Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools
Abstract
1. Introduction
2. Current BrCa Therapeutics
3. Alternative Splicing in Cancer
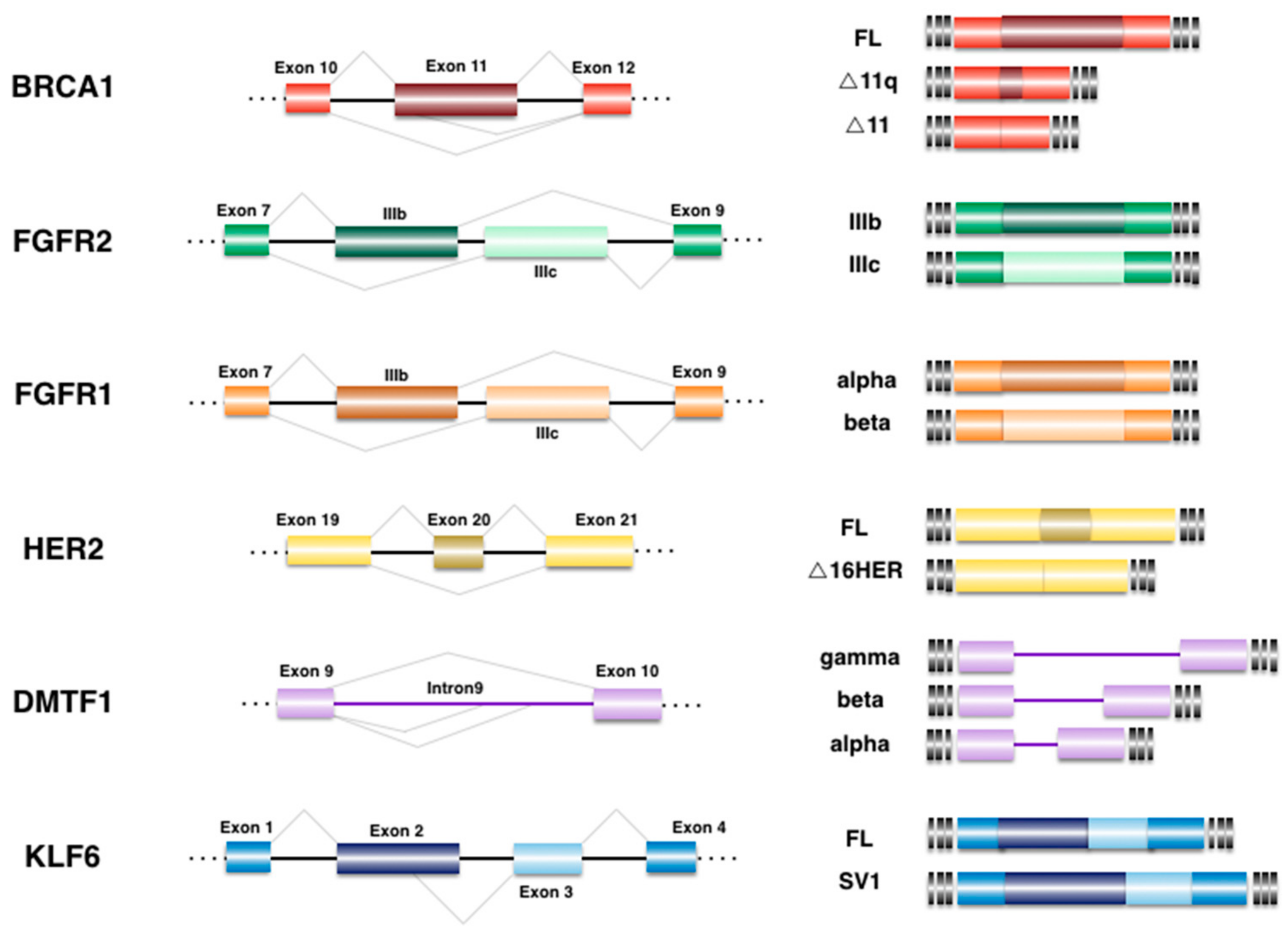
4. Alternative Splicing Events Associated to BrCa
4.1. Breast Cancer 1 (BRCA1)
4.2. Cyclin D-Binding myb-like Transcription Factor 1 (DMTF1)
4.3. Epidermal Growth Factor Receptor 2 (HER2)
4.4. Fibroblast Growth Factor Receptor (FGFR)
4.5. Krüppel-like Zinc Finger Factor 6 (KLF6)
4.6. Survivin
4.7. TP53
5. Prognostic Value of AS Variants in Breast Cancer
6. Modification of Splicing Events as a Therapeutic Approach
7. AS events and Chemotherapeutic Response
8. AS and Endocrine Therapy in BrCa
9. Conclusions
Conflicts of Interest
References
- Anaya-Ruiz, M.; Vallejo-Ruiz, V.; Flores-Mendoza, L.; Perez-Santos, M. Female Breast Cancer Incidence and Mortality in Mexico. Asian Pac. J. Cancer Prev. 2014, 15, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Dogan, N.; Toprak, D. Female breast cancer mortality rates in Turkey. Asian Pac. J. Cancer Prev. 2014, 15, 7569–7573. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Au, W.W.; Wu, K.S.; Chen, L.X.; Lin, K. Mortality characteristics and prediction of female breast cancer in China from 1991 to 2011. Asian Pac. J. Cancer Prev. 2014, 15, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, M.; Bertuccio, P.; Levi, F.; La Vecchia, C.; Negri, E. European cancer mortality predictions for the year 2014. Ann. Oncol. 2014, 25, 1650–1656. [Google Scholar] [CrossRef] [PubMed]
- Youlden, D.R.; Cramb, S.M.; Yip, C.H.; Baade, P.D. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol. Med. 2014, 11, 101–115. [Google Scholar] [PubMed]
- Taghavi, A.; Fazeli, Z.; Vahedi, M.; Baghestani, A.R.; Pourhoseingholi, A.; Barzegar, F.; Pourhoseingholi, M.A. Increased trend of breast cancer mortality in Iran. Asian Pac. J. Cancer Prev. 2012, 13, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, U.; Ismail, M.; Mehmood, N. Epidemiology, major risk factors and genetic predisposition for breast cancer in the Pakistani population. Asian Pac. J. Cancer Prev. 2013, 14, 5625–5629. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Leong, S.P.; Shen, Z.Z.; Liu, T.J.; Agarwal, G.; Tajima, T.; Paik, N.S.; Sandelin, K.; Derossis, A.; Cody, H.; Foulkes, W.D. Is breast cancer the same disease in Asian and Western countries? World J. Surg. 2010, 34, 2308–2324. [Google Scholar] [CrossRef] [PubMed]
- Kohler, R.E.; Goyal, R.K.; Lich, K.H.; Domino, M.E.; Wheeler, S.B. Association between medical home enrollment and health care utilization and costs among breast cancer patients in a state Medicaid program. Cancer 2015, 121, 3975–3981. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Rentería, E.; Conway, D.I.; Bray, F.; Van Ourti, T.; Soerjomataram, I. Inequalities in cancer incidence and mortality across medium to highly developed countries in the twenty-first century. Cancer Causes Control 2016, 27, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, B.; Shimokata, T.; Honda, K.; Tsukuura, H.; Ando, Y. Should low-income countries invest in breast cancer screening? Cancer Causes Control 2016, 27, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Ghoncheh, M.; Pournamdar, Z.; Salehiniya, H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac. J. Cancer Prev. 2015, 17, 43–46. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.; Perou, C.; Livasy, C.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.H.; Edmiston, S.; et al. Race, breast cancer sub-types, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.; Dees, E.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative para-dox: Primary tumor chemosensitivity of breast cancer sub-types. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.I.; Malone, K.E.; Porter, P.L.; Daling, J.R.; Li, C.I. Reproductive and hormonal risk factors for postmenopausal luminal, HER-2-overexpressing, and triple-negative breast cancer. Cancer 2008, 113, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- O’Shaughnessy, J.; Osborne, C.; Pippen, J.E.; Yoffe, M.; Patt, D.; Rocha, C.; Chou Koo, I.; Sherman, B.M.; Bradley, C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N. Engl. J. Med. 2011, 364, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Campone, M.; Piccart, M.; Steger, G. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Crown, J.P.; Lang, I.; Boer, K.; Bondarenko, I.M.; Kulyk, S.O.; Ettl, J.; Patel, R.; Pinter, T.; Schmidt, M.; et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor- positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015, 16, 25–35. [Google Scholar] [PubMed]
- Dhankhar, R.; Vyas, S.P.; Jain, A.K.; Arora, S.; Rath, G.; Goyal, A.K. Advances in novel drug delivery strategies for breast cancer therapy. Artif. Cells Blood Subst. Immobil. Biotechnol. 2010, 38, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Wuerstlein, R.; Harbeck, N. Neoadjuvant therapy for HER2-positive breast cancer. Rev. Recent Clin. Trials 2017. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Ramalho, S.; Gonçalves, R.; Barrios, C.H.; Graudenz, M.S.; Bines, J. Multidisciplinary Approach to Neoadjuvant Endocrine Therapy in Breast Cancer: A Comprehensive Review. Rev. Bras. Ginecol. Obstet. 2016, 38, 615–622. [Google Scholar] [PubMed]
- Castrellon, A.B.; Pidhorecky, I.; Valero, V.; Raez, L.E. The role of carboplatin in the neoadjuvant chemotherapy treatment of triple negative breast cancer. Oncol. Rev. 2017, 11, 324. [Google Scholar] [CrossRef] [PubMed]
- Brufsky, A. nab-Paclitaxel for the treatment of breast cancer: An update across treatment settings. Exp. Hematol. Oncol. 2017, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Puhalla, S.; Brufsky, A.; Davidson, N. Adjuvant endocrine therapy for premenopausal women with breast cancer. Breast 2009, 18, S122–S130. [Google Scholar] [CrossRef]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Medeiros Alencar, V.H.; Badran, A.; Bonfill, X.; et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013, 381, 805–816. [Google Scholar] [CrossRef]
- Yardley, D.A. Combining mTOR inhibitors with chemotherapy and other targeted therapies in advanced breast cancer: Rationale, clinical experience, and future directions. Breast Cancer 2013, 7, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Cadoo, K.A.; Gucalp, A.; Traina, T.A. Palbociclib: An evidence-based review of its potential in the treatment of breast cancer. Breast Cancer 2014, 6, 123–133. [Google Scholar] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Law, A.M.; Lim, E.; Ormandy, C.J.; Gallego-Ortega, D. The innate and adaptive infiltrating immune systems as targets for breast cancer immunotherapy. Endocr. Relat. Cancer 2017, 24, R123–R144. [Google Scholar] [CrossRef] [PubMed]
- McCrudden, C.M.; McCarthy, H.O. Current status of gene therapy for breast cancer: Progress and challenges. Appl. Clin. Genet. 2014, 7, 209. [Google Scholar] [PubMed]
- Shamsi, M.; Islamian, J.P. Breast cancer: Early diagnosis and effective treatment by drug delivery tracing. Nucl. Med. Rev. Cent. East Eur. 2017, 20, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Thirukkumaran, C.; Morris, D.G. Oncolytic viral therapy using reovirus. Methods Mol. Biol. 2015, 1317, 187–223. [Google Scholar] [PubMed]
- Gholami, S.; Marano, A.; Chen, N.G.; Aguilar, R.J.; Frentzen, A.; Chen, C.H.; Lou, E.; Fujisawa, S.; Eveno, C.; Belin, L.; et al. A novel vaccinia virus with dual oncolytic and anti-angiogenic therapeutic effects against triple-negative breast cancer. Breast Cancer Res. Treat. 2014, 148, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Huang, Q.; Qiu, F.; Sui, M. Non-viral delivery systems for the application in p53 cancer gene therapy. Curr. Med. Chem. 2015, 22, 4118–4136. [Google Scholar] [CrossRef] [PubMed]
- Templeton, N.S. Nonviral delivery for genomic therapy of cancer. World J. Surg. 2009, 33, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Huang, L. Non-viral nanocarriers for siRNA delivery in breast cancer. J. Control Release 2014, 190, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.B.; Yao, M.; Brummer, G.; Acevedo, D.; Alhakamy, N.; Berkland, C.; Cheng, N. Targeted gene silencing of CCL2 inhibits triple negative breast cancer progression by blocking cancer stem cell renewal and M2 macrophage recruitment. Oncotarget 2016, 7, 49349. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Ma, W.H.; Ge, Y.L.; Xue, M.L.; Zhang, Z.; Zhang, J.Y.; Hou, L.; Mu, R.H. RNAi-mediated gene silencing of vascular endothelial growth factor C suppresses growth and induces apoptosis in mouse breast cancer in vitro and in vivo. Oncol. Lett. 2016, 12, 3896–3904. [Google Scholar] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar]
- Hu, X.C.; Zhang, J.; Xu, B.H.; Cai, L.; Ragaz, J.; Wang, Z.H.; Wang, B.Y.; Teng, Y.E.; Tong, Z.S.; Pan, Y.Y.; et al. Cisplatin plus gemcitabine versus paclitaxel plus gemcitabine as first-line therapy for metastatic triple-negative breast cancer (CBCSG006): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2015, 16, 436–446. [Google Scholar] [PubMed]
- Rugo, H.S.; Olopade, O.I.; DeMichele, A.; Yau, C.; van ’t Veer, L.J.; Buxton, M.B.; Hogarth, M.; Hylton, N.M.; Paoloni, M.; Perlmutter, J.; et al. Adaptive randomization of veliparib-carboplatin treatment in breast cancer. N. Engl. J. Med. 2016, 375, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Boér, K. Impact of palbociclib combinations on treatment of advanced estrogen receptor-positive/human epidermal growth factor 2-negative breast cancer. Onco Targets Ther. 2016, 9, 6119. [Google Scholar] [CrossRef] [PubMed]
- Kaczyńska, A.; Herman-Antosiewicz, A. Combination of lapatinib with isothiocyanates overcomes drug resistance and inhibits migration of HER2 positive breast cancer cells. Breast Cancer 2016, 24, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Matlin, A.J.; Clark, F.; Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Ladomery, M. Aberrant Alternative Splicing Is Another Hallmark of Cancer. Int. J. Cell Biol. 2013, 2013, 463786. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Montiel, N.; Rosas-Murrieta, N.H.; Martínez-Contreras, R. Alternative splicing regulation: Implications in cancer diagnosis and treatment. Med. Clin. 2015, 144, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Montiel, N.; Rosas-Murrieta, N.H.; Martínez-Montiel, M.; Gaspariano-Cholula, M.P.; Martínez-Contreras, R.D. Microbial and natural metabolites that inhibit splicing: A powerful alternative for cancer treatment. BioMed Res. Int. 2016, 3681094. [Google Scholar] [CrossRef] [PubMed]
- Sebestyen, E.; Zawisza, M.; Eyras, E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015, 43, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef] [PubMed]
- Silipo, M.; Gautrey, H.; Tyson-Capper, A. Deregulation of splicing factors and breast cancer development. J. Mol. Cell. Biol. 2015, 7, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Vanharanta, S.; Marney, C.B.; Shu, W.; Valiente, M.; Zou, Y.; Mele, A.; Darnell, R.B.; Massagué, J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife 2014, 3, e02734. [Google Scholar] [CrossRef] [PubMed]
- Anczuków, O.; Akerman, M.; Cléry, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Takeda, J.-I.; Suzuki, Y.; Sakate, R.; Sato, Y.; Seki, M.; Irie, T.; Takeuchi, N.; Ueda, T.; Nakao, M.; Sugano, S.; et al. Low conservation and species-specific evolution of alternative splicing in humans and mice: Comparative genomics analysis using well-annotated full-length cDNAs. Nucleic Acids Res. 2008, 36, 6386–6395. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Schad, E.; Tompa, P.; Hegyi, H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef] [PubMed]
- Venkitaraman, A.R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 2014, 343, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.X. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006, 34, 1416–1426. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Zhang, H.B.; Peng, Y.; Le, H.; Carroll, B.; Ward, T.; Yao, J.; Farid, L.M.; Couch, F.J.; Wilson, R.B.; et al. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 1997, 17, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Cao, L.; Lim, S.C.; Le, H.; Carroll, B.; Ward, T.; Yao, J.; Farid, L.M.; Couch, F.J.; Wilson, R.B.; et al. Hyperplasia and spontaneous tumor development in the gynecologic system in mice lacking the BRCA1-Delta11 isoform. Mol. Cell. Biol. 2006, 26, 6983–6992. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.; Gajardo-Meneses, P.; Ortega-Hernández, V.; Herrera-Cares, C.; Díaz, S.; Fernández, W.; Cornejo, V.; Gamboa, J.; Tapia, T.; Alvarez, C.; et al. BRCA1 and BARD1 colocalize mainly in the cytoplasm of breast cancer tumors, and their isoforms show differential expression. Breast Cancer Res. Treat. 2015, 153, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Blok, M.J.; Whiley, P.; Santamariña, M.; Gutiérrez-Enríquez, S.; Romero, A.; Garre, P.; Becker, A.; Smith, L.D.; De Vecchi, G.; et al. Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: A report from the ENIGMA consortium. Hum. Mol. Genet. 2014, 23, 3666–3680. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Tancredi, M.; Falaschi, E.; Aretini, P.; Caligo, M.A. Characterization of three alternative transcripts of the BRCA1 gene in patients with breast cancer and a family history of breast and/or ovarian cancer who tested negative for pathogenic mutations. Int. J. Mol. Med. 2015, 35, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Maglic, D.; Stovall, D.B.; Cline, J.M.; Fry, E.A.; Mallakin, A.; Taneja, P.; Caudell, D.L.; Willingham, M.C.; Sui, G.; Inoue, K. DMP1, a splice isoform of the tumour suppressor DMP1 locus, induces proliferation and progression of breast cancer. J. Pathol. 2015, 236, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Tschan, M.P.; Federzoni, E.A.; Haimovici, A.; Britschgi, C.; Moser, B.A.; Jin, J.; Reddy, V.A.; Sheeter, D.A.; Fischer, K.M.; Sun, P.; et al. Human DMTF1 antagonizes DMTF1 regulation of the p14(ARF) tumor suppressor and promotes cellular proliferation. Biochim. Biophys. Acta 2015, 1849, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.; Li, J.; Shi, J.; Sui, G. From General Aberrant Alternative Splicing in Cancers and Its Therapeutic Application to the Discovery of an Oncogenic DMTF1 Isoform. Int. J. Mol. Sci. 2017, 18, 191. [Google Scholar] [CrossRef] [PubMed]
- Fuller, S.J.; Sivarajah, K.; Sugden, P.H. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J. Mol. Cell. Cardiol. 2008, 44, 831–854. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.; Ullrich, A.; McGuire, W. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Kwong, K.Y.; Hung, M.C. A novel splice variant of HER2 with increased transformation activity. Mol. Carcinog. 1998, 23, 62–68. [Google Scholar] [CrossRef]
- Marchini, C.; Gabrielli, F.; Iezzi, M.; Zenobi, S.; Montani, M.; Pietrella, L.; Kalogris, C.; Rossini, A.; Ciravolo, V.; Castagnoli, L.; et al. The human splice variant Delta16HER2 induces rapid tumor onset in a reporter transgenic mouse. PLoS ONE 2011, 6, e18727. [Google Scholar] [CrossRef] [PubMed]
- Alajati, A.; Sausgruber, N.; Aceto, N.; Duss, S.; Sarret, S.; Voshol, H.; Bonenfant, D.; Bentires-Alj, M. Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res. 2013, 73, 5320–5327. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, F.; Tagliabue, E.; Campiglio, M.; Pupa, S.M.; Balsari, A.; Menard, S. Role of exon-16-deleted HER2 in breast carcinomas. Endocr. Relat. Cancer 2006, 13, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, F.; Ghedini, G.C.; Koschorke, A.; Triulzi, T.; Dugo, M.; Gasparini, P.; Casalini, P.; Palladini, A.; Iezzi, M.; Lamolinara, A.; et al. Pathobiological implications of the d16HER2 splice variant for stemness and aggressiveness of HER2-positive breast cancer. Oncogene 2017, 36, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Druillennec, S.; Dorard, C.; Eychène, A. Alternative splicing in oncogenic kinases: From physiological functions to cancer. J. Nucleic Acids 2012, 2012, 639062. [Google Scholar] [CrossRef] [PubMed]
- Cittelly, D.M.; Das, P.M.; Salvo, V.A.; Fonseca, J.P.; Burow, M.E.; Jones, F.E. Oncogenic HER2Δ16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis 2010, 31, 2049–2057. [Google Scholar] [CrossRef] [PubMed]
- Huynh, F.C.; Jones, F.E. MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2Δ16 mediated breast tumorigenesis and reverse trastuzumab resistance. PLoS ONE 2014, 22, E114419. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Fry, E.A. Aberrant Splicing of Estrogen Receptor, HER2, and CD44 Genes in Breast Cancer. Genet. Epigenet. 2015, 7, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Lu, J.; Chen, H.; Werner, S.; Williams, L.T. The human fibroblast growth factor receptor genes: A common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol. Cell. Biol. 1991, 11, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Madden, S.F.; Clarke, C.; Gaule, P.; Aherne, S.T.; O’Donovan, N.; Clynes, M.; Crown, J.; Gallagher, W.M. BreastMark: An integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013, 15, R52. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, J.; Dutta, K.; Ilghari, D.; Beenken, A.; Goetz, R.; Eliseenkova, A.V.; Cowburn, D.; Mohammadi, M. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure 2012, 20, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kan, M.; Yan, G.; Xu, J.; McKeehan, W.L. Alternately spliced NH2-terminal immunoglobulin-like loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 1995, 270, 10231–10235. [Google Scholar] [CrossRef] [PubMed]
- Luqmani, Y.A.; Mortimer, C.; Yiangou, C.; Johnston, C.L.; Bansal, G.S.; Sinnett, D.; Law, M.; Coombes, R.C. Expression of 2 variant forms of fibroblast growth factor receptor 1 in human breast. Int. J. Cancer 1995, 64, 274–279. [Google Scholar] [CrossRef] [PubMed]
- DiFeo, A.; Martignetti, J.A.; Narla, G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist. Updates 2009, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vetter, D.; Cohen-Naftaly, M.; Villanueva, A.; Lee, Y.A.; Kocabayoglu, P.; Hannivoort, R.; Narla, G.; M Llovet, J.; Thung, S.N.; Friedman, S.L. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology 2012, 56, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Hatami, R.; Sieuwerts, A.M.; Izadmehr, S.; Yao, Z.; Qiao, R.F.; Papa, L.; Look, M.P.; Smid, M.; Ohlssen, J.; et al. Levine AC KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci. Transl. Med. 2013, 5, 169ra12. [Google Scholar] [CrossRef] [PubMed]
- Olson, O.A.; Joyce, J.A. A Splicing Twist on Metastasis. Sci. Transl. Med. 2013, 5, 169fs2. [Google Scholar] [CrossRef] [PubMed]
- Mita, A.C.; Mita, M.M.; Nawrocki, S.T.; Giles, F.J. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin. Cancer Res. 2008, 14, 5000–5005. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.; O’Donovan, N.; Browne, B.; O’Shea, C.; Crown, J.; Hill, A.D.; McDermott, E.; O’Higgins, N.; Duffy, M.J. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br. J. Cancer 2005, 92, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Toyoda, M.; Shinohara, H.; Okuda, J.; Watanabe, I.; Yamamoto, T.; Tanaka, K.; Tenjo, T.; Tanigawa, N. Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumorigenesis. Cancer 2001, 91, 2026–2032. [Google Scholar] [CrossRef]
- Pavlidou, A.; Kroupis, C.; Dimas, K. Association of survivin splice variants with prognosis and treatment of breast cancer. World J. Clin. Oncol. 2014, 5, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Bennit, H.F.; Turay, D. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef] [PubMed]
- Blasberg, R. PET imaging of gene expression. Eur. J. Cancer 2002, 38, 2137–2146. [Google Scholar]
- Aylon, Y.; Oren, M. New plays in the p53 theater. Curr. Opin. Genet. Dev. 2011, 21, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H.; Norman, J.C. p53 and its mutants in tumor cell migration and invasion. J. Cell. Biol. 2011, 192, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Goh, A.M.; Coffill, C.R.; Lane, D.P. The role of mutant p53 in human cancer. J. Pathol. 2011, 223, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Fry, E.A. Aberrant splicing of the DMP1-ARF-MDM2-p53 pathway in cancer. Int. J. Cancer 2016, 139, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Yoshida, H.; Kitagishi, Y.; Nishimura, Y.; Matsuda, S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem. Biophys. Res. Commun. 2011, 413, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [PubMed]
- Ding, L.; Ellis, M.J.; Li, S.; Larson, D.E.; Chen, K.; Wallis, J.W.; Harris, C.C.; McLellan, M.D.; Fulton, R.S.; Fulton, L.L.; et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010, 464, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Venables, J.P.; Klinck, R.; Bramard, A.; Inkel, L.; Dufresne-Martin, G.; Koh, C.; Gervais-Bird, J.; Lapointe, E.; Froehlich, U.; Durand, M.; et al. Identification of Alternative Splicing Markers for Breast Cancer. Cancer Res. 2008, 68, 9525. [Google Scholar] [CrossRef] [PubMed]
- Martensen, P.M.; Oka, K.; Christensen, L.; Rettenberger, P.M.; Petersen, H.H.; Christensen, A.; Chan, L.; Heegaard, C.W.; Andreasen, P.A. Breast carcinoma epithelial cells express a very low-density lipoprotein receptor variant lacking the O-linked glycosylation domain encoded by exon 16, but with full binding activity for serine proteinase/serpin complexes and Mr-40,000 receptor-associated protein. Eur. J. Biochem. 1997, 248, 583–591. [Google Scholar] [PubMed]
- Sciacca, L.; Mineo, R.; Pandini, G.; Murabito, A.; Vigneri, R.; Belfiore, A. In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene 2002, 21, 8240–8250. [Google Scholar] [CrossRef] [PubMed]
- Lapuk, A.; Marr, H.; Jakkula, L.; Pedro, H.; Bhattacharya, S.; Purdom, E.; Hu, Z.; Simpson, K.; Pachter, L.; Durinck, S.; et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol. Cancer Res. 2010, 8, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hoadley, K.A.; Parker, J.S.; Perou, C.M. Identification of mRNA isoform switching in breast cancer. BMC Genom. 2016, 17, 181. [Google Scholar] [CrossRef] [PubMed]
- Bauman, J.; Jearawiriyapaisarn, N.; Kole, R. Therapeutic potential of splice switching oligonucleotides. Oligonucleotides 2009, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zanetta, C.; Nizzardo, M.; Simone, C.; Monguzzi, E.; Bresolin, N.; Comi, G.P.; Corti, S. Molecular Therapeutic Strategies for Spinal Muscular Atrophies: Current and Future Clinical Trials. Clin. Ther. 2014, 36, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Zaharieva, E.; Chipman, J.K.; Soller, M. Alternative splicing interference by xenobiotics. Toxicology 2012, 296, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.; Misteli, T. Small Molecule Modulators of Pre-mRNA Splicing in Cancer Therapy. Trends Mol. Med. 2016, 22, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.; Ranson, M.; Kelso, M.J. Anti-tumour/metastasis effects of the potassium-sparing diuretic amiloride: An orally active anti-cancer drug waiting for its call-of-duty? Int. J. Cancer 2011, 129, 2051–2061. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.P.; Chao, C.C. Cancer cells acquire resistance to anticancer drugs: An update. Biomed. J. 2012, 35, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Shkreta, L.; Froehlich, U.; Paquet, E.R.; Toutant, J.; Elela, S.A.; Chabot, B. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol. Cancer Ther. 2008, 7, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, M.; Delforge, Y.; Deward, A.; Habraken, Y.; Hennuy, B.; Piette, J.; Klinck, R.; Chabot, B.; Colige, A.; Lambert, C. Role of the splicing factor SRSF4 in cisplatin induced modifications of pre-mRNA splicing and apoptosis. BMC Cancer 2015, 15, 227. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Ruan, S.; Wang, M.; Habraken, Y.; Hennuy, B.; Piette, J.; Klinck, R.; Chabot, B.; Colige, A.; Lambert, C. A novel chemical, STF-083010, reverses tamoxifen-related drug resistance in breast cancer by inhibiting IRE1/XBP1. Oncotarget 2015, 6, 40692–40703. [Google Scholar] [CrossRef] [PubMed]
- Margolese, R.G.; Fisher, B.; Hortobagyi, G.N.; Buchholz, T.A. Neoplasms of the breast. In Cancer Medicine, 6th ed.; Section 32, Chapter 18; Kufe, D.W., Pollock, R.E., Weichselbaum, R.R., Bast, R.C., Gansler, T.S., Holland, J.F., Frei, E., Eds.; BC Decker: Hamilton, ON, Canada, 2003. [Google Scholar]
- Stahlberg, C.; Pedersen, A.T.; Lynge, E.; Andersen, Z.J.; Keiding, N.; Hundrup, Y.A.; Obel, E.B.; Ottesen, B. Increased risk of breast cancer following different regimens of hormone replacement therapy frequently used in Europe. Int. J. Cancer 2004, 109, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Brown, M. Advances in estrogen receptor biology: Prospects for improvements in targeted breast cancer therapy. Breast Cancer Res. 2004, 6, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Chlebowski, R.T.; Anderson, G.L. Menopausal hormone therapy and cancer: Changing clinical observations of target site specificity. Steroids 2014, 90, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Begam, A.J.; Jubie, S.; Nanjan, M.J. Estrogen receptor agonists/antagonists in breast cancer therapy: A critical review. Bioorg. Chem. 2017, 71, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, L.H.; Williams, A.R.; Critchley, H.O. Selective progesterone receptor modulators. Curr. Opin. Obstet. Gynecol. 2014, 26, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Dorssers, L.C.; Van der Flier, S.; Brinkman, A.; van Agthoven, T.; Veldscholte, J.; Berns, E.M.; Klijn, J.G.; Beex, L.V.; Foekens, J.A. Tamoxifen resistance in breast cancer: Elucidating mechanisms. Drugs 2001, 61, 1721–1733. [Google Scholar] [CrossRef] [PubMed]
- Droog, M.; Beelen, K.; Linn, S.; Zwart, W. Tamoxifen resistance: From bench to bedside. Eur. J. Pharmacol. 2013, 717, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Aya, L.F.; Gonzalez-Angulo, A.M. Adjuvant systemic therapies in breast cancer. Surg. Clin. N. Am. 2013, 93, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T. Clinical application of drug delivery systems in cancer chemotherapy: Review of the efficacy and side effects of approved drugs. Biol. Pharm. Bull. 2013, 36, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R.; Arpino, G.; Lee, A.S.; Hilsenbeck, V.G. Endocrine responsiveness: Understanding how progesterone receptor can be used to select endocrine therapy. Breast 2005, 14, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Iwase, H.; Yamamoto, Y. Clinical benefit of sequential use of endocrine therapies for metastatic breast cancer. Int. J. Clin. Oncol. 2015, 20, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Ribnikar, D.; Sousa, B.; Cufer, T.; Cardoso, F. Extended adjuvant endocrine therapy—A standard to all or some? Breast 2017, 32, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.C.; Cho, W.C.; Lin, P.W.; Lin, S.L.; Lee, W.Y.; Young, K.C. Quantitative profile of estrogen receptor variants/isoforms in Taiwanese women with breast cancer. Eur. J. Surg. Oncol. 2006, 32, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Ho, S.; Tarapore, P.; Chung, I.; Leung, Y.K. Estrogen Receptor β Isoform 5 Confers Sensitivity of Breast Cancer Cell Lines to Chemotherapeutic Agent-Induced Apoptosis through Interaction with Bcl2L12. Neoplasia 2013, 15, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Montiel, N.; Anaya-Ruiz, M.; Pérez-Santos, M.; Martínez-Contreras, R.D. Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools. Genes 2017, 8, 217. https://doi.org/10.3390/genes8100217
Martínez-Montiel N, Anaya-Ruiz M, Pérez-Santos M, Martínez-Contreras RD. Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools. Genes. 2017; 8(10):217. https://doi.org/10.3390/genes8100217
Chicago/Turabian StyleMartínez-Montiel, Nancy, Maricruz Anaya-Ruiz, Martín Pérez-Santos, and Rebeca D. Martínez-Contreras. 2017. "Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools" Genes 8, no. 10: 217. https://doi.org/10.3390/genes8100217
APA StyleMartínez-Montiel, N., Anaya-Ruiz, M., Pérez-Santos, M., & Martínez-Contreras, R. D. (2017). Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools. Genes, 8(10), 217. https://doi.org/10.3390/genes8100217






