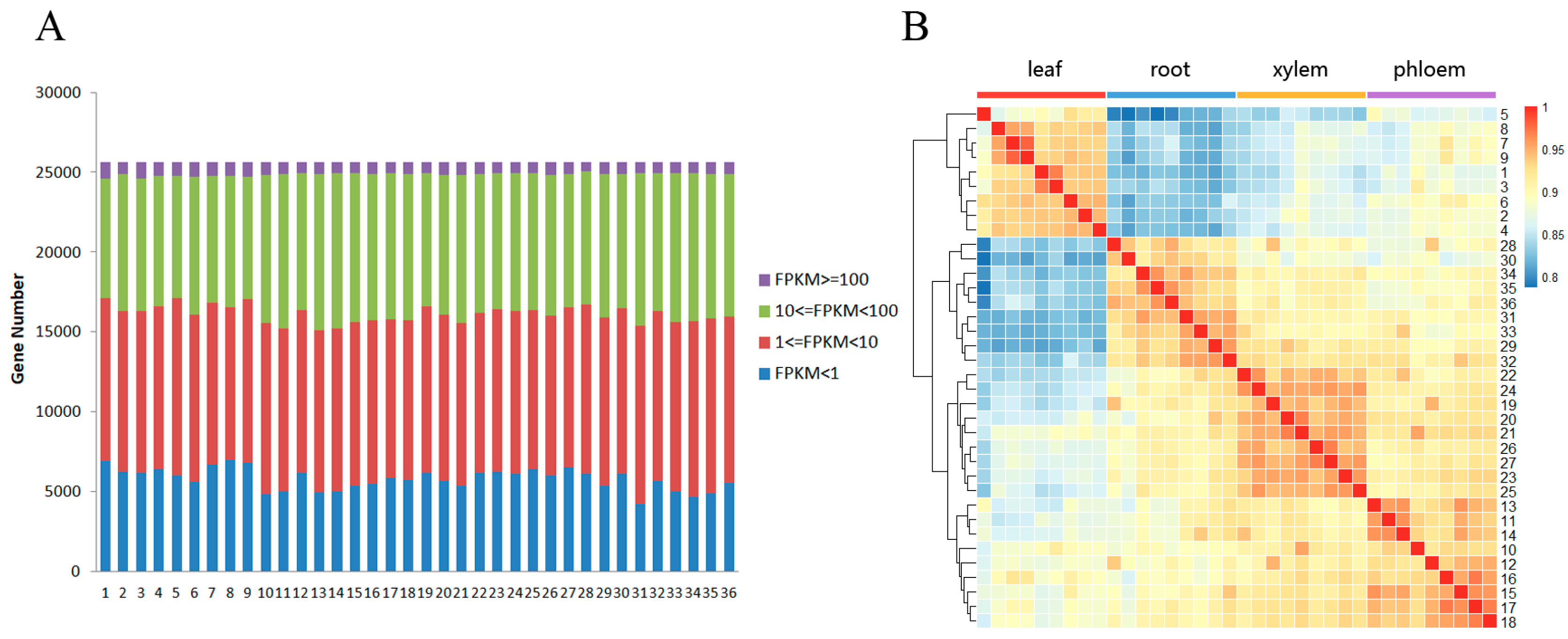
3.1. RNA-Seq and Mapping P. euphratica Sequences
3.2. Identification of DEGs from Plants Grown under Different Salt Concentrations
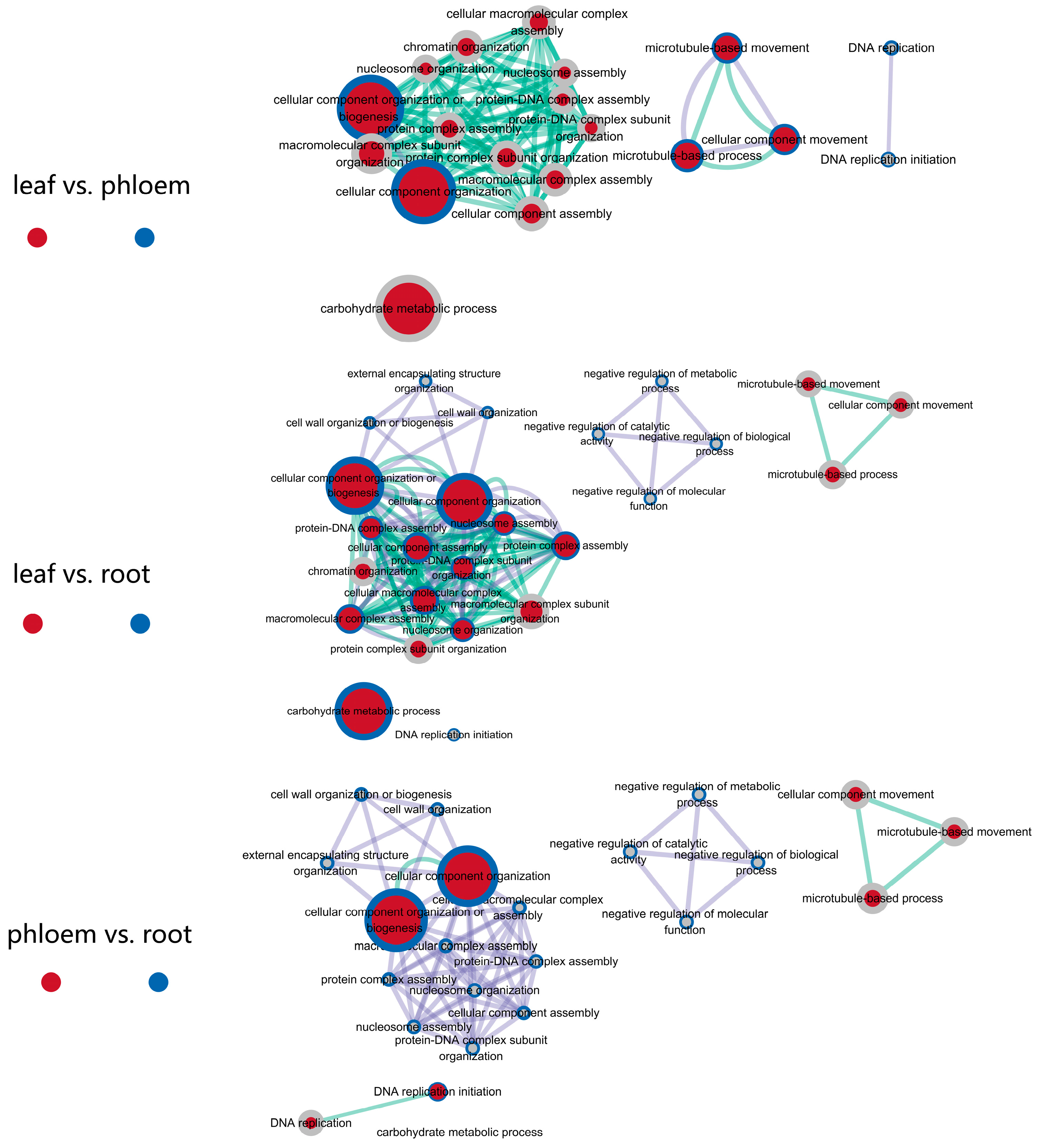
3.3. The Same Gene Functions and Expression Patterns Were Observed in Response to Salt Stress in Different Tissues
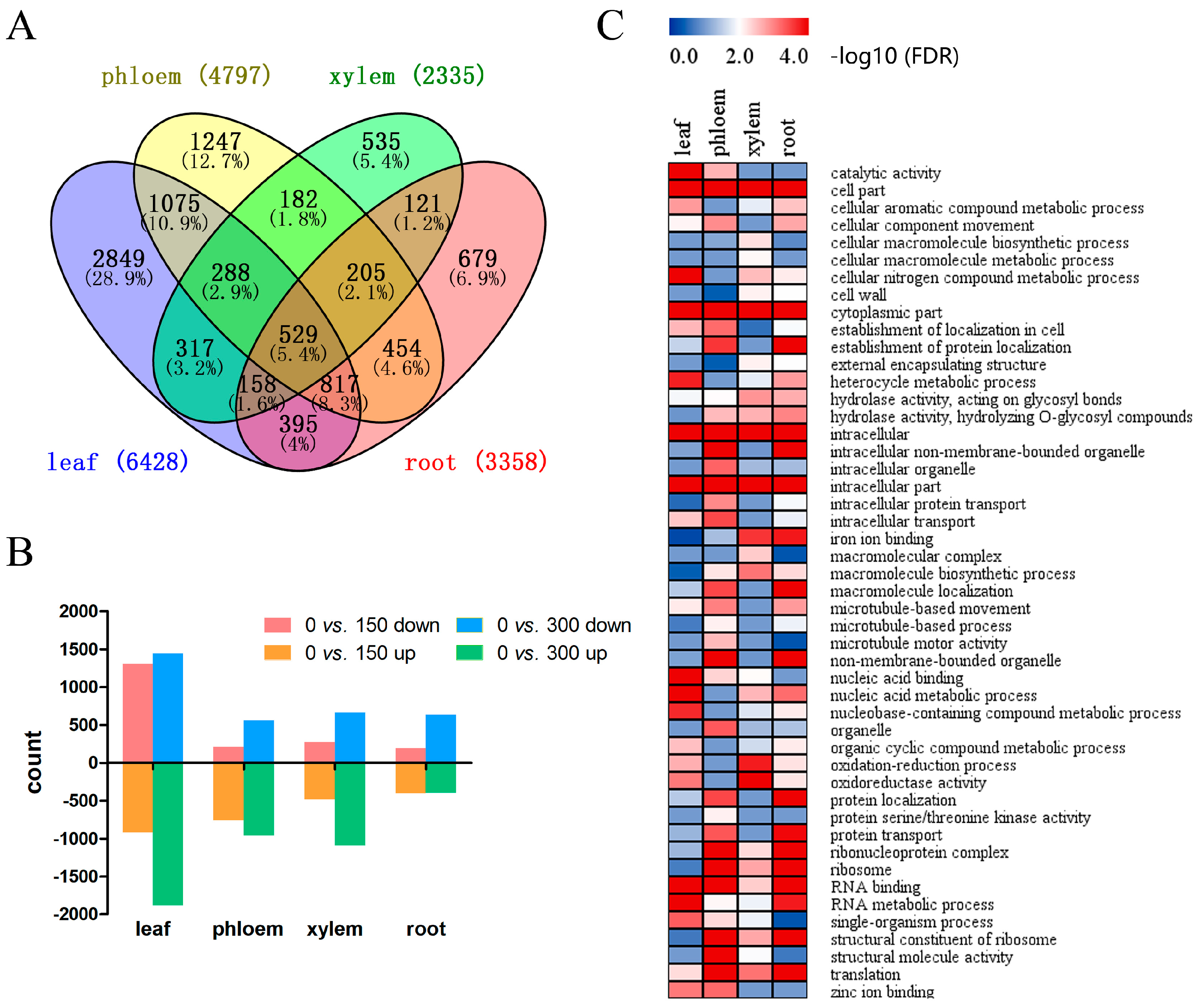
3.4. Tissue-Specific DEGs in P. euphratica under Salt Stress
3.5. Full-Tissue DEGs in Response to Salt Stress: Diversified Expression Patterns in Different Tissues
3.6. Transcription Factors in P. euphratica Involved in Response to Salinity Stress
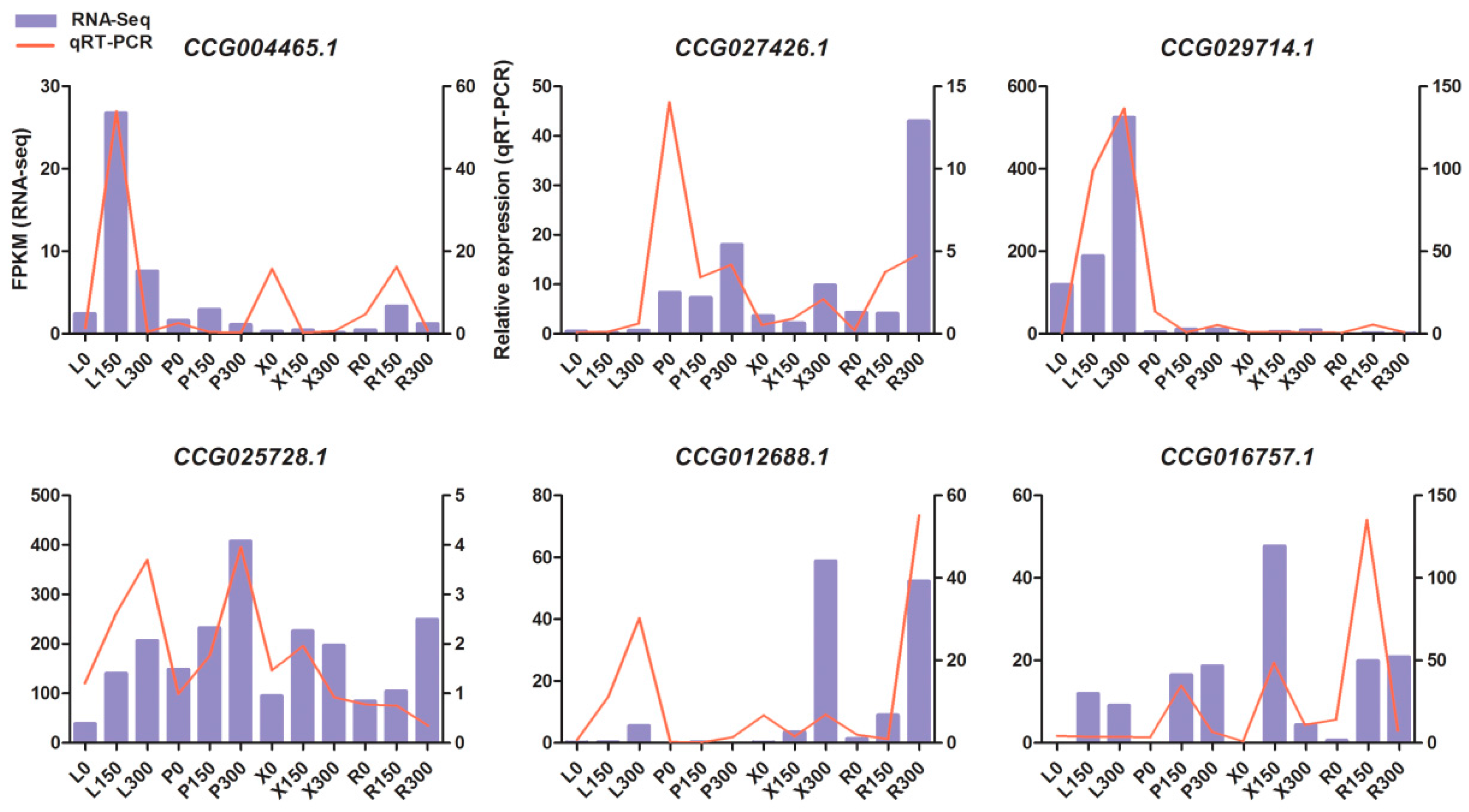
3.7. Validation of Gene Expression Patterns by qRT-PCR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A. Predicting the interaction between the effects of salinity and climate change on crop plants. Sci. Hortic. 1998, 78, 159–174. [Google Scholar] [CrossRef]
- Ziemann, M.; Kamboj, A.; Hove, R.M.; Loveridge, S.; El-Osta, A.; Bhave, M. Analysis of the barley leaf transcriptome under salinity stress using mRNA-Seq. Acta Physiol. Plant. 2013, 35, 1915–1924. [Google Scholar] [CrossRef]
- Rus, A.M.; Bressan, R.A.; Hasegawa, P.M. Unraveling salt tolerance in crops. Nat. Genet. 2005, 37, 1029–1030. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Niemietz, C.M.; Bramley, H. Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002, 25, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Langridge, P.; Paltridge, N.; Fincher, G. Functional genomics of abiotic stress tolerance in cereals. Brief. Funct. Genom. Proteom. 2006, 4, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Urano, K.; Kurihara, Y.; Seki, M.; Shinozaki, K. ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 2010, 13, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P.; Blumwald, E. Na+ transport in plants. FEBS Lett. 2007, 581, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Venema, K.; Belver, A.; Marín-Manzano, M.C.; Rodríguez-Rosales, M.P.; Donaire, J.P. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 2003, 278, 22453–22459. [Google Scholar] [CrossRef] [PubMed]
- Pardo, J.M.; Cubero, B.; Leidi, E.O.; Quintero, F.J. Alkali cation exchangers: Roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 2006, 57, 1181–1199. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Polle, A.; Chen, S.-L. On the salty side of life: Molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 2015, 38, 1794–1816. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, L.; Liu, M.; Wang, M.; Ding, M.; Deng, S.; Lu, C.; Zhou, X.; Shen, X.; Zheng, X.; et al. Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell Tissue Org. Cult. 2010, 103, 205–215. [Google Scholar] [CrossRef]
- Sun, J.; Wang, M.J.; Ding, M.Q.; Deng, S.R.; Liu, M.Q.; Lu, C.F.; Zhou, X.Y.; Shen, X.; Zheng, X.J.; Zhang, Z.K.; et al. H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant Cell Environ. 2010, 33, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Du, N.; Liu, X.; Li, Y.; Chen, S.; Zhang, J.; Ha, D.; Deng, W.; Sun, C.; Zhang, Y.; Pijut, P.M. Genetic transformation of Populus tomentosa to improve salt tolerance. Plant Cell Tissue Org. Cult. 2012, 108, 181–189. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kojima, K.; Ide, Y.; Sasaki, S. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Org. Cult. 2000, 63, 199. [Google Scholar] [CrossRef]
- Sixto, H.; Grau, J.M.; Alba, N.; Alía, R. Response to sodium chloride in different species and clones of genus populus L. Forestry 2005, 78, 93–104. [Google Scholar] [CrossRef]
- Ottow, E.A.; Brinker, M.; Teichmann, T.; Fritz, E.; Kaiser, W.; Brosché, M.; Kangasjärvi, J.; Jiang, X.; Polle, A. Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiol. 2005, 139, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Ottow, E.A.; Polle, A.; Brosché, M.; Kangasjärvi, J.; Dibrov, P.; Zörb, C.; Teichmann, T. Molecular characterization of PeNhaD1: The first member of the NhaD Na+/H+ antiporter family of plant origin. Plant Mol. Biol. 2005, 58, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Hou, P.; Shen, X.; Wang, M.; Deng, S.; Sun, J.; Xiao, F.; Wang, R.; Zhou, X.; Lu, C.; et al. Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol. Biol. 2010, 73, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J.; et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef] [PubMed]
- KY, Y.; DR, H.; WL, R. Validating clustering for gene expression data. Bioinformatics 2001, 17, 309–318. [Google Scholar]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa. Science 2006, 313, 1596. [Google Scholar] [CrossRef] [PubMed]
- Marchant, A.; Mougel, F.; Almeida, C.; Jacquin-Joly, E.; Costa, J.; Harry, M. De novo transcriptome assembly for a non-model species, the blood-sucking bug Triatoma Brasiliensis, a vector of chagas disease. Genetica 2015, 143, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Ma, T.; Hu, Q.; Liu, B.; Wu, Y.; Zhou, H.; Wang, Q.; Wang, J.; Liu, J. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011, 31, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Feng, J.; Lu, J.; Yang, Y.; Zhang, X.; Wan, D.; Liu, J. Transcriptome differences between two sister desert poplar species under salt stress. BMC Genom. 2014, 15, 337. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lu, J.; Xu, J.; Duan, B.; He, X.; Liu, J. Genome-wide identification of WRKY genes in the desert poplar Populus euphratica and adaptive evolution of the genes in response to salt stress. Evol. Bioinform. 2015, 11 (Suppl. 1), 47–55. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; He, X.; Bai, X.; Niu, Z.; Duan, B.; Chen, N.; Shao, X.; Wan, D. Genome-wide survey reveals transcriptional differences underlying the contrasting trichome phenotypes of two sister desert poplars. Genes 2016, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.-J.; Zhang, C.; Zou, Q.; Ji, Z.-L. TiSGeD: A database for tissue-specific genes. Bioinformatics 2010, 26, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.-S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Shabala, L.; Cuin, T.A.; Huang, X.; Zhou, M.; Munns, R.; Shabala, S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 2016, 67, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002, 30, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.-Y.; Zhang, H.-C.; Chen, J.-H.; Xia, X.-L.; Yin, W.-L. Molecular characterization of putative vacuolar NHX-type Na+/H+ exchanger genes from the salt-resistant tree Populus euphratica. Physiol. Plant. 2009, 137, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Luo, W.; Lin, W.; Ma, L.; Kabir, M.H. Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol. Proced. Online 2015, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lou, Y.; Sun, H.; Li, L.; Wang, L.; Dong, L.; Gao, Z. Transcriptome and comparative gene expression analysis of Phyllostachys edulis in response to high light. BMC Plant Biol. 2016, 16, 34. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Belfield, E.J.; Mithani, A.; Visscher, A.; Ragoussis, J.; Mott, R.; Smith, J.A.C.; Harberd, N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012, 31, 4359–4370. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B.; Mohanty, P. Defense potentials to NaCl in a mangrove, Bruguiera parviflora: Differential changes of isoforms of some antioxidative enzymes. J. Plant Physiol. 2004, 161, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Brosché, M.; Vinocur, B.; Alatalo, E.R.; Lamminmäki, A.; Teichmann, T.; Ottow, E.A.; Djilianov, D.; Afif, D.; Bogeat-Triboulot, M.-B.; Altman, A.; et al. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Boil. 2005, 6, R101. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, J.-K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.-K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Y.; Wang, J.-P.; He, Y. A Populus euphratica NAC protein regulating Na+/K+ homeostasis improves salt tolerance in Arabidopsis thaliana. Gene 2013, 521, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-C.; Kim, J.-Y.; Ko, J.-H.; Kang, H.; Han, K.-H. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 2014, 85, 589–599. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Ma, J.; Niu, Z.; Bai, X.; Lei, W.; Shao, X.; Chen, N.; Zhou, F.; Wan, D. Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings. Genes 2017, 8, 372. https://doi.org/10.3390/genes8120372
Yu L, Ma J, Niu Z, Bai X, Lei W, Shao X, Chen N, Zhou F, Wan D. Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings. Genes. 2017; 8(12):372. https://doi.org/10.3390/genes8120372
Chicago/Turabian StyleYu, Le, Jianchao Ma, Zhimin Niu, Xiaotao Bai, Wenli Lei, Xuemin Shao, Ningning Chen, Fangfang Zhou, and Dongshi Wan. 2017. "Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings" Genes 8, no. 12: 372. https://doi.org/10.3390/genes8120372
APA StyleYu, L., Ma, J., Niu, Z., Bai, X., Lei, W., Shao, X., Chen, N., Zhou, F., & Wan, D. (2017). Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings. Genes, 8(12), 372. https://doi.org/10.3390/genes8120372



