Full Genome Sequence Analysis of Two Isolates Reveals a Novel Xanthomonas Species Close to the Sugarcane Pathogen Xanthomonas albilineans
Abstract
:1. Introduction
2. Results and Discussion
2.1. Taxonomic Characterization of MUS 060 and GPE 39 Isolates
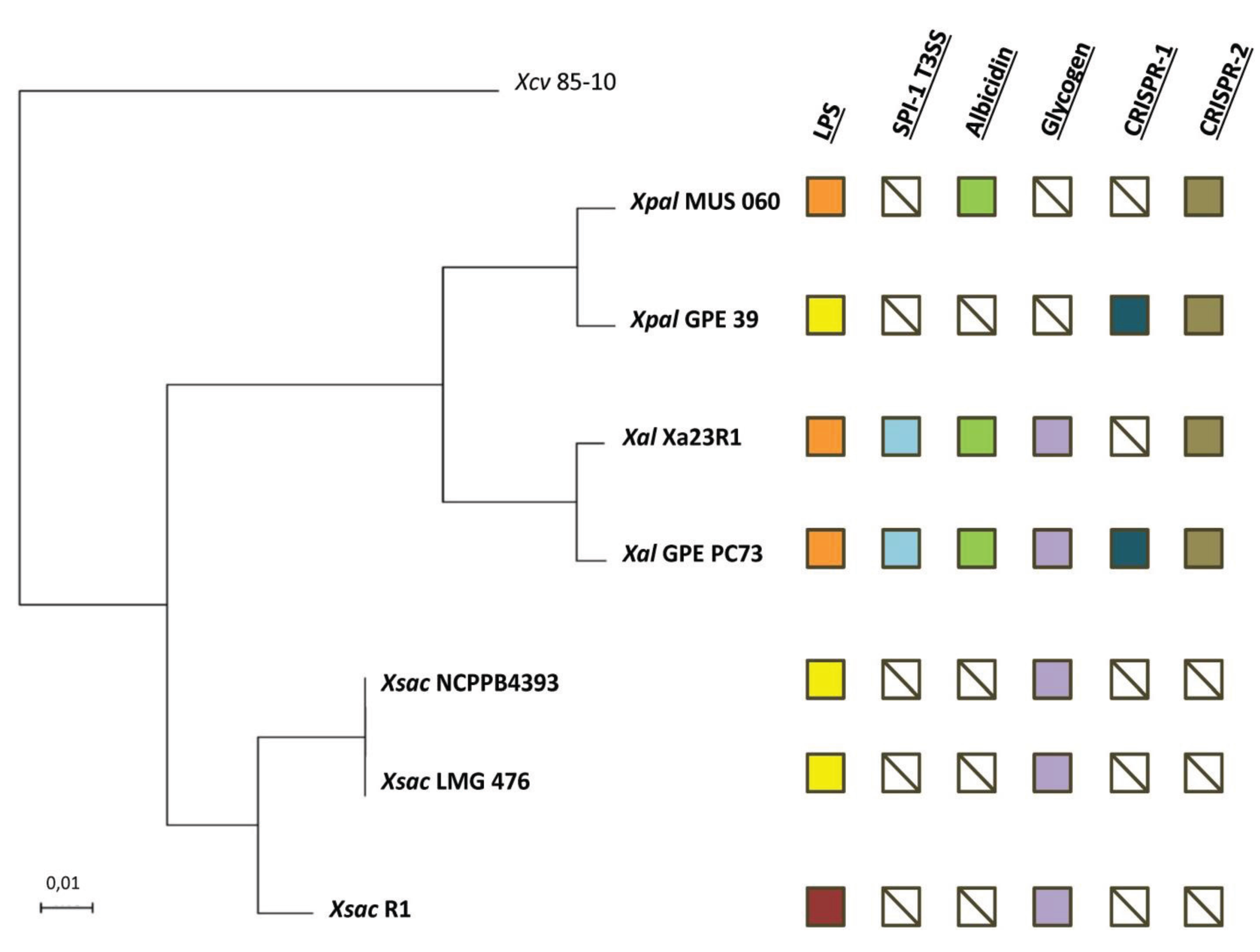
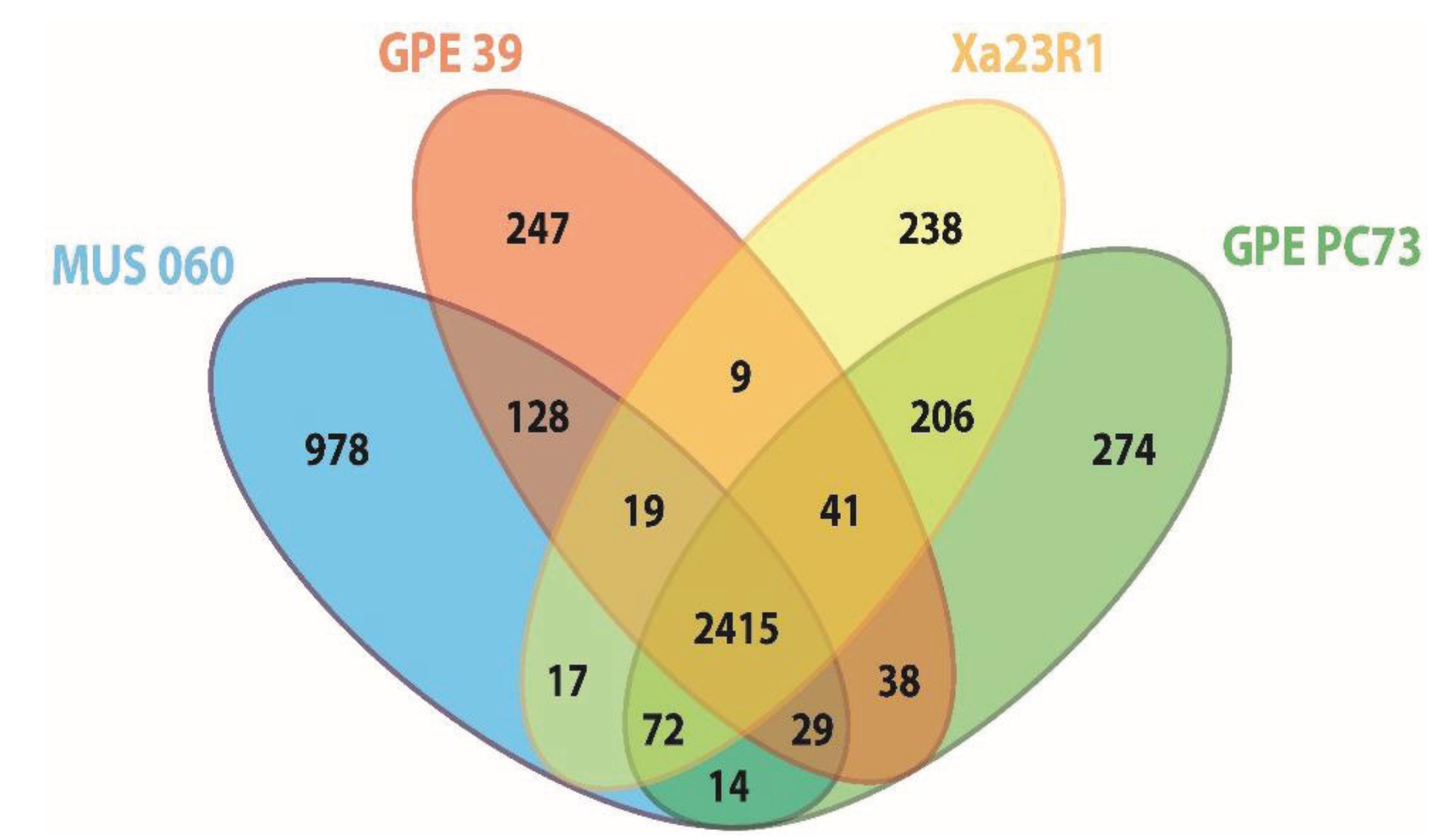
2.2. Orthology Analysis
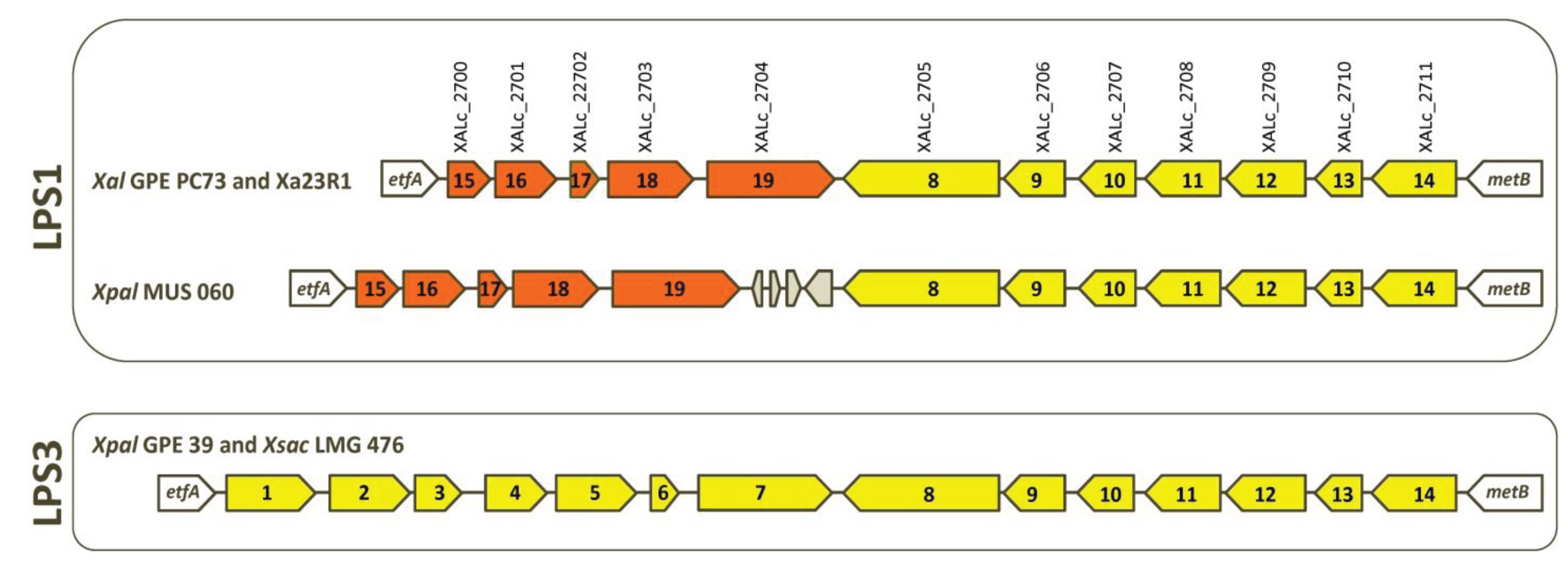
2.3. Serotypes and Variability of LPS Loci
2.4. The Albicidin Locus and other NRPS Loci
2.5. Genes of X. albilineans Linked to a Xylem-Invading Lifestyle
2.6. Variability of the Locus Encoding the Type IV Pilus
2.7. CRISPR and Restriction-Modification System Loci
2.8. Other Genes or Loci Specific to X. albilineans
2.9. Other Genes or Loci Specific to “X. pseudalbilineans”
2.10. Genome Erosion
3. Experimental Section
3.1. Bacterial Strains and DNA Preparation
| Organism | Geographical Origin | Date of Isolation | Biological Origin | GenBank Accession n° | Reference |
|---|---|---|---|---|---|
| X. albilineans GPE PC73 | Caribbean/Guadeloupe | 2006 | Sugarcane (Saccharum spp.) | GCA_000087965.1 | [6] |
| X. albilineans Xa23R1 | USA/Florida | 1993 | Sugarcane (Saccharum spp.) | JZIK00000000 | [65] |
| “X. pseudalbilineans” MUS 060 | Indian ocean/Mauritius | 1940 | Dallis grass (Paspalum dilatatum Poir.) | JZIM00000000 | [21] |
| “X. pseudalbilineans” GPE 39 | Caribbean/Guadeloupe | 1997 | Water droplets collected on leaves of sugarcane | JZHZ00000000 | [19] |
| X. sacchari LMG 476 | Caribbean/Guadeloupe | 1980 | Sugarcane (Saccharum spp.) | JXQE00000000 | [25] |
| X. sacchari NCPPB4393 | Africa/Tanzania | 2007 | Insect collected on a banana plant (Musa spp.) | AGDB00000000 | [26] |
| X. sacchari R1 | China/Heilongjiang | 2011 | Asymptomatic seed of rice (Oriza sativa) | GCA_000815185.1 | [28] |
3.2. DNA Sequencing and Assembly
3.3. Genotyping with 16S–23S rRNA ITS and 16S rRNA Gene Sequence
3.4. Phylogenetic Core Genome Analysis: MLSA
3.5. Whole-Genome Sequences Analyses: ANI Calculation
3.6. OrthoMCL Analyses
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- North, D.S.; Colonial Sugar Refining Company. Leaf-Scald: A Bacterial Disease of Sugar Cane; Colonial Sugar Refining Company: Sydney, Australia, 1926. [Google Scholar]
- Martin, J.; Robinson, P. Leaf scald. In Sugar-Cane Diseases of the World; Martin, J., Abbott, E., Hughes, C., Eds.; Elsevier Publishing Company: Amsterdam, The Netherlands, 1961; Volume I, pp. 79–101. [Google Scholar]
- Rott, P.; Davis, M. Leaf scald. In A Guide to Sugarcane Diseases; Rott, P., Bailey, R., Comstock, J., Croft, B., Saumtally, A., Eds.; CIRAD-ISSCT: Montpellier, France, 2000; p. 339. [Google Scholar]
- Klett, P.; Rott, P. Inoculum sources for the spread of leaf scald disease of sugarcane caused by Xanthomonas albilineans in Guadeloupe. Phytopathology 1994, 142, 283–291. [Google Scholar] [CrossRef]
- Naushad, S.; Adeolu, M.; Wong, S.; Sohail, M.; Schellhorn, H.E.; Gupta, R.S. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: Proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 2015, 107, 467–485. [Google Scholar] [PubMed]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Cociancich, S.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.-A.; et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.-A.; Lauber, E.; et al. Genomic insights into strategies used by Xanthomonas albilineans with its reduced artillery to spread within sugarcane xylem vessels. BMC Genomics 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Marguerettaz, M.; Pieretti, I.; Gayral, P.; Puig, J.; Brin, C.; Cociancich, S.; Poussier, S.; Rott, P.; Royer, M. Genomic and evolutionary features of the SPI-1 type III secretion system that is present in Xanthomonas albilineans but is not essential for xylem colonization and symptom development of sugarcane leaf scald. Mol. Plant Microbe Interact. 2011, 24, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Royer, M.; Koebnik, R.; Marguerettaz, M.; Barbe, V.; Robin, G.; Brin, C.; Carrere, S.; Gomez, C.; Hügelland, M.; Völler, G.; et al. Genome mining reveals the genus Xanthomonas to be a promising reservoir for new bioactive non-ribosomally synthesized peptides. BMC Genomics 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Birch, R.G. Xanthomonas albilineans and the antipathogenesis approach to disease control. Mol. Plant Pathol. 2001, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cociancich, S.; Pesic, A.; Petras, D.; Uhlmann, S.; Kretz, J.; Schubert, V.; Vieweg, L.; Duplan, S.; Marguerettaz, M.; Noëll, J.; et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat. Chem. Biol. 2015, 11, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.M.; Wall, M.K.; Smith, A.B.; Maxwell, A.; Birch, R.G. The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 2007, 51, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Rott, P.; Warmuth, C.; Chatenet, M.; Baudin, P. Intraspecific genomic variation within Xanthomonas albilineans, the sugarcane leaf scald pathogen. Phytopathology 1997, 87, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Rott, P.; Arnaud, M.; Baudin, P. Serological and lysotypical variability of Xanthomonas albilineans (Ashby) Dowson, Causal agent of sugarcane leaf scald disease. J. Phytopathol. 1986, 116, 201–211. [Google Scholar] [CrossRef]
- Rott, P.; Davis, M.J.; Baudin, P. Serological variability in Xanthomonas albilineans, causal agent of leaf scald disease of sugarcane. Plant Pathol. 1994, 43, 344–349. [Google Scholar] [CrossRef]
- Alvarez, A.M.; Schenck, S.; Benedict, A.A. Differentiation of Xanthomonas albilineans strains with monoclonal antibody reaction pattern and DNA fingerprints. Plant Pathol. 1996, 45, 358–366. [Google Scholar] [CrossRef]
- Ricaud, C.; Ryan, C. Leaf scald. In Diseases of Sugarcane: Major Diseases; Ricaud, C., Egan, B., Gillaspie, A., Hugues, C., Eds.; Elsevier: Amsterdam, The Netherlands, 1989; pp. 39–53. [Google Scholar]
- Saumtally, S.; Medan, H.; Autrey, L. Evolution of Aerial Infection of Leaf Scald Caused by Xanthomonas albilineans (Ashby) Dowson in Sugarcane. In Proceedings of the XXII Congress, Cartagena, Colombia, 11–15 September 1995; Cock, J., Brekelbaum, T., Eds.; International Society of Sugarcane Technologists: Cartagena, Colombia, 1996; pp. 493–497.
- Daugrois, J.H.; Dumont, V.; Champoiseau, P.; Costet, L.; Boisne-Noc, R.; Rott, P. Aerial contamination of sugarcane in Guadeloupe by two strains of Xanthomonas albilineans. Eur. J. Plant Pathol. 2003, 109, 445–458. [Google Scholar] [CrossRef]
- Champoiseau, P.; Rott, P.; Daugrois, J.H. Epiphytic populations of Xanthomonas albilineans and subsequent sugarcane stalk infection are linked to rainfall in Guadeloupe. Plant Dis. 2009, 93, 339–346. [Google Scholar] [CrossRef]
- Orian, G. A disease of Paspalum dilatatum in Mauritius caused by a species of bacterium closely resembling Xanthomonas albilineans (Ashby) Dowson. Rev. Agric. Sucr. Ile Maurice 1962, 41, 7–20. [Google Scholar]
- Daugrois, J.H.; Boisne-Noc, R.; Champoiseau, P.; Rott, P. The revisited infection cycle of Xanthomonas albilineans, the causal agent of leaf scald of sugarcane. Func. Plant Sci. Biotech. 2012, 6, 91–97. [Google Scholar]
- Gonçalves, E.R.; Rosato, Y.B. Phylogenetic analysis of Xanthomonas species based upon 16S–23S rDNA intergenic spacer sequences. Int. J. Syst. Evol. Microbiol. 2002, 52, 355–361. [Google Scholar] [PubMed]
- Young, J.M.; Park, D.C.; Shearman, H.M.; Fargier, E. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 2008, 31, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Pieretti, I.; Bolot, S.; Carrère, S.; Barbe, V.; Cociancich, S.; Rott, P.; Royer, M. Draft genome sequence of Xanthomonas sacchari Strain LMG 476. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Wasukira, A.; Paszkiewicz, K.; Aritua, V.; Thwaites, R.; Smith, J.; Grant, M. Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas group 1 clade. Genes 2011, 2, 1050–1065. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Wasukira, A.; Paszkiewicz, K.; Aritua, V.; Thwaites, R.; Smith, J.; Grant, M. Correction: Studholme et al., Draft Genome Sequences of Xanthomonas sacchari and Two Banana-Associated Xanthomonads Reveal Insights into the Xanthomonas Group 1 clade. Genes 2011, 2, 1050–1065. Genes 2012, 3, 88–89. [Google Scholar] [CrossRef]
- Fang, Y.; Lin, H.; Wu, L.; Ren, D.; Ye, W.; Dong, G.; Zhu, L.; Guo, L. Genome sequence of Xanthomonas sacchari R1, a biocontrol bacterium isolated from the rice seed. J. Biotechnol. 2015, 206, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [PubMed]
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.-P.; Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000, 38, 3623–3630. [Google Scholar] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Papke, R.T. Molecular Phylogeny of Microorganisms; Horizon Scientific Press: Norfolk, UK, 2010. [Google Scholar]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Jalan, N.; Aritua, V.; Kumar, D.; Yu, F.; Jones, J.B.; Graham, J.H.; Setubal, J.C.; Wang, N. Comparative genomic analysis of Xanthomonas axonopodis pv. citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity. J. Bacteriol. 2011, 193, 6342–6357. [Google Scholar] [PubMed]
- Darrasse, A.; Carrere, S.; Barbe, V.; Boureau, T.; Arrieta-Ortiz, M.; Bonneau, S.; Briand, M.; Brin, C.; Cociancich, S.; Durand, K.; et al. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 2013, 14. [Google Scholar] [CrossRef]
- Desaki, Y.; Miya, A.; Venkatesh, B.; Tsuyumu, S.; Yamane, H.; Kaku, H.; Minami, E.; Shibuya, N. Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 2006, 47, 1530–1540. [Google Scholar] [CrossRef] [PubMed]
- Petrocelli, S.; Tondo, M.L.; Daurelio, L.D.; Orellano, E.G. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS ONE 2012, 7, e40051. [Google Scholar] [PubMed]
- Patil, P.; Bogdanove, A.; Sonti, R. The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Molinaro, A.; Sturiale, L.; Dow, J.M.; Erbs, G.; Lanzetta, R.; Newman, M.-A.; Parrilli, M. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J. Biol. Chem. 2005, 280, 33660–33668. [Google Scholar] [CrossRef] [PubMed]
- Vorhölter, F.J.; Niehaus, K.; Pühler, A. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: A cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol. Genet. Genomics 2001, 266, 79–95. [Google Scholar]
- Strieker, M.; Tanovic, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Postle, K.; Kadner, R.J. Touch and go: Tying TonB to transport. Mol. Microbiol. 2003, 49, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Kawai, F.; Kitajima, S.; Oda, K.; Higasa, T.; Charoenpanich, J.; Hu, X.; Mamoto, R. Polyvinyl alcohol and polyethylene glycol form polymer bodies in the periplasm of sphingomonads that are able to assimilate them. Arch. Microbiol. 2013, 195, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Rott, P.; Fleites, L.; Marlow, G.; Royer, M.; Gabriel, D.W. Identification of new candidate pathogenicity factors in the xylem-invading pathogen Xanthomonas albilineans by transposon mutagenesis. Mol. Plant Microbe Interact. 2011, 24, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Fronzes, R.; Remaut, H.; Waksman, G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008, 27, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Rangaraj, N.; Sonti, R.V. Multiple adhesin-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Mol. Plant Microbe Interact. 2009, 22, 73–85. [Google Scholar] [PubMed]
- Van Sluys, M.A.; Monteiro-Vitorello, C.B.; Camargo, L.E.A.; Menck, C.F.M.; da Silva, A.C.R.; Ferro, J.A.; Oliveira, M.C.; Setubal, J.C.; Kitajima, J.P.; Simpson, A.J. Comparative genomic analysis of plant-associated bacteria. Annu. Rev. Phytopathol. 2002, 40, 169–189. [Google Scholar] [CrossRef] [PubMed]
- Darsonval, A.; Darrasse, A.; Durand, K.; Bureau, C.; Cesbron, S.; Jacques, M.-A. Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans. Mol. Plant Microbe Interact. 2009, 22, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.-J.; Chung, I.-Y.; Choi, K.B.; Lau, G.W.; Cho, Y.-H. Genome sequence comparison and superinfection between two related Pseudomonas aeruginosa phages, D3112 and MP22. Microbiology 2007, 153, 2885–2895. [Google Scholar] [CrossRef] [PubMed]
- Mhedbi-Hajri, N.; Darrasse, A.; Pigne, S.; Durand, K.; Fouteau, S.; Barbe, V.; Manceau, C.; Lemaire, C.; Jacques, M.-A. Sensing and adhesion are adaptive functions in the plant pathogenic xanthomonads. BMC Evol. Biol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Wasukira, A.; Coulter, M.; al-Sowayeh, N.; Thwaites, R.; Paszkiewicz, K.; Kubiriba, J.; Smith, J.; Grant, M.; Studholme, D. Genome sequencing of Xanthomonas vasicola pathovar vasculorum reveals variation in plasmids and genes encoding lipopolysaccharide synthesis, type-IV pilus and type-III secretion effectors. Pathogens 2014, 3, 211–237. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.; Nagornykh, M.; Pyatnitskiy, M.; Artamonova, I.I.; Severinov, K. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol. Lett. 2009, 296, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Koebnik, R.; Lu, H.; Furutani, A.; Angiuoli, S.V.; Patil, P.B.; van Sluys, M.-A.; Ryan, R.P.; Meyer, D.F.; Han, S.-W.; et al. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 2011, 193, 5450–5464. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.P.; Wood, M.W.; Taylor, L.A.; Monaghan, P.; Hawes, P.; Jones, P.W.; Wallis, T.S.; Galyov, E.E. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 2002, 46, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Schikora, A.; Virlogeux-Payant, I.; Bueso, E.; Garcia, A.V.; Nilau, T.; Charrier, A.l.; Pelletier, S.; Menanteau, P.; Baccarini, M.; Velge, P.; et al. Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE 2011, 6, e24112. [Google Scholar] [CrossRef] [PubMed]
- Almagro, G.; Viale, A.M.; Montero, M.; Rahimpour, M.; Muñoz, F.J.; Baroja-Fernández, E.; Bahaji, A.; Zúñiga, M.; González-Candelas, F.; Pozueta-Romero, J. Comparative genomic and phylogenetic analyses of Gammaproteobacterial glg genes traced the origin of the Escherichia coli glycogen glgBXCAP operon to the last common ancestor of the sister orders Enterobacteriales and Pasteurellales. PLoS ONE 2015, 10, e0115516. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Deleury, E.; Coutinho, P.M. Glycogen metabolism loss: A common marker of parasitic behaviour in bacteria? Trends Genet. 2002, 18, 437–440. [Google Scholar] [CrossRef]
- Murphy, J.; Mahony, J.; Ainsworth, S.; Nauta, A.; van Sinderen, D. Bacteriophage orphan DNA methyltransferases: Insights from their bacterial origin, function, and occurrence. Appl. Environ. Microbiol. 2013, 79, 7547–7555. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 2002, 108, 583–586. [Google Scholar] [CrossRef]
- Moya, A.; Pereto, J.; Gil, R.; Latorre, A. Learning how to live together: Genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 2008, 9, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Rott, P.; Abel, M.; Soupa, D.; Feldman, P.; Letourmy, P. Population dynamics of Xanthomonas albilineans in sugarcane plants as determined with an antibiotic-resistant mutant. Plant Dis. 1994, 78, 241–247. [Google Scholar] [CrossRef]
- Rott, P.C.; Costet, L.; Davis, M.J.; Frutos, R.; Gabriel, D.W. At least two separate gene clusters are involved in albicidin production by Xanthomonas albilineans. J. Bacteriol. 1996, 178, 4590–4596. [Google Scholar] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA Sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Humana Press: New York, NY, USA, 2009; Volume 537, pp. 39–64. [Google Scholar]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Patil, P.; van Sluys, M.-A.; White, F.F.; Ryan, R.P.; Dow, J.M.; Rabinowicz, P.; Salzberg, S.L.; Leach, J.E.; Sonti, R.; et al. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS ONE 2008, 3, e3828. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pieretti, I.; Cociancich, S.; Bolot, S.; Carrère, S.; Morisset, A.; Rott, P.; Royer, M. Full Genome Sequence Analysis of Two Isolates Reveals a Novel Xanthomonas Species Close to the Sugarcane Pathogen Xanthomonas albilineans. Genes 2015, 6, 714-733. https://doi.org/10.3390/genes6030714
Pieretti I, Cociancich S, Bolot S, Carrère S, Morisset A, Rott P, Royer M. Full Genome Sequence Analysis of Two Isolates Reveals a Novel Xanthomonas Species Close to the Sugarcane Pathogen Xanthomonas albilineans. Genes. 2015; 6(3):714-733. https://doi.org/10.3390/genes6030714
Chicago/Turabian StylePieretti, Isabelle, Stéphane Cociancich, Stéphanie Bolot, Sébastien Carrère, Alexandre Morisset, Philippe Rott, and Monique Royer. 2015. "Full Genome Sequence Analysis of Two Isolates Reveals a Novel Xanthomonas Species Close to the Sugarcane Pathogen Xanthomonas albilineans" Genes 6, no. 3: 714-733. https://doi.org/10.3390/genes6030714
APA StylePieretti, I., Cociancich, S., Bolot, S., Carrère, S., Morisset, A., Rott, P., & Royer, M. (2015). Full Genome Sequence Analysis of Two Isolates Reveals a Novel Xanthomonas Species Close to the Sugarcane Pathogen Xanthomonas albilineans. Genes, 6(3), 714-733. https://doi.org/10.3390/genes6030714






