Gene Regulatory Network Rewiring in the Immune Cells Associated with Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Regulatory Network Construction
2.3. Analyzing of the TFsand Hub Regulators in the Networks
2.4. Network Visualization and Comparison
3. Results
3.1. Characterize Gene Regulatory Networks of Key Immune Cells Associated with Cancer Immunotherapy
3.2. Melanoma Cells Shut Down Many Network Activities of the CD8 T Cells
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, E.; Lenferink, A.; Connor-McCourt, M.O. Cancer systems biology: Exploring cancer-associated genes on cellular networks. Cell. Mol. Life Sci. 2007, 64, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Rieckmann, J.C.; Geiger, R.; Hornburg, D.; Wolf, T.; Kveler, K.; Jarrossay, D.; Sallusto, F.; Shen-Orr, S.S.; Lanzavecchia, A.; Mann, M.; et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017, 18, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Levine, J.H.; Cogdill, A.P.; Zhao, Y.; Anang, N.A.A.S.; Andrews, M.C.; Sharma, P.; Wang, J.; Wargo, J.A.; Pe’er, D.; et al. Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell 2017, 170, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Carreno, L.J.; Prados-Rosales, R.; Lopez, M.; Baena, A.; Gonzalez, P.A. Targeting Innate Immune Cells for Immunotherapy. J. Immunol. Res. 2017, 2017, 4271384. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Buenostro, J.D.; Wu, B.J.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Neph, S.; Vierstra, J.; Stergachis, A.B.; Reynolds, A.P.; Haugen, E.; Vernot, B.; Thurman, R.E.; John, S.; Sandstrom, R.; Johnson, A.K.; et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 2012, 489, 83–90. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.R.; Tibiche, C.; Trifiro, M.; Wang, E. Network Analysis Reveals A Signaling Regulatory Loop in PIK3CA-mutated Breast Cancer Predicting Survival Outcome. Genom. Proteom. Bioinf. 2017, 15, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Zaman, N.; Mcgee, S.; Milanese, J.S.; Masoudi-Nejad, A.; O’Connor-McCourt, M. Predictive genomics: A cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin. Cancer Biol. 2015, 30, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Zaman, N.; Li, L.; Jaramillo, M.L.; Sun, Z.P.; Tibiche, C.; Banville, M.; Collins, C.; Trifiro, M.; Paliouras, M.; Nantel, A.; et al. Signaling Network Assessment of Mutations and Copy Number Variations Predict Breast Cancer Subtype-Specific Drug Targets. Cell Rep. 2013, 5, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tibiche, C.; Fu, C.; Kaneko, T.; Moran, M.F.; Schiller, M.R.; Li, S.S.C.; Wang, E. The human phosphotyrosine signaling network: Evolution and hotspots of hijacking in cancer. Genome Res. 2012, 22, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, M.; Wang, E. Dynamic modeling and analysis of cancer cellular network motifs. Integr. Biol. 2011, 3, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Li, J.; Wang, E. Signaling network analysis of ubiquitin-mediated proteins suggests correlations between the 26S proteasome and tumor progression. Mol. Biosyst. 2009, 5, 1809–1816. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Zou, J.; Zaman, N.; Beitel, L.K.; Trifiro, M.; Paliouras, M. Cancer systems biology in the genome sequencing era: Part 2, evolutionary dynamics of tumor clonal networks and drug resistance. Semin. Cancer Biol. 2013, 23, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Zou, J.; Zaman, N.; Beitel, L.K.; Trifiro, M.; Paliouras, M. Cancer systems biology in the genome sequencing era: Part 1, dissecting and modeling of tumor clones and their networks. Semin. Cancer Biol. 2013, 23, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013, 340, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Feingold, E.A.; Good, P.J.; Guyer, M.S.; Kamholz, S.; Liefer, L.; Wetterstrand, K.; Collins, F.S.; Gingeras, T.R.; Kampa, D.; Sekinger, E.A.; et al. The ENCODE (ENCyclopedia of DNA elements) Project. Science 2004, 306, 636–640. [Google Scholar]
- Philip, M.; Fairchild, L.; Sun, L.; Horste, E.L.; Amara, S.C.; Shakiba, M.; Scott, A.C.; Viale, A.; Lauer, P.; Erghoub, T.M.; et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017, 545, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of intersecting sets. IEEE Transt. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Pulko, V.; Davies, J.S.; Martinez, C.; Lanteri, M.C.; Busch, M.P.; Diamond, M.S.; Knox, K.; Bush, E.C.; Sims, P.A.; Sinari, S. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 2016, 17, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Engle, S.; Whalen, S.; Joshi, A.; Pollard, K.S. Unboxing cluster heatmaps. BMC Bioinformatics 2017, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Page, L.; Brin, S.; Motwani, R.; Winograd, T. The PageRank Citation Ranking: Bringing Order to the Web; Stanford InfoLab, Stanford University: Stanford, CA, USA, 1999. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Icwsm 2009, 8, 361–362. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformat. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Purisima, E. Network motifs are enriched with transcription factors whose transcripts have short half-lives. Trends Genet. 2005, 21, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Garín, M.I.; Chu, C.-C.; Golshayan, D.; Cernuda-Morollón, E.; Wait, R.; Lechler, R.I. Galectin-1: A key effector of regulation mediated by CD4+ CD25+ T cells. Blood 2007, 109, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Hong, P.W.; Lee, B.; Baum, L.G. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. USA 2011, 108, 10650–10655. [Google Scholar] [CrossRef] [PubMed]
- Pauken, K.E.; Sammons, M.A.; Odorizzi, P.M.; Manne, S.; Godec, J.; Khan, O.; Drake, A.M.; Chen, Z.; Sen, D.; Kurachi, M. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 2016, 354, 1160–1165. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ge, Y.; Xiao, M.; Lopez-Coral, A.; Li, L.; Roesch, A.; Huang, C.; Alexander, P.; Vogt, T.; Xu, X. SECTM1 produced by tumor cells attracts human monocytes via CD7-mediated activation of the PI3K pathway. J. Invest. Dermatol. 2014, 134, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
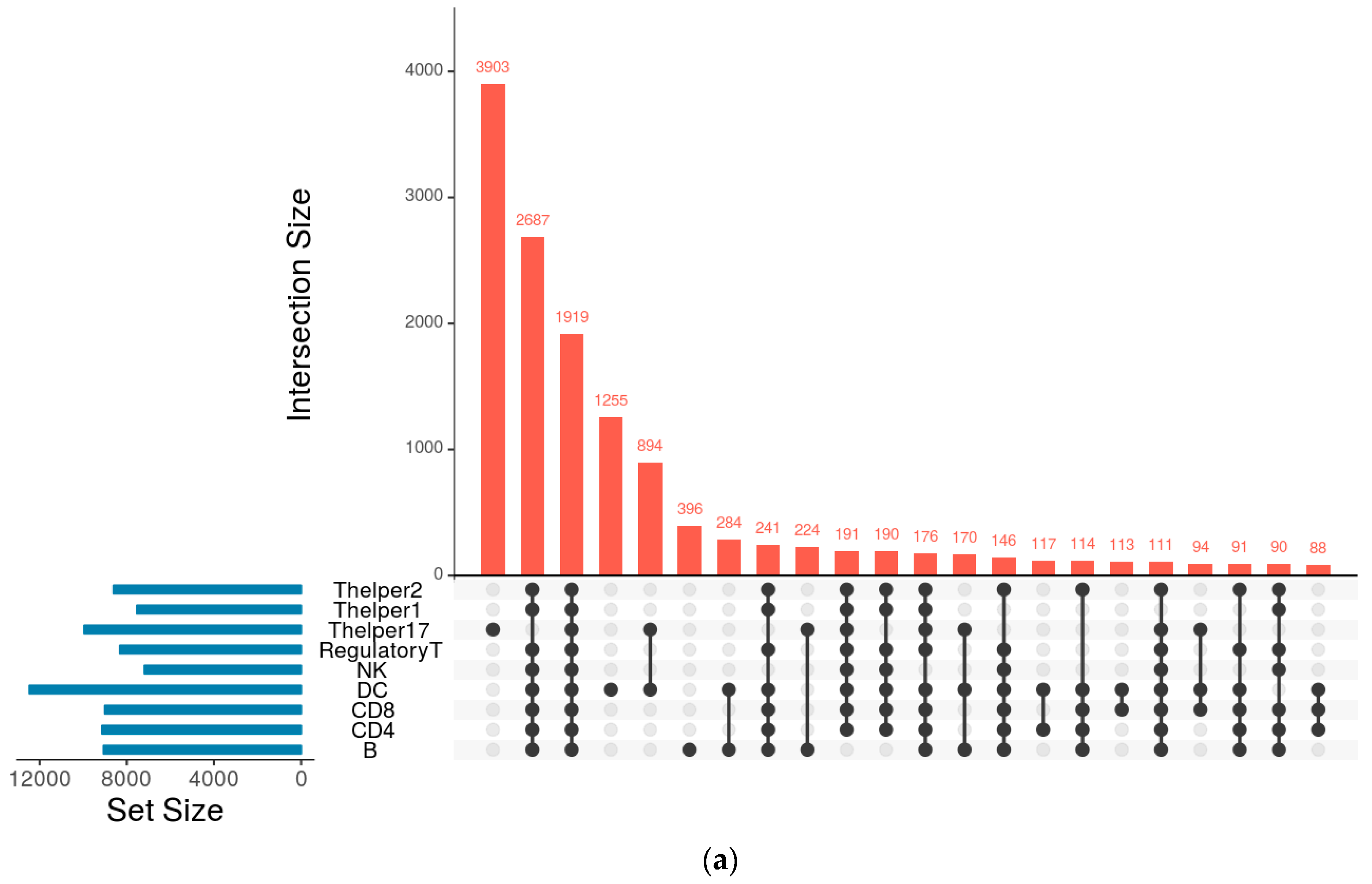
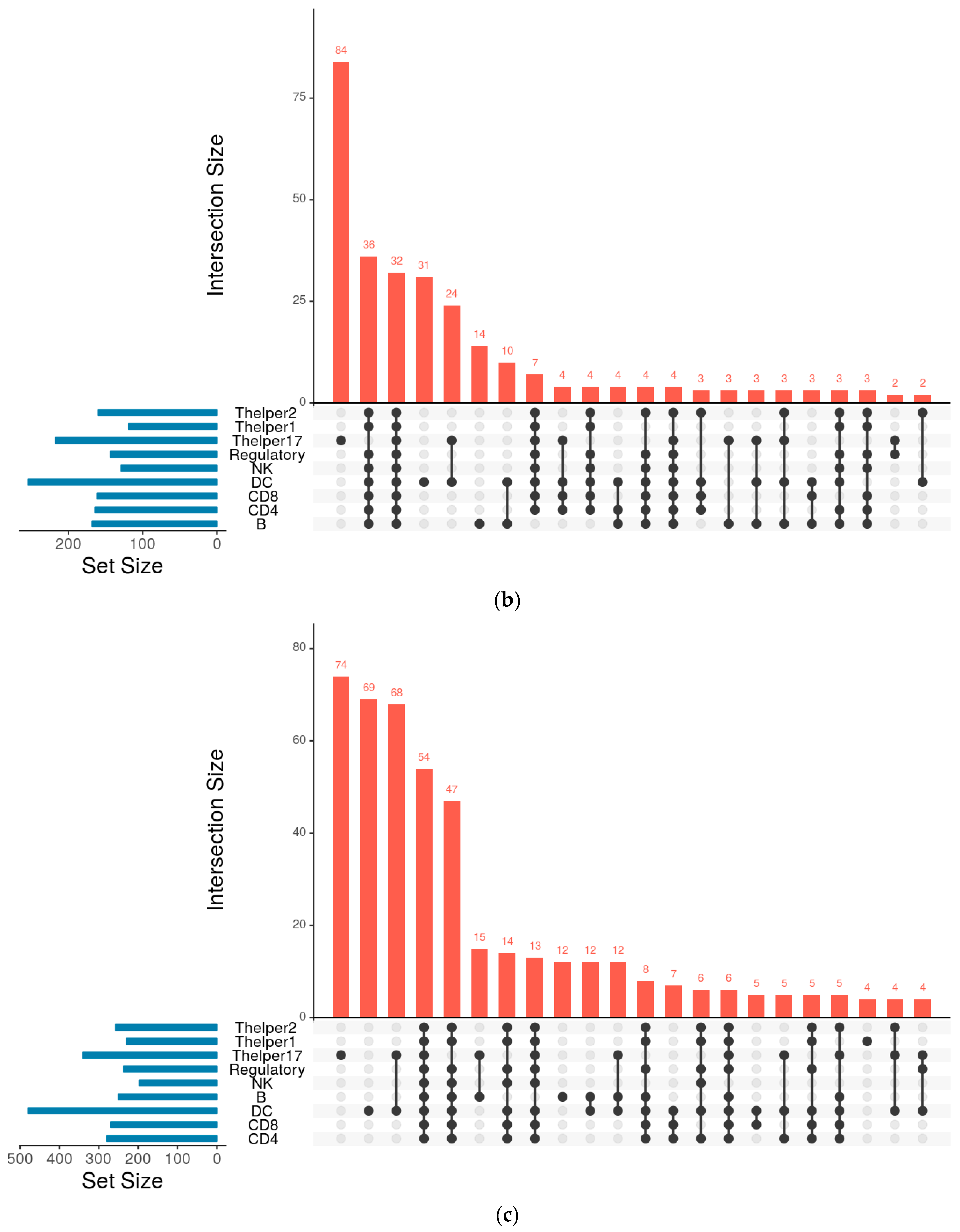
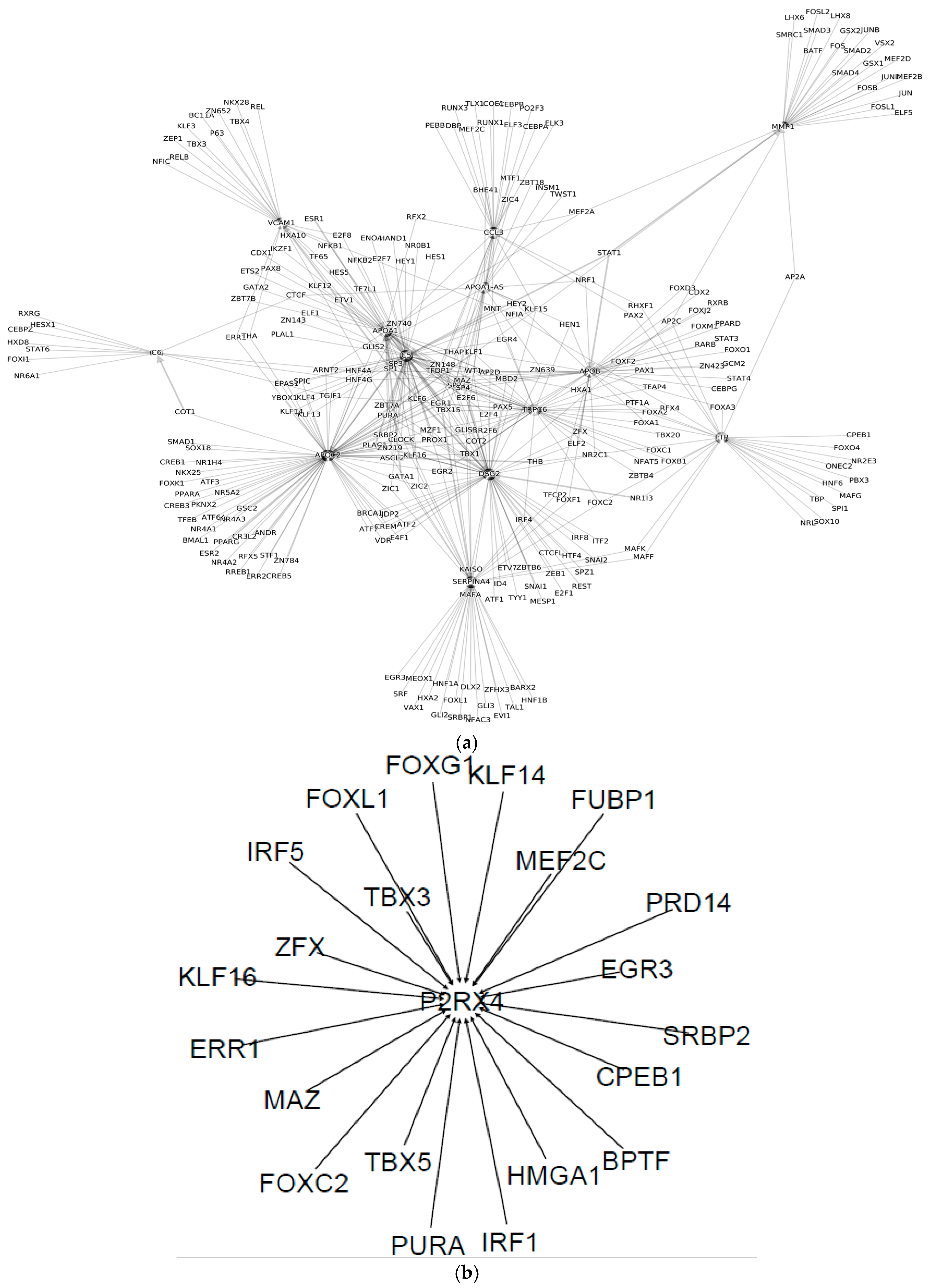


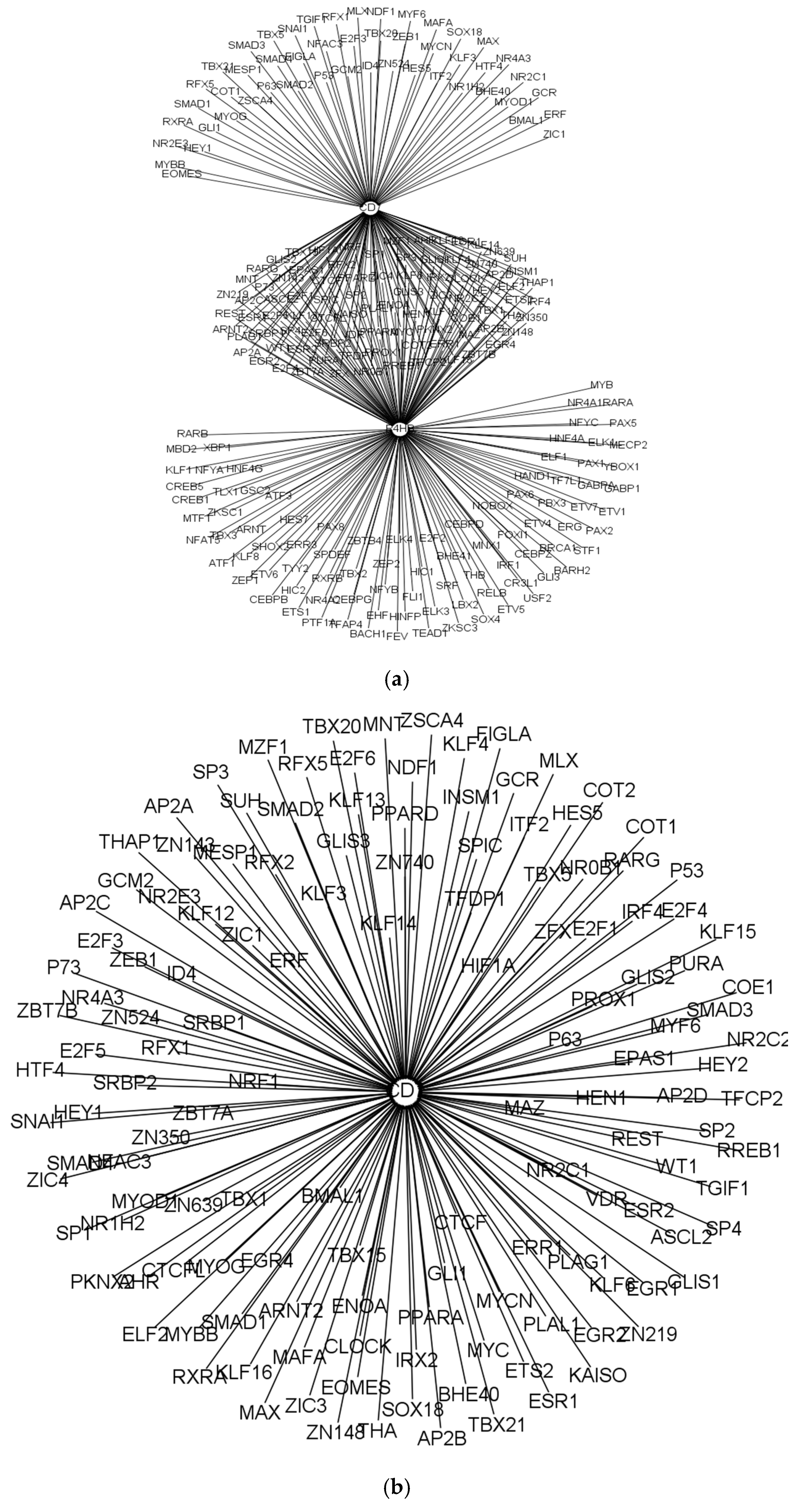
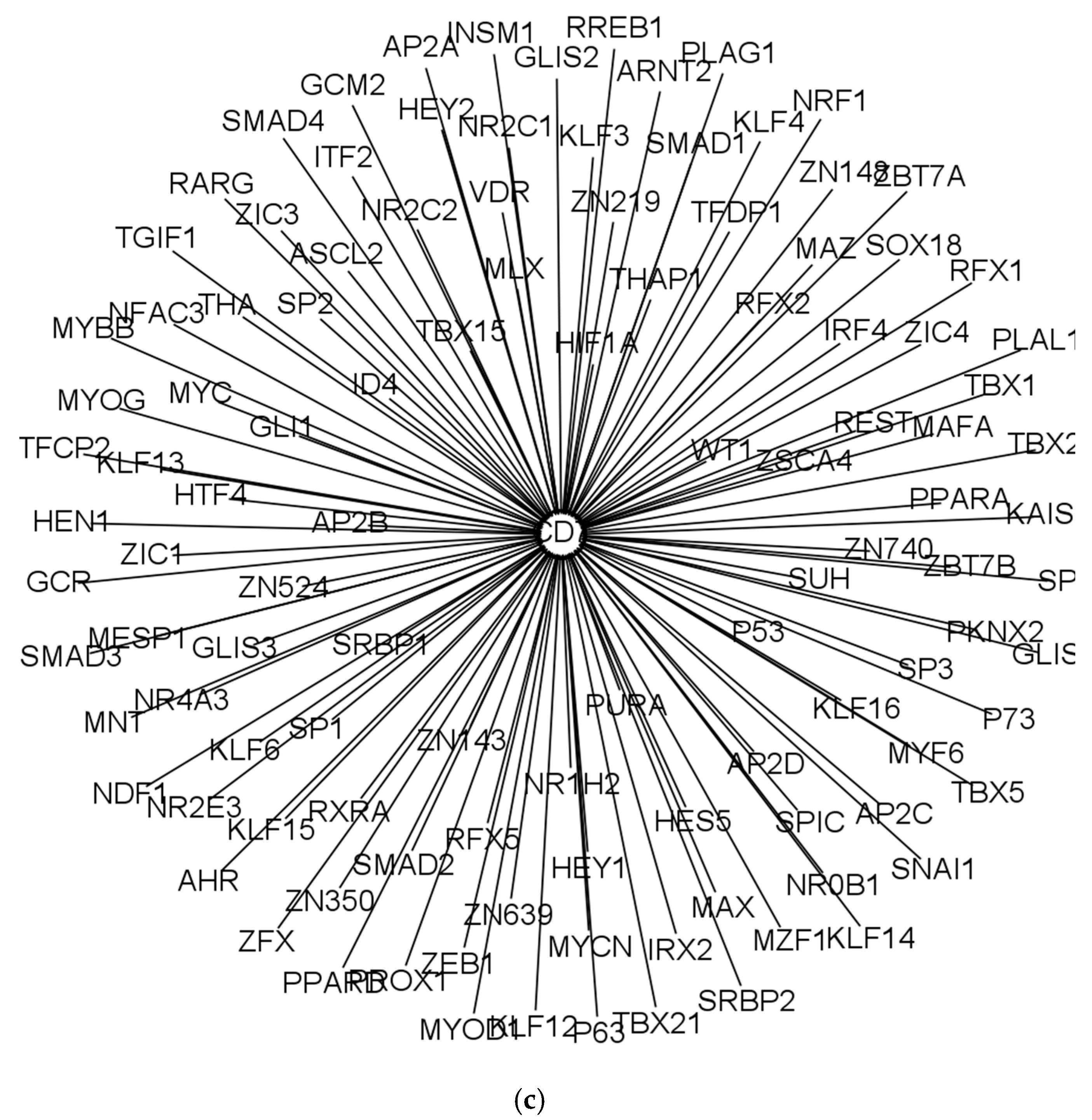
| Cell Type | Numbers of Annotated Genes | Number of Genes that Are Unique to Each Cell Type | Numbers of Receptors | Numbers of Receptor that Are Unique to Each Cell Type | Numbers of TFs | Numbers of TFs that Are Unique to Each Cell Type |
|---|---|---|---|---|---|---|
| B | 9058 | 396 | 250 | 12 | 168 | 14 |
| CD4 | 9128 | 68 | 280 | 1 | 164 | 2 |
| CD8 | 8997 | 71 | 269 | 3 | 161 | 1 |
| DC | 12,459 | 1255 | 480 | 69 | 254 | 34 |
| NK | 7186 | 39 | 197 | 0 | 129 | 1 |
| Regulatory T | 8309 | 72 | 238 | 1 | 143 | 1 |
| Thelper1 | 7533 | 41 | 229 | 4 | 119 | 1 |
| Thelper2 | 8606 | 36 | 257 | 0 | 160 | 1 |
| Thelper17 | 9947 | 3903 | 341 | 74 | 217 | 84 |
| B-Cell | |
| Pathway Name | p-Value |
| TPO Signaling Pathway | 1.95 × 10−4 |
| IL-2 Receptor Beta Chain in T Cell Activation | 4.45 × 10−5 |
| PDGF Signaling Pathway | 1.16 × 10−3 |
| Role of Calcineurin-dependent NFAT (Nuclear factor of activated T-cells) signaling in lymphocytes | 1.23792 × 10−4 |
| Phosphoinositides and their downstream targets | 5.69 × 10−4 |
| CD4 | |
| Pathway Name | p-Value |
| ErbB1 downstream signaling | 3.10 × 10−10 |
| mTOR signaling pathway | 6.80 × 10−9 |
| Ras Pathway | 6.80 × 10−9 |
| IL2-mediated signaling events | 3.17 × 10−8 |
| PDGFR (Platelet-derived growth factor receptors)-beta signaling pathway | 1.13 × 10−7 |
| CD8 | |
| Pathway Name | p-Value |
| FoxO family signaling | 1.25 × 10−8 |
| Fanconi anemia pathway | 3.60 × 10−6 |
| E2F transcription factor network | 4.26 × 10−6 |
| Dendritic Cell | |
| Pathway Name | p-Value |
| BCR signaling pathway | 0.81 × 10−8 |
| CCKR signaling map | 6.83 × 10−7 |
| TCR signaling in naive CD4+ T cells | 1.455 × 10−6 |
| CXCR4-mediated signaling events | 1.66 × 10−6 |
| FoxO family signaling | 4.05 × 10−6 |
| Class I PI3K signaling events | 4.05 × 10−6 |
| NK cells | |
| Pathway Name | p-Value |
| ATR signaling pathway | 3.25 × 10−8 |
| FoxO family signaling | 1.97 × 10−7 |
| CCKR (cholecystokinin receptor) signaling map signal transduction | 4.29 × 10−7 |
| Regulatory T cell | |
| Pathway Name | p-Value |
| ATR signaling pathway | 6.72 × 10−7 |
| Cell Type | Computational Method Employed | Key_Regulators |
|---|---|---|
| B | Network-analysis | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, TBX15, WT1, KLF4, ZN148, EGR4, TBX1, PLAG1, KLF6, ZN639, ZFX, KLF14 |
| PageRank algorithm | SP1, EGR1, TFDP1, SP4, SP2, MAZ, SP3, THAP1, KLF16, WT1, TBX15, KLF4, ZN148, EGR4, TBX1, ZFX, KLF6, ZN639, ELF2, PLAG1 | |
| CD4 | Network-analysis | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, TBX15, WT1, EGR4, KLF4, ZN148, TBX1, KLF6, ZN639, PLAG1, ZFX, ELF2, |
| PageRank algorithm | SP1, EGR1, TFDP1, MAZ, SP2, SP4, SP3, THAP1, KLF16, TBX15, WT1, EGR4, KLF4, ZN148, TBX1, KLF6ZN639, ZFX, PLAG1, ELF2 | |
| CD8 | Network-analysis | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, WT1, TBX15, EGR4, KLF4, ZN148, KLF6, TBX1, PLAG1, ZN639, ZFX, KLF14 |
| PageRank algorithm | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, WT1, TBX15, EGR4, KLF4, ZN148, KLF6, TBX1, ZN639, PLAG1, ZFX, ELF2 | |
| DC | Network-analysis | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, TBX15, WT1, EGR4, ZN148, TBX1, KLF6, PLAG1, KLF4, ZFX, ZN639, AP2D |
| PageRank algorithm | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, TBX15, WT1, EGR4, TBX1, ZN148, KLF6, KLF4, PLAG1, ZN639, ZFX, ELF2 | |
| NK | Network-analysis | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, TBX15, WT1, KLF4, EGR4, ZN148, TBX1, KLF6, ELF2, ZN639, PLAG1, ZFX |
| PageRank algorithm | SP1, EGR1, TFDP1, MAZ, SP2, SP4, SP3, THAP1, KLF16, TBX15, WT1, KLF4, EGR4, TBX1, ZN639, ZN148, KLF6, ELF2, ZFX, PLAG1 | |
| Regulatory T | Network-analysis | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, TBX15, WT1, KLF4, ZN148, EGR4, TBX1, KLF6, PLAG1, ZN639, ELF2, ZFX |
| PageRank algorithm | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, TBX15, WT1, KLF4, EGR4, ZN148, TBX1, ZN639, KLF6, ELF2, ZFX, PLAG1 | |
| Thelper17 | Network-analysis | SP1, EGR1, MAZ, SP2, TFDP1, SP4, SP3, TBX15, PLAG1, KLF16, TBX1, PAX5, PURA, ZN148, THAP1, WT1, KLF15, MNT, ZFX, AP2D |
| PageRank algorithm | SP1, EGR1, MAZ, SP2, TFDP1, SP4, SP3, TBX15, PLAG1, KLF16, TBX1, PAX5, PURA, ZN148, THAP1, MNT, WT1, KLF15, ZFX, AP2D | |
| Thelper1 | Network-analysis | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, TBX15, WT1, KLF4, ZN148, TBX1, ELF2, KLF6, EGR4, ZN639, KLF14, ZFX |
| PageRank algorithm | SP1, EGR1, TFDP1, SP2, MAZ, SP4, SP3, THAP1, KLF16, TBX15, WT1, KLF4, ELF2, ZN148, ZN639, KLF6, EGR4, TBX1, ZFX, KLF14 | |
| Thelper2 | Network-analysis | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, WT1, TBX15, KLF4, EGR4, ZN148, KLF6, TBX1, ZN639, KLF14, ELF2, PLAG1 |
| PageRank algorithm | SP1, EGR1, TFDP1, SP2, SP4, MAZ, SP3, THAP1, KLF16, WT1, TBX15, KLF4, EGR4, ZN639, KLF6, ZN148, TBX1, ELF2, ZFX, KLF14 |
| Cell Type | Name | p-Value |
|---|---|---|
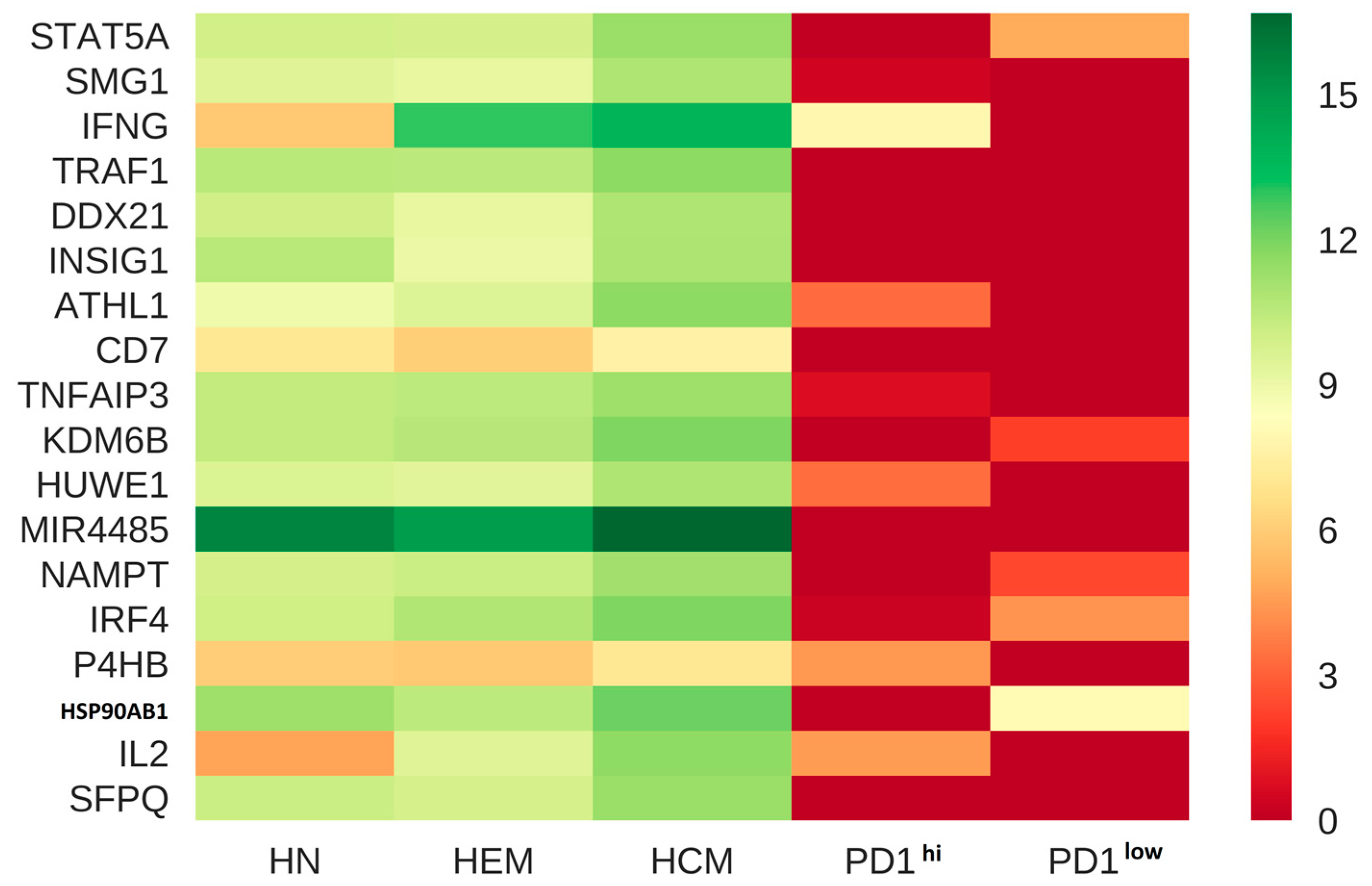
| HCM vs PD1lo | Calcineurin-regulated NFAT (Nuclear factor of activated T-cells) -dependent transcription in lymphocytes | 1.443 × 10−12 |
| IL2 signaling events mediated by STAT5 | 1.34 × 10−12 | |
| Downstream signaling in naive CD8+ T cells | 1.036 × 10−8 | |
| IL12-mediated signaling events | 2.724 × 10−8 | |
| FoxO family signaling | 3.688 × 10−8 | |
| HCM vs PD1hi | Calcineurin-regulated NFAT-dependent transcription in lymphocytes | 9.083 × 10−13 |
| IL2 signaling events mediated by STAT5 | 4.072 × 10−11 | |
| GMCSF-mediated signaling events | 8.323 × 10−9 | |
| IL2-mediated signaling events | 2.378 × 10−8 | |
| AP-1 transcription factor network | 5.012 × 10−7 | |
| HEM vs PD1lo | Calcineurin-regulated NFAT-dependent transcription in lymphocytes | 6.401 × 10−16 |
| IL2 signaling events mediated by STAT5 | 1.157 × 10−12 | |
| Downstream signaling in naive CD8+ T cells | 6.909 × 10−11 | |
| IL12-mediated signaling events | 4.682 × 10−10 | |
| AP-1 transcription factor network | 2.142 × 10−8 | |
| HEM vs PD1hi | Calcineurin-regulated NFAT-dependent transcription in lymphocytes | 2.304 × 10−14 |
| AP-1 transcription factor network | 1.869 × 10−9 | |
| IL2 signaling events mediated by STAT5 | 1.363 × 10−10 | |
| IL2-mediated signaling events | 4.521 × 10−8 | |
| IL12-mediated signaling events | 1.329 × 10−7 | |
| HN vs PD1lo | Validated targets of C-MYC transcriptional activation | 5.009 × 10−7 |
| Glucocorticoid receptor regulatory network | 5.60 × 10−5 | |
| FoxO family signaling | 4.64 × 10−5 | |
| Role of Calcineurin-dependent NFAT signaling in lymphocytes | 9.98 × 10−5 | |
| IL12-mediated signaling events | 3.25 × 10−4 | |
| HN vs PD1hi | Calcineurin-regulated NFAT-dependent transcription in lymphocytes | 8.443 × 10−8 |
| AP-1 transcription factor network | 3.14 × 10−6 | |
| IL2 signaling events | 6.686 × 10−7 | |
| IL5-mediated signaling events | 2.65 × 10−5 | |
| IL2-mediated signaling events | 4.72 × 10−5 | |
| PD1hi vs PD1lo | IL12 signaling mediated by STAT4 | 5.04 × 10−4 |
| IL12-mediated signaling events | 3.60 × 10−3 | |
| TCR signaling in naive CD4+ T cells | 4.00 × 10−3 | |
| Glucocorticoid receptor regulatory network | 8.30 × 10−3 | |
| ATF-2 transcription factor network | 7.50 × 10−2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, P.; Gopalakrishnan, C.; Yu, H.; Wang, E. Gene Regulatory Network Rewiring in the Immune Cells Associated with Cancer. Genes 2017, 8, 308. https://doi.org/10.3390/genes8110308
Han P, Gopalakrishnan C, Yu H, Wang E. Gene Regulatory Network Rewiring in the Immune Cells Associated with Cancer. Genes. 2017; 8(11):308. https://doi.org/10.3390/genes8110308
Chicago/Turabian StyleHan, Pengyong, Chandrasekhar Gopalakrishnan, Haiquan Yu, and Edwin Wang. 2017. "Gene Regulatory Network Rewiring in the Immune Cells Associated with Cancer" Genes 8, no. 11: 308. https://doi.org/10.3390/genes8110308
APA StyleHan, P., Gopalakrishnan, C., Yu, H., & Wang, E. (2017). Gene Regulatory Network Rewiring in the Immune Cells Associated with Cancer. Genes, 8(11), 308. https://doi.org/10.3390/genes8110308





