Hierarchical Control of Nitrite Respiration by Transcription Factors Encoded within Mobile Gene Clusters of Thermus thermophilus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Nucleic Acid Manipulation and Transformation
2.3. Construction of Deletion Mutants
2.4. Promoter Activity Assays
2.5. Directed Mutagenesis
2.6. Recombinant Protein Overexpression and Purification
2.7. Western Blot
2.8. Electrophoretic Mobility Shift Assays
3. Results
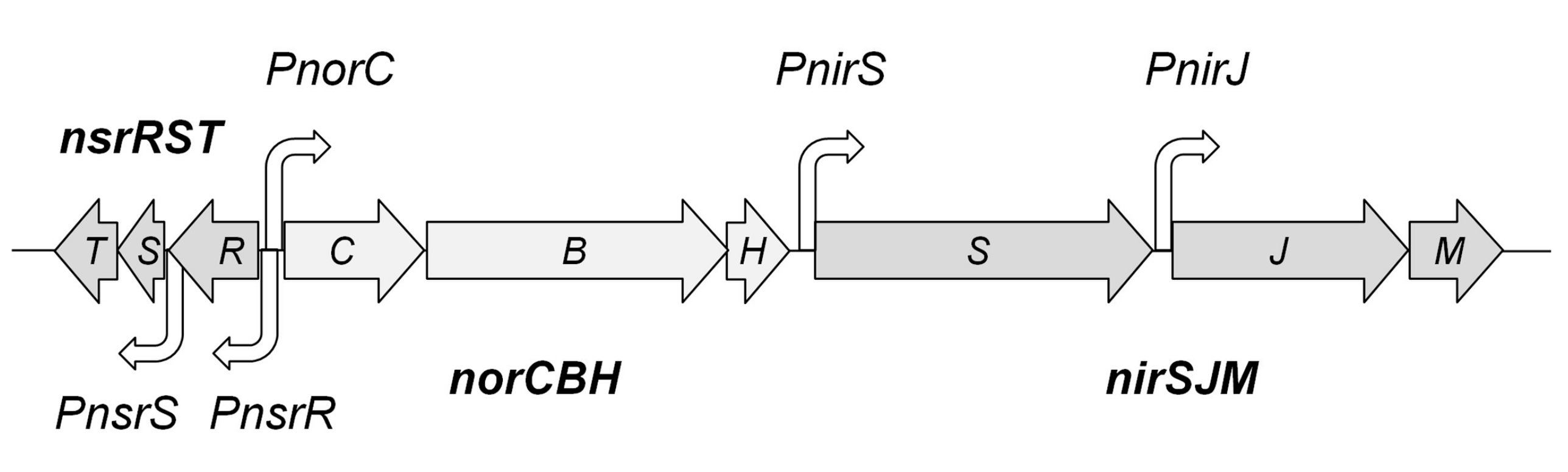
3.1. Genes and Putative Promoters of the Nic Cluster
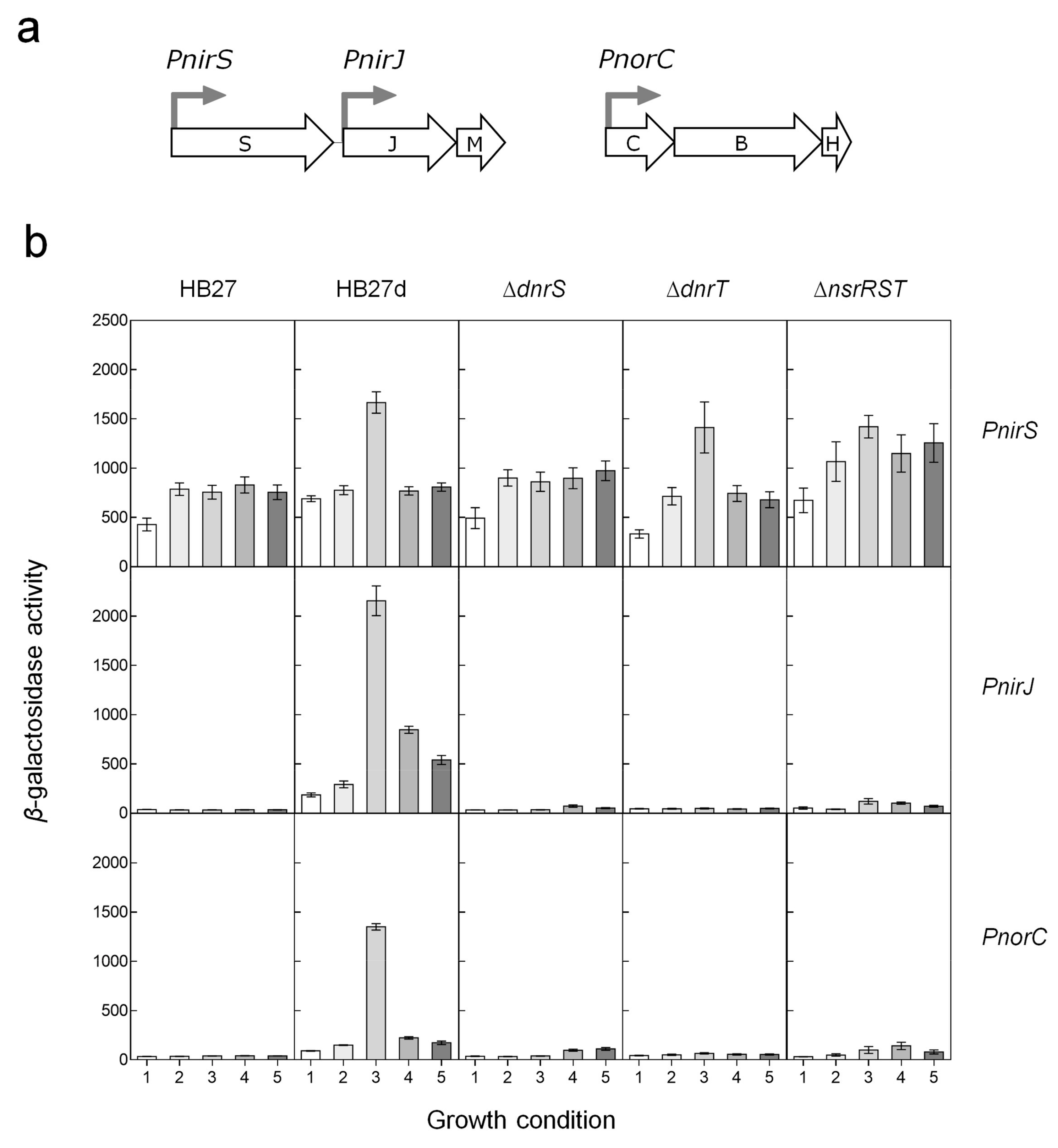
3.2. Regulation of the Nitrite and NO Reductases Gene Promoters
3.3. NsrRTh is a Transcription Factor
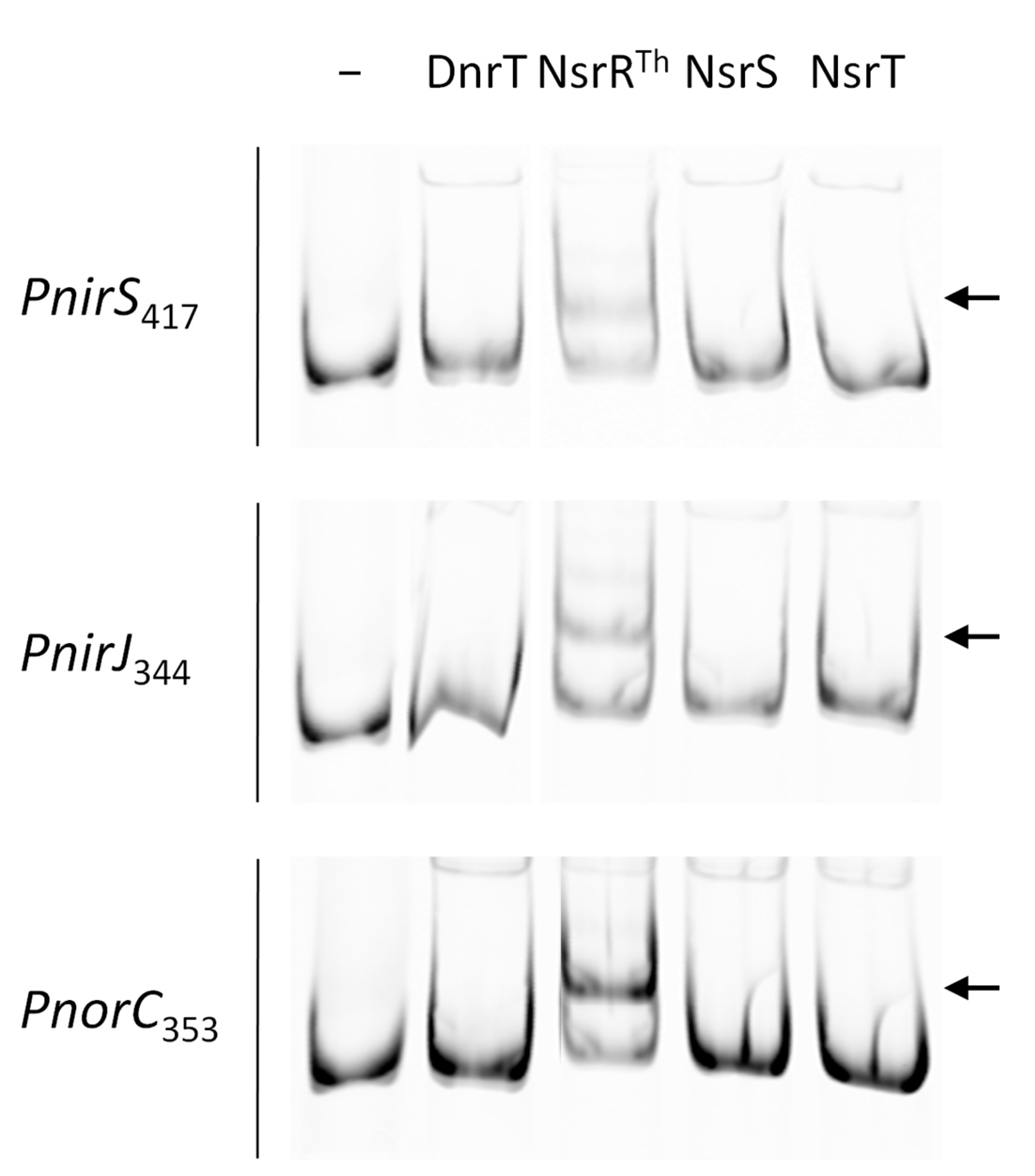
3.4. NsrRTh Binds to the Main Nic Promoters
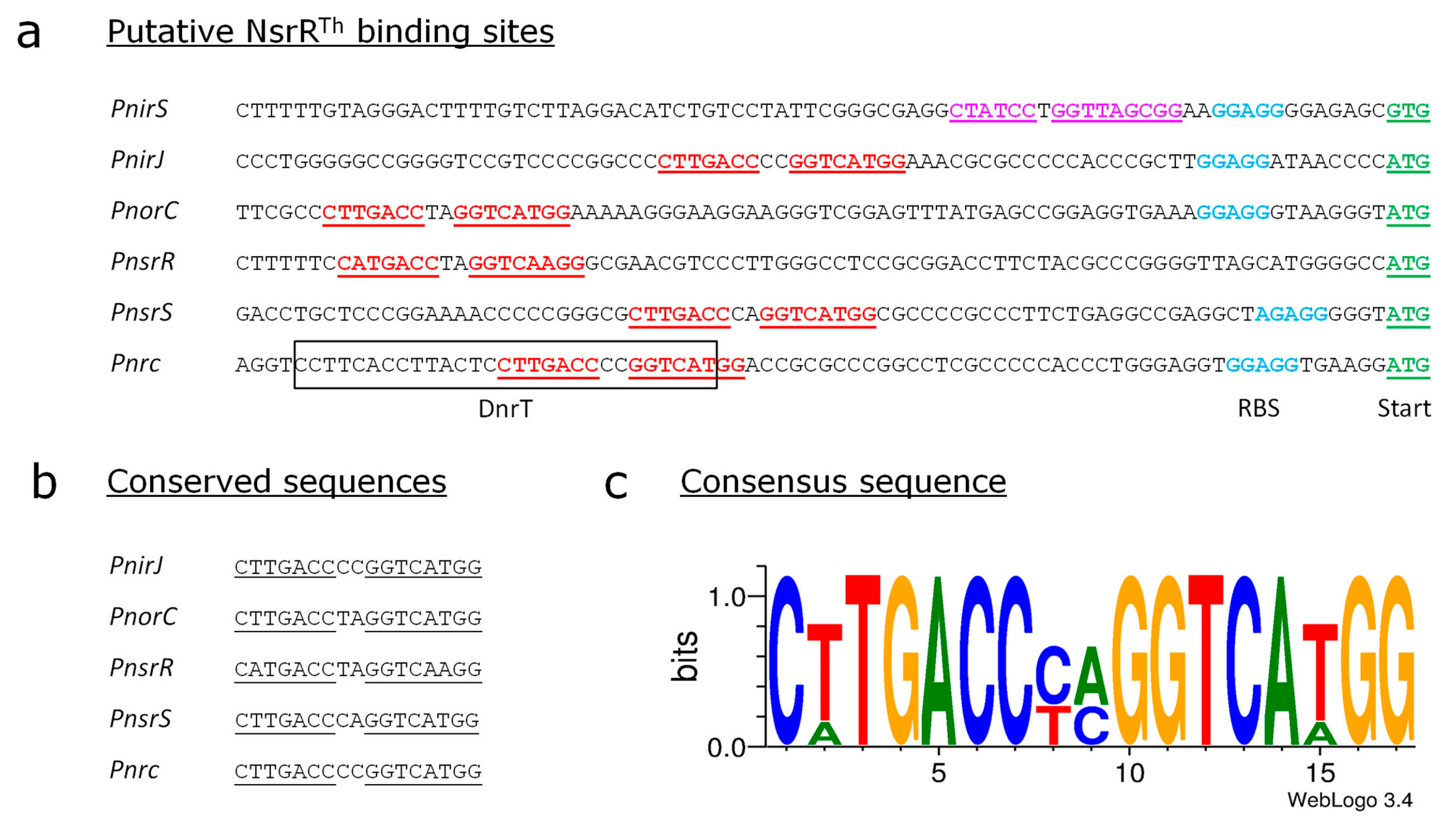
3.5. Identification of the Putative Binding Sites of NsrRTh
4. Discussion
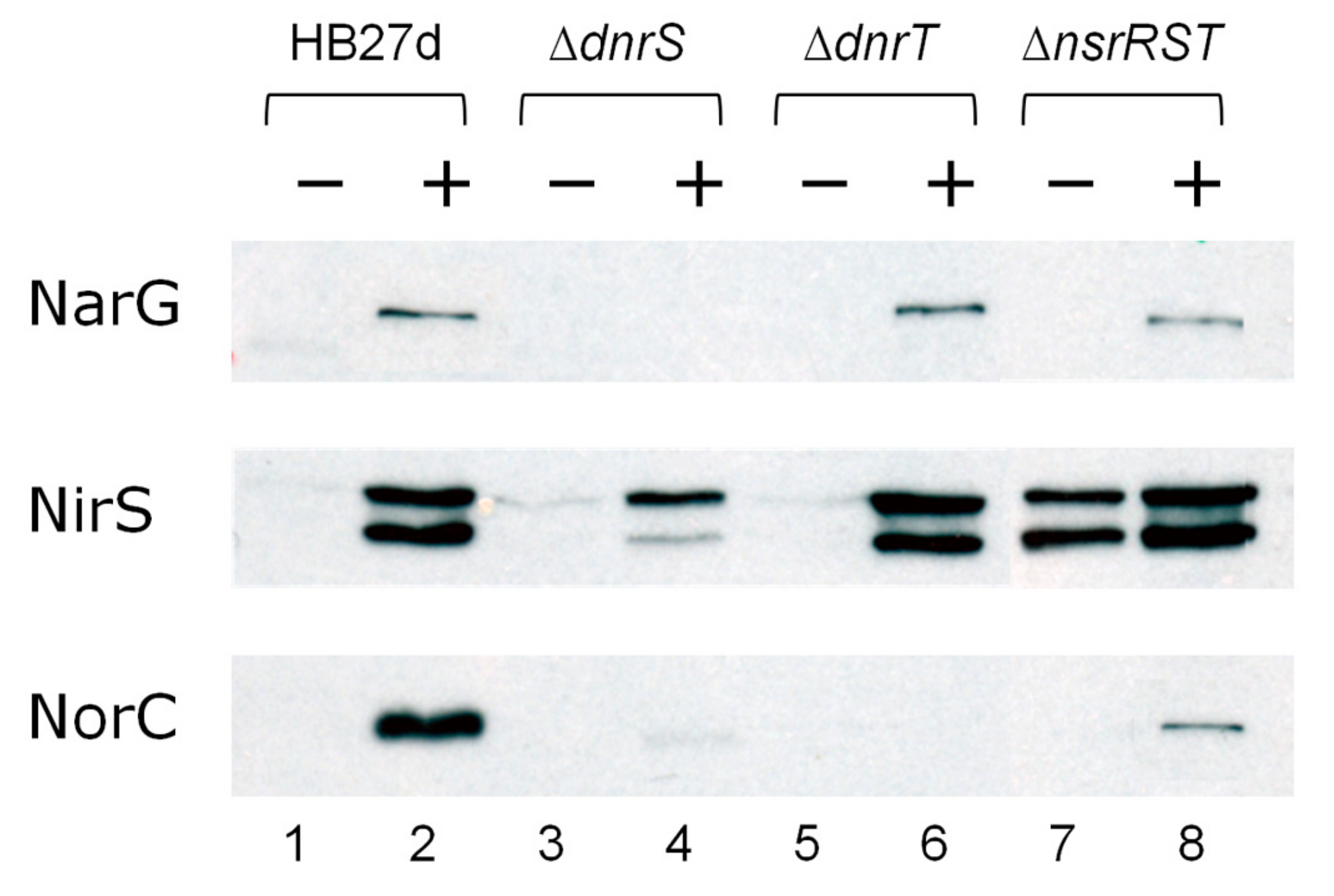
4.1. DnrS and DnrT as Master Regulators of Denitrification.
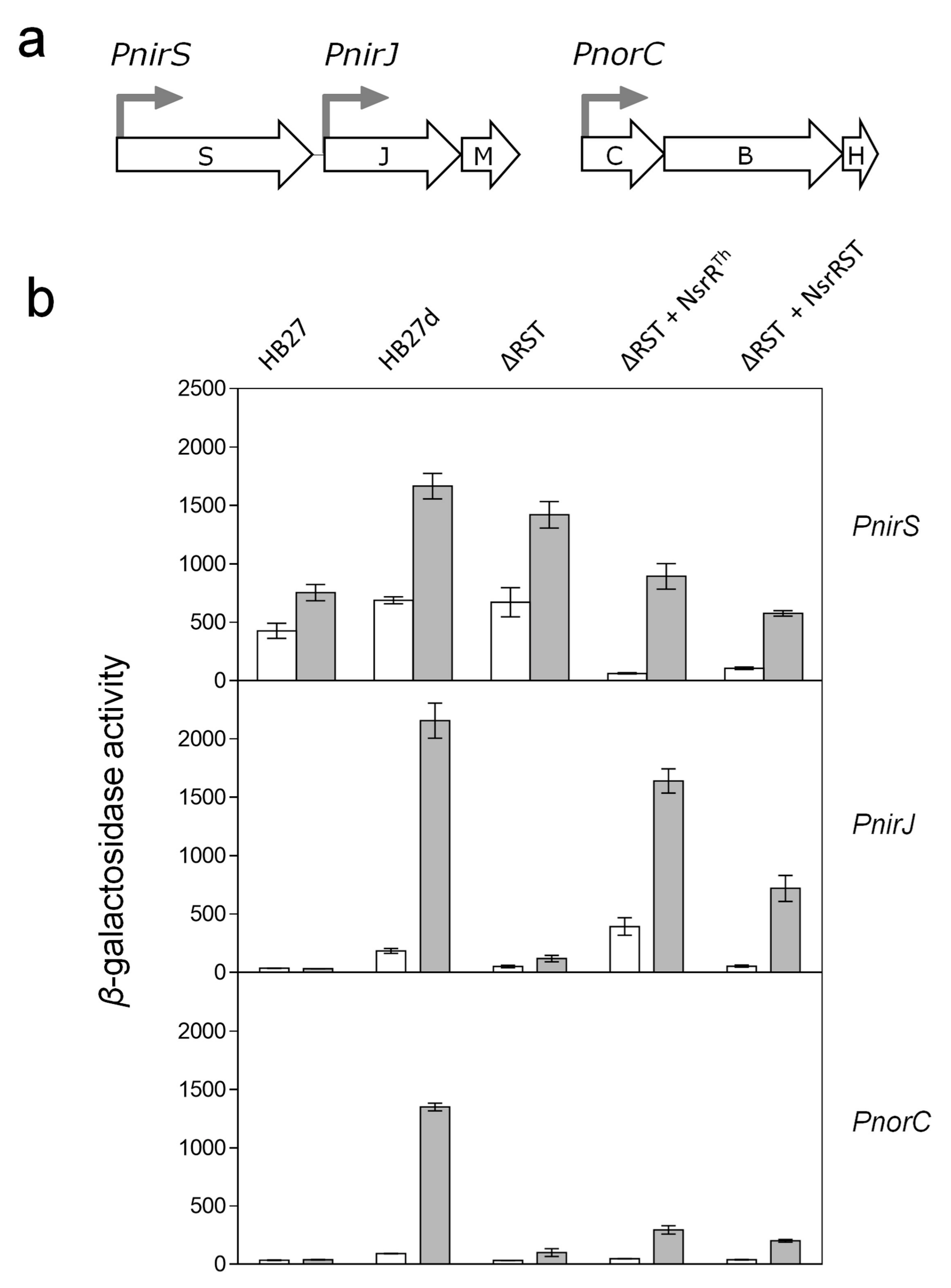
4.2. The Role of NsrRTh
4.3. The Role of NsrS and NsrT
4.4. A Regulatory Model for Nitrite Respiration
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Simon, J.; Klotz, M.G. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Bichim. Biophys. Acta 2013, 1827, 114–135. [Google Scholar] [CrossRef] [PubMed]
- Betlach, M.R. Evolution of bacterial denitrification and denitrifier diversity. Antonie van Leeuwenhoek 1982, 48, 585–607. [Google Scholar] [CrossRef] [PubMed]
- Knowles, R. Denitrification. Microbiol. Rev. 1982, 46, 43–70. [Google Scholar] [PubMed]
- Tiedje, J.M. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In Biology of Anaerobic Microorganisms; John Wiley & Sons: New York, NY, USA, 1988; pp. 179–244. [Google Scholar]
- Philippot, L.; Mirleau, P.; Mazurier, S.; Siblot, S.; Hartmann, A.; Lemanceau, P.; Germon, J.C. Characterization and transcriptional analysis of Pseudomonas fluorescens denitrifying clusters containing the nar, nir, nor and nos genes. Biochim. Biophys. Acta 2001, 1517, 436–440. [Google Scholar] [CrossRef]
- Vollack, K.U.; Xie, J.; Hartig, E.; Romling, U.; Zumft, W.G. Localization of denitrification genes on the chromosomal map of Pseudomonas aeruginosa. Microbiology 1998, 144, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [PubMed]
- Zumft, W.G.; Korner, H. Enzyme diversity and mosaic gene organization in denitrification. Antonie van Leeuwenhoek 1997, 71, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration–genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Spiro, S. Nitrous oxide production and consumption: Regulation of gene expression by gas-sensitive transcription factors. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012, 367, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Giardina, G.; Rinaldo, S.; Johnson, K.A.; Di Matteo, A.; Brunori, M.; Cutruzzola, F. NO sensing in Pseudomonas aeruginosa: Structure of the Transcriptional Regulator DNR. J. Mol. Biol. 2008, 378, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, N.; Rinaldo, S.; Giardina, G.; Cutruzzola, F. The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 2009, 155, 2838–2844. [Google Scholar] [CrossRef] [PubMed]
- Crack, J.C.; Munnoch, J.; Dodd, E.L.; Knowles, F.; Al Bassam, M.M.; Kamali, S.; Holland, A.A.; Cramer, S.P.; Hamilton, C.J.; Johnson, M.K.; et al. NsrR from Streptomyces coelicolor is a nitric oxide-sensing [4Fe-4S] cluster protein with a specialized regulatory function. J. Biol. Chem. 2015, 290, 12689–12704. [Google Scholar] [CrossRef] [PubMed]
- Averhoff, B. Shuffling genes around in hot environments: The unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 2009, 33, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Sazanov, L.A.; Hinchliffe, P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 2006, 311, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Efremov, R.G.; Baradaran, R.; Sazanov, L.A. The architecture of respiratory complex I. Nature 2010, 465, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Kolaj-Robin, O.; O’Kane, S.R.; Nitschke, W.; Leger, C.; Baymann, F.; Soulimane, T. Biochemical and biophysical characterization of succinate: Quinone reductase from Thermus thermophilus. Biochim. Biophys. Acta 2011, 1807, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Mooser, D.; Maneg, O.; Corvey, C.; Steiner, T.; Malatesta, F.; Karas, M.; Soulimane, T.; Ludwig, B. A four-subunit cytochrome bc1 complex complements the respiratory chain of Thermus thermophilus. Biochim. Biophys. Acta 2005, 1708, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Mooser, D.; Maneg, O.; MacMillan, F.; Malatesta, F.; Soulimane, T.; Ludwig, B. The menaquinol-oxidizing cytochrome bc complex from Thermus thermophilus: Protein domains and subunits. Biochim. Biophys. Acta 2006, 1757, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Soulimane, T.; Buse, G.; Bourenkov, G.P.; Bartunik, H.D.; Huber, R.; Than, M.E. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000, 19, 1766–1776. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Zafra, O.; da Costa, M.S.; Berenguer, J. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ. Microbiol. 2008, 10, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Bricio, C.; Gomez, M.J.; Berenguer, J. Lateral transfer of the denitrification pathway genes among Thermus thermophilus strains. Appl. Environ. Microbiol. 2011, 77, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Bricio, C.; Blesa, A.; Hidalgo, A.; Berenguer, J. Transferable denitrification capability of Thermus thermophilus. Appl. Environ. Microbiol. 2014, 80, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Cesar, C.E.; Averhoff, B.; Berenguer, J. Noncanonical cell-to-cell DNA transfer in Thermus spp. is insensitive to argonaute-mediated interference. J. Bacteriol. 2015, 197, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Baquedano, I.; Quintans, N.G.; Mata, C.P.; Caston, J.R.; Berenguer, J. The transjugation machinery of Thermus thermophilus: Identification of TdtA, an ATPase involved in DNA donation. PLoS Genet. 2017, 13, e1006669. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Arcos, S.; Fernandez-Herrero, L.A.; Marin, I.; Berenguer, J. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 1998, 180, 3137–3143. [Google Scholar] [PubMed]
- Alvarez, L.; Bricio, C.; Hidalgo, A.; Berenguer, J. Parallel pathways for nitrite reduction during anaerobic growth in Thermus thermophilus. J. Bacteriol. 2014, 196, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Bricio, C.; Alvarez, L.; San Martin, M.; Schurig-Briccio, L.A.; Gennis, R.B.; Berenguer, J. A third subunit in ancestral cytochrome c-dependent nitric oxide reductases. Appl. Environ. Microbiol. 2014, 80, 4871–4878. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Laptenko, O.; Borukhov, S.; Chahlafi, Z.; Blas-Galindo, E.; Gomez-Puertas, P.; Berenguer, J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 2007, 64, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Korner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Ramírez-Arcos, S.; Fernandez-Herrero, L.A.; Berenguer, J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1998, 1396, 215–227. [Google Scholar] [CrossRef]
- Hanahan, D. Techniques for transformation of E. Coli. In DNA Cloning: A Practical Approach; Glover, D.M., Ed.; IRL Press: Oxford, UK, 1985; pp. 109–135. [Google Scholar]
- Rosenberg, A.H.; Lade, B.N.; Chui, D.S.; Lin, S.W.; Dunn, J.J.; Studier, F.W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 1987, 56, 125–135. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Koyama, Y.; Hoshino, T.; Tomizuka, N.; Furukawa, K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 1986, 166, 338–340. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Messing, J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 1982, 19, 259–268. [Google Scholar] [CrossRef]
- Bricio, C.; Alvarez, L.; Gomez, M.J.; Berenguer, J. Partial and complete denitrification in Thermus thermophilus: Lessons from genome drafts. Biochem. Soc. Trans. 2011, 39, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.; Wu, H.; Akerboom, J.; Turnbull, A.P.; de Vos, W.M.; van der Oost, J. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 2005, 280, 11422–11431. [Google Scholar] [CrossRef] [PubMed]
- Lasa, I.; Caston, J.R.; Fernandez-Herrero, L.A.; de Pedro, M.A.; Berenguer, J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol. 1992, 6, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.W.; Tao, W.; Bedzyk, L.; Young, T.; Chen, M.; Li, L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 2000, 182, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, H.J.; Lens, S.I.; Reijnders, W.N.; Westerhoff, H.V.; van Spanning, R.J. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 2004, 54, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Bodenmiller, D.M.; Spiro, S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 2006, 188, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.M.; Geng, H.; Nakano, S.; Kobayashi, K. The nitric oxide-responsive regulator NsrR controls resDE-dependent gene expression. J. Bacteriol. 2006, 188, 5878–5887. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.D.; Thomson, M.J.; Read, R.C.; Moir, J.W. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 2007, 189, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Overton, T.W.; Justino, M.C.; Li, Y.; Baptista, J.M.; Melo, A.M.; Cole, J.A.; Saraiva, L.M. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 2008, 190, 2004–2013. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. Weblogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Justino, M.C.; Baptista, J.M.; Saraiva, L.M. Di-iron proteins of the Ric family are involved in iron-sulfur cluster repair. Biomet. Int. J. Role Met. Ions Biol. Biochem. Med. 2009, 22, 99–108. [Google Scholar] [CrossRef] [PubMed]

| Name | Genotype/Phenotype | Reference |
|---|---|---|
| DH5α | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80 lacZΔM15 Δ(lacZYA-argF)U169, hsdR17 (rK- mK+), λ- | [33] |
| BL21 (DE3) | F- ompT gal dcm lon hsdSB (rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | [34] |
| T. thermophilus HB27 | Wild type | Dr. Koyama |
| T. thermophilus HB27d | Denitrifying strain | [23] |
| HB27d ∆dnrS::kat | dnrS mutant | This work |
| HB27d ∆dnrT::kat | dnrT mutant | This work |
| HB27d ∆nsrRST::kat | nsrRST mutant | This work |
| Name | Sequence | Amplicon/Purpose |
|---|---|---|
| PnsrRXbaIdir | AAAATCTAGAGGAGTCCACCGTCAGGT | PnsrR/promoter probe plasmid |
| PnsrRNdeIrev | AAAACATATGCCCATGCTAACCCCGG | PnsrR/promoter probe plasmid |
| PnsrSXbaIdir | AAAATCTAGAGTGGAAAGCCGCGTGG | PnsrS/promoter probe plasmid |
| PnsrSNdeIrev | AAAACATATGCCCTCTAGCCTCGGC | PnsrS/promoter probe plasmid |
| katXbaIdir | AAAATCTAGACCCGGGAGTATAACAGA | kat/resistance cassette |
| katXbaIrev | AAAATCTAGACGTTCAAAATGGTATGCGTTTTGA | kat/resistance cassette |
| MutSa | AAAAGAATTCGCGTTTGGCGGCGTC | dnrS/deletion mutant |
| MutSb | AAAATCTAGAGCGAGGGCCTCCC | dnrS/deletion mutant |
| MutSc | AAAATCTAGAGGGGGTGAGGCCGT | dnrS/deletion mutant |
| MutSd | AAAAAAGCTTGGCGTGGCAGCGG | dnrS/deletion mutant |
| MutTa | AAAAGAATTCACCGCCCTGCGGC | dnrT/deletion mutant |
| MutTb | AAAATCTAGACTCCACGGCCTCACC | dnrT/deletion mutant |
| MutTc | AAATTCTAGATAAACGCGAGCGGTTCT | dnrT/deletion mutant |
| MutTd | AAAAAAGCTTCGCGCCTCCTCGG | dnrT/deletion mutant |
| nsrR 3’ dir | AAAATCTAGAGGCCCCATGCTAACCCCG | nsrRST/deletion mutant |
| nsrR 3’ rev | AAAAGTCGACTTGCGGCTCTGCAGGGTCAT | nsrRST/deletion mutant |
| nsrT 5’ dir | AAAAGAATTCAGGGGCTCGAGGGTGAAC | nsrRST/deletion mutant |
| nsrT 5’ rev | AAAATCTAGATGATCCCCGCAAGCCGCC | nsrRST/deletion mutant |
| nsrRrbsXbadir | AAAATCTAGAGGAGGATAGATGGCCCTTCGGAGC | nsrR/complementation plasmid |
| nsrRNdeIdir | AAAACATATGGCCCTTCGGAGCCTTC | nsrR/overexpression plasmid |
| nsrR+stopEcorev | AAATGAATTCTCAAGCGCCCGGGGG | nsrR/complementation plasmid |
| nsrR-stopEcorev | AAAAGAATTCGCGCCCGGGGG | nsrR/overexpression plasmid |
| nsrSNdeIdir2 | AAAACATATGGAAGGCCTCACCCTAAG | nsrS/overexpression plasmid |
| nsrS+stopEcorev | AAATGAATTCTCATGCCCTTCCCTCC | nsrS/overexpression plasmid |
| nsrTNdeIdir | AAAACATATGAACCTCTTGGAAAAGGC | nsrT/overexpression plasmid |
| nsrT+stopEcoRIrev | AAAAGAATTCATGGCCTCGGGGTGAT | nsrT/overexpression plasmid |
| dnrTNdeIdir | AAAACATATGGAGCTCGCCCAG | dnrT/overexpression plasmid |
| dnrTEcoRIrev | AATTGAATTCTTAGCGGATCAGGGC | dnrT/overexpression plasmid |
| PnirXbaIdir | AAAATCTAGAGCGCGACCTTATGCTCTACG | PnirS/EMSA PnirS417 |
| PnirDY782_rev | [DY782]AAAACATATGCTCCCCTCCTTCCGCTAAC | PnirS/EMSA PnirS417 (fluorescent label) |
| Pnir2XbaIdir | AAAATCTAGACTTGAAGTCACCAAGTGCTGG | PnirJ/EMSA PnirJ344 |
| Pnir2DY782_rev | [DY782]AAAACATATGGTTATCCTCCAAGCGGGTGG | PnirJ/EMSA PnirJ344 (fluorescent label) |
| PnorXbaIdir | AAAATCTAGACTTGGGCCACACCCCTC | PnorC/EMSA PnorC353 |
| PnorDY782_rev | [DY782]AAAACATATGCTTACCCTCCTTTCACCTCCG | PnorC/EMSA PnorC353 (fluorescent label) |
| Pnor_s -98_dir | CGCGGAGGCCCAAG | EMSA PnorC -98 |
| Pnor_s -85_dir | GGGACGTTCGCCCTTGAC | EMSA PnorC -85 |
| Pnor_s -75_dir | CCCTTGACCTAG | EMSA PnorC -75 |
| Pnor_s -64_dir | GGTCATGGAAAAA | EMSA PnorC -64 |
| Pnor -56_dir | AAAAAGGGAAGGAAGGGTCGGAGTTTATGAGCCGGAGGTGAAAGGAGGGTAAG | EMSA PnorC -56 |
| Pnor -56_rev | CTTACCCTCCTTTCACCTCCGGCTCATAAACTCCGACCCTTCCTTCCCTTTTT | EMSA PnorC -56 |
| Pnor -51_dir | GGAAGGGTCGGAGTTTATGAGCCGGAGGTGAAAGGAGGGTAAG | EMSA PnorC -51 |
| Pnor -51_rev | CTTACCCTCCTTTCACCTCCGGCTCATAAACTCCGACCCTTCC | EMSA PnorC -51 |
| NsrR C93A Fw | GGACCTCGCCGCCACC | nsrR/mutation |
| NsrR C93A Rv | GGTGGCGGCGAGGTCC | nsrR/mutation |
| Plasmid | Use | Reference |
|---|---|---|
| pMHbgaA | Empty promoter probe vector. HygR. | [30] |
| pMHPnsrRbgaA | PnsrR promoter probe vector. HygR. | This work |
| pMHPnsrSbgaA | PnsrS promoter probe vector. HygR. | This work |
| pUC19 | Cloning vector. AmpR | [38] |
| pUC19 ∆dnrS::kat | dnrS mutant construction. AmpR, KanR. | This work |
| pUC19 ∆dnrT::kat | dnrT mutant construction. AmpR, KanR. | This work |
| pUC19 nsrRST::kat | nsrRST mutant construction. AmpR, KanR. | This work |
| pMHPnirSbgaA | PnirS promoter probe vector. HygR. | [28] |
| pMHPnirJbgaA | PnirJ promoter probe vector. HygR. | [28] |
| pMHPnorCbgaA | PnorC promoter probe vector. HygR. | [39] |
| pWUR112/77-1 | Expression vector in T. thermophilus.BleoR. | [40] |
| pWURnsrR | Complementation of NsrRTh. BleoR. | This work |
| pWURnsrRST | Complementation of NsrRThST. BleoR. | This work |
| pET22b(+) | Expression vector in E. coli. AmpR. | Novagen, Merck KGaA, Darmstadt, Germany |
| pET28b(+) | Expression vector in E. coli. KanR. | Novagen, Merck KGaA, Darmstadt, Germany |
| pET28 dnrT | Overexpression of His-DnrT. KanR. | This work |
| pET22 nsrR | Overexpression of NsrRTh -His. AmpR. | This work |
| pET28 nsrS | Overexpression of His-NsrS. KanR. | This work |
| pET28 nsrT | Overexpression of His-NsrT. KanR. | This work |
| pET22 nsrRC93A | Overexpression of NsrRC93A-His. AmpR | This work |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez, L.; Quintáns, N.G.; Blesa, A.; Baquedano, I.; Mencía, M.; Bricio, C.; Berenguer, J. Hierarchical Control of Nitrite Respiration by Transcription Factors Encoded within Mobile Gene Clusters of Thermus thermophilus. Genes 2017, 8, 361. https://doi.org/10.3390/genes8120361
Alvarez L, Quintáns NG, Blesa A, Baquedano I, Mencía M, Bricio C, Berenguer J. Hierarchical Control of Nitrite Respiration by Transcription Factors Encoded within Mobile Gene Clusters of Thermus thermophilus. Genes. 2017; 8(12):361. https://doi.org/10.3390/genes8120361
Chicago/Turabian StyleAlvarez, Laura, Nieves G. Quintáns, Alba Blesa, Ignacio Baquedano, Mario Mencía, Carlos Bricio, and José Berenguer. 2017. "Hierarchical Control of Nitrite Respiration by Transcription Factors Encoded within Mobile Gene Clusters of Thermus thermophilus" Genes 8, no. 12: 361. https://doi.org/10.3390/genes8120361
APA StyleAlvarez, L., Quintáns, N. G., Blesa, A., Baquedano, I., Mencía, M., Bricio, C., & Berenguer, J. (2017). Hierarchical Control of Nitrite Respiration by Transcription Factors Encoded within Mobile Gene Clusters of Thermus thermophilus. Genes, 8(12), 361. https://doi.org/10.3390/genes8120361





