MicroRNA-224 is Readily Detectable in Urine of Individuals with Diabetes Mellitus and is a Potential Indicator of Beta-Cell Demise
Abstract
:1. Introduction
2. Experimental Section
2.1. Patient Cohorts
2.2. Phenotyping of the Cohort
2.3. Oral Glucose Tolerance Test (OGTT)
2.4. Biochemical Assays
2.5. RNA Isolation from Human Urine
| Parameter | Normal Controls | T1DM | T2DM | HNF1A-MODY | Statistical Analysis p Value (Bonferroni Corrected) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | 0 | 1 | 2 | 3 | 0 vs. 1 | 0 vs. 2 | 0 vs. 3 | 1 vs. 2 | 1 vs. 3 | 2 vs. 3 |
| N | 26 | 44 | 36 | 38 | ||||||
| Duration of diabetes (years) | na | 19 (4–28) | 3 (1–8) | 8 (2–24.25) | n.a. | n.a. | n.a. | 0.0009 (*) | n.s. | n.s. |
| HbA1c (mmol/mol)/ (%) | 32.5 (31–35)/9.2 (9.1–9.4) | 63 (55–75)/8 (7.2–9) | 53 (49–65)/7 (6.6–8.1) | 54 (44–63)/7.1 (6.2–7.9) | <0.0001 (*) | <0.0001 (*) | <0.0001 (*) | 0.0044 (n.s.) | 0.0014 (n.s.) | n.s. |
| SBP (mmHg) | 121 (113–128) | 125 (119.5–130) | 135 (130–149.5) | 120.5 (113.5–129) | n.s. | <0.0001 (*) | n.s. | 0.0003 (*) | n.s. | 0.0001 (*) |
| DBP (mmHg) | 73 (69–79) | 76.5 (69–80) | 80 (74–85) | 71 (66.5–77) | n.s. | 0.0156 (n.s.) | n.s. | 0.0441 (n.s.) | n.s. | 0.0002 (*) |
| T. Cholesterol (mmol/L) | 4.8 (4.13–5.5) | 4.25 (3.8–4.7) | 4 (3.4–4.68) | 4.35 (3.7–5.1) | 0.0465 (n.s.) | 0.0079 (n.s.) | n.s. | n.s. | n.s. | n.s. |
| Creatinine (mmol/L) | 79 (71.25–95) | 70 (65.5–83) | 76 (70–85.75) | 68 (58–76.25) | 0.0282 (n.s.) | n.s. | 0.0014 (n.s.) | n.s. | n.s. | 0.0136 (n.s.) |
| ACR (g/mol) | 0 (0–0.8) | 0.5 (0–1.33) | 0.9 (0.5–2.7) | 0.6 (0.4–0.96) | n.s. | 0.0001 (*) | 0.0130 (n.s.) | 0.0101 (n.s.) | n.s. | 0.0403 (n.s.) |
| GFR (mL/min/1.73 m2) | 74 (70–83) | 89 (78–113) | 85.5 (68–103) | 88.5 (75–112) | 0.0005 (*) | n.s. | 0.0024 (n.s.) | n.s. | n.s. | n.s. |
| Fasting C-Peptide (pmol/L) | 367 (236.5–581) | <66 | 924 (775–1295) | 533 (365–681) | n.a. | 0.0009 (*) | n.s. | <0.0001 (*) | n.a. | 0.0001 (*) |
2.6. Reverse Transcription and Quantitative Real-Time-PCR of Mature miRNA
2.7. Absolute Quantification of miRNA and Data Normalization against Spiked-in Synthetic C. elegans miRNA Control
2.8. Statistical Analysis
3. Results and Discussion
3.1. Clinical Characteristics of All Groups
3.2. miR-224 is Detectable in Urine and is Highly Expressed in HNF1A-MODY Mutation Carriers and Participants with T1DM
3.3. miR-103 is Detectable in Urine and is Highly Expressed in Patients with Diabetes

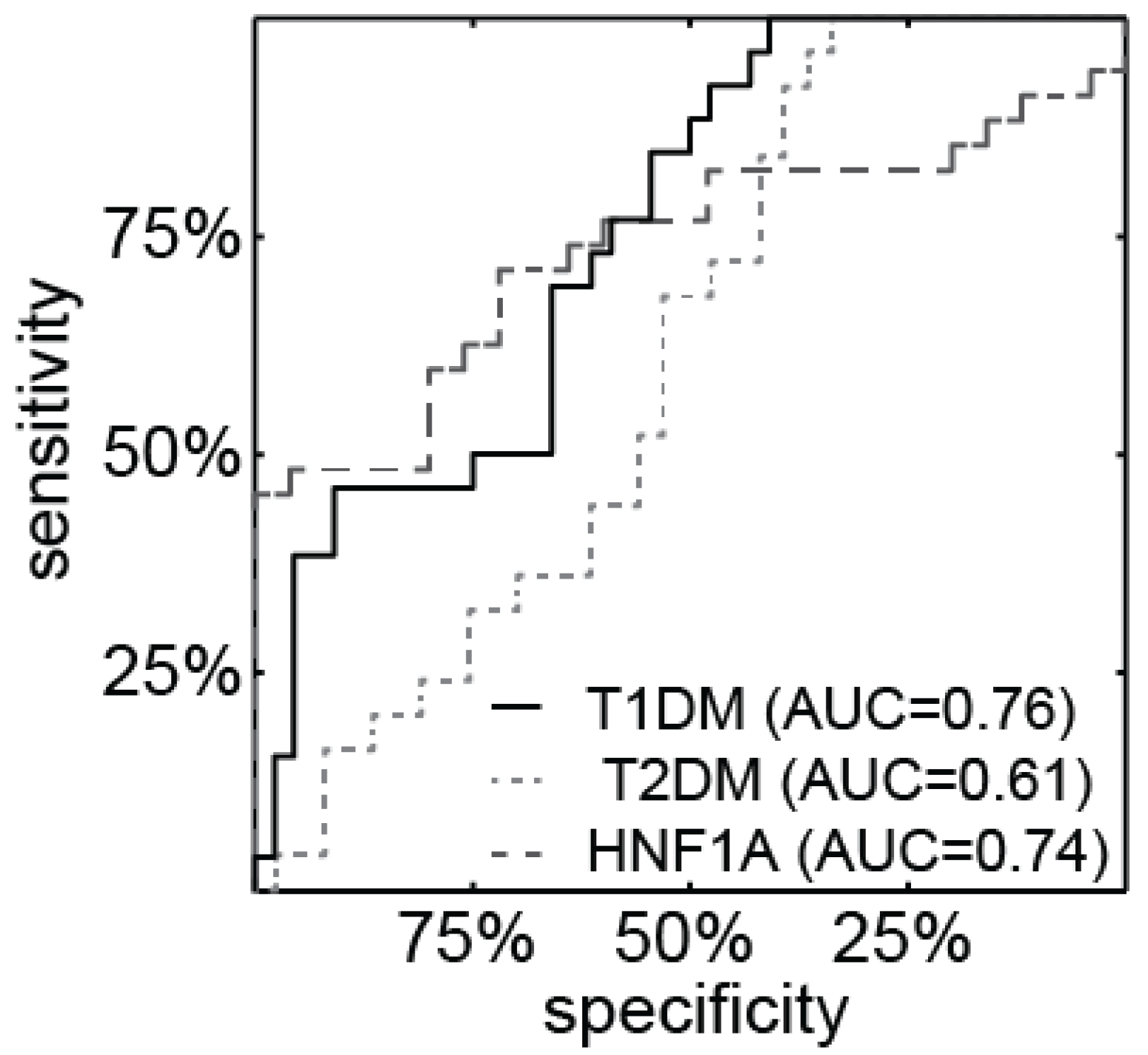
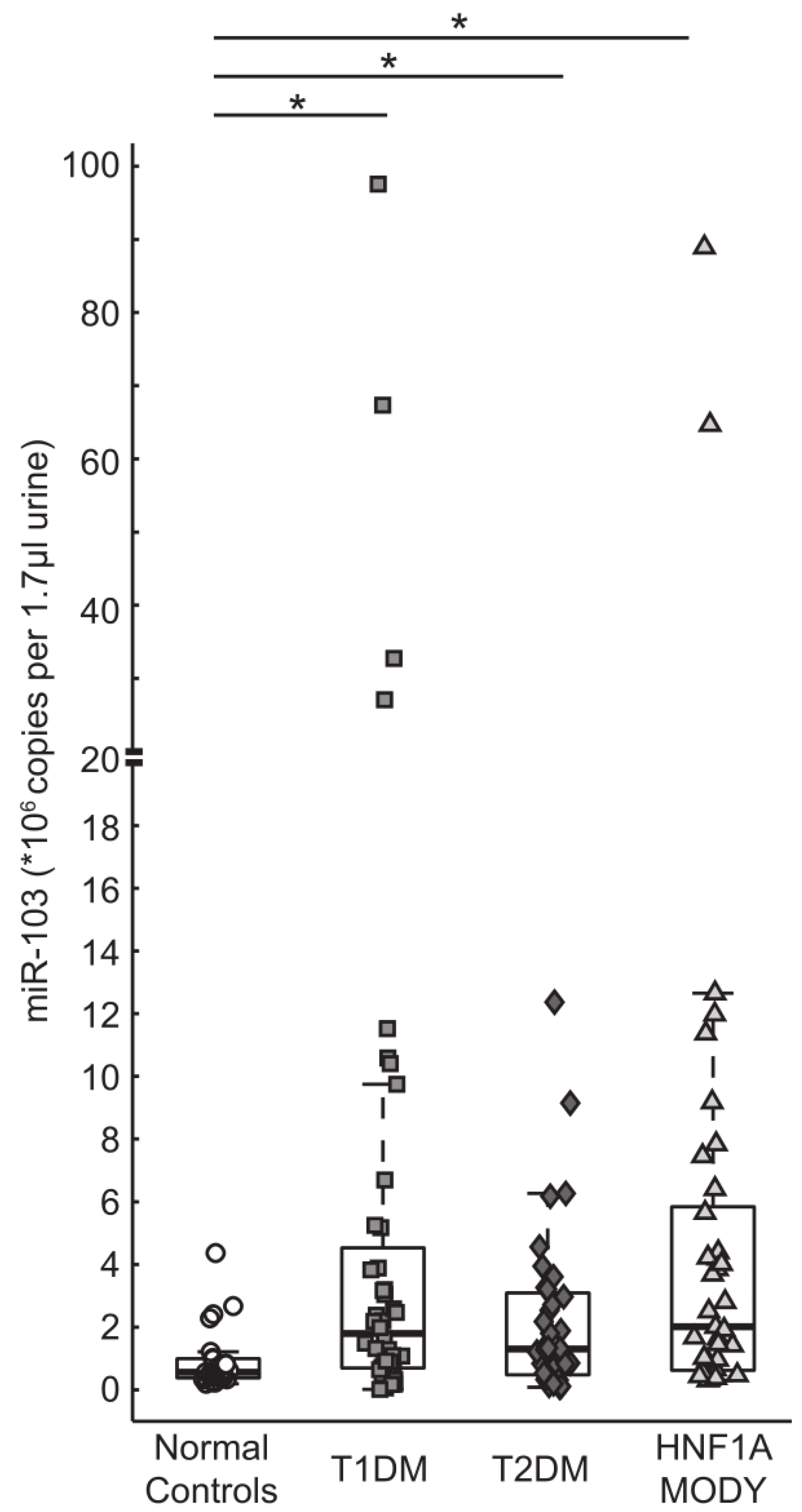
3.4. Correlation of Urine miR-224 and miR-103 Levels with Clinical and Biochemical Characteristics


4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Funding
References
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.M.; Clark, E.A.; Goodman, R.H. Regulation of microRNA-375 by cAMP in pancreatic beta-cells. Mol. Endocrinol. 2012, 26, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Latreille, M.; Hausser, J.; Stutzer, I.; Zhang, Q.; Hastoy, B.; Gargani, S.; Kerr-Conte, J.; Pattou, F.; Zavolan, M.; Esguerra, J.L.; et al. MicroRNA-7a regulates pancreatic beta cell function. J. Clin. Investig. 2014, 124, 2722–2735. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Chen, S.; McArthur, K.; Wu, Y.; Sen, S.; Ding, Q.; Feldman, R.D.; Chakrabarti, S. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 2011, 60, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, K.; Mi, Q.S.; Dong, Z. microRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am. J. Physiol. Ren. Physiol. 2011, 300, F602–F610. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Fasanaro, P.; Castelvecchio, S.; D’Alessandra, Y.; Arcelli, D.; di Donato, M.; Malavazos, A.; Capogrossi, M.C.; Menicanti, L.; Martelli, F. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes 2012, 61, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed]
- Osipova, J.; Fischer, D.C.; Dangwal, S.; Volkmann, I.; Widera, C.; Schwarz, K.; Lorenzen, J.M.; Schreiver, C.; Jacoby, U.; Heimhalt, M.; et al. Diabetes-associated microRNAs in paediatric patients with type 1 diabetes mellitus: A cross-sectional cohort study. J. Clin. Endocrinol. Metab. 2014, 99, E1661–E1665. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Pescador, N.; Perez-Barba, M.; Ibarra, J.M.; Corbaton, A.; Martinez-Larrad, M.T.; Serrano-Rios, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.B.; Wang, C.; Sorensen, K.; Bang-Berthelsen, C.H.; Hansen, L.; Andersen, M.L.; Hougaard, P.; Juul, A.; Zhang, C.Y.; Pociot, F.; et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: Evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp. Diabetes Res. 2012. [Google Scholar] [CrossRef]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Luo, X.; Lin, H.; Zhang, Y.; Lu, Y.; Wang, N.; Zhang, Y.; Yang, B.; Wang, Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J. Biol. Chem. 2007, 282, 12363–12367. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lv, D.; Chen, P.; Xu, T.; Fu, S.; Li, J.; Bei, Y. MicroRNAs in diabetic cardiomyopathy and clinical perspectives. Front. Genet. 2014. [Google Scholar] [CrossRef] [PubMed]
- Baldeon, R.L.; Weigelt, K.; de Wit, H.; Ozcan, B.; van Oudenaren, A.; Sempertegui, F.; Sijbrands, E.; Grosse, L.; Freire, W.; Drexhage, H.A.; et al. Decreased serum level of miR-146a as sign of chronic inflammation in type 2 diabetic patients. PLoS ONE 2014, 9, e115209. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Bulamn, M.P.; Ellard, S.; Appleton, M.; Dronsfield, M.J.; Mackie, A.D.; Baird, J.D.; Kaisaki, P.J.; Yamagata, K.; Bell, G.I.; et al. Mutations in the hepatocyte nuclear factor-1alpha gene are a common cause of maturity-onset diabetes of the young in the U.K. Diabetes 1997, 46, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.; Bacon, S.; Concannon, C.G.; Rizvi, S.R.; Baquie, M.; Farrelly, A.M.; Kilbride, S.M.; Dussmann, H.; Ward, M.W.; Boulanger, C.M.; et al. INS-1 cells undergoing caspase-dependent apoptosis enhance the regenerative capacity of neighboring cells. Diabetes 2010, 59, 2799–2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servitja, J.M.; Pignatelli, M.; Maestro, M.A.; Cardalda, C.; Boj, S.F.; Lozano, J.; Blanco, E.; Lafuente, A.; McCarthy, M.I.; Sumoy, L.; et al. Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol. Cell. Biol. 2009, 29, 2945–2959. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Maechler, P.; Hagenfeldt, K.A.; Wollheim, C.B. Dominant-negative suppression of HNF-1α function results in defective insulin gene transcription and impaired metabolism-secretion coupling in a pancreatic beta-cell line. EMBO J. 1998, 17, 6701–6013. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.; Nyhan, K.C.; Bacon, S.; Kyithar, M.P.; Schmid, J.; Concannon, C.G.; Bray, I.M.; Stallings, R.L.; Prehn, J.H.; Byrne, M.M. Identification of circulating microRNAs in HNF1A-MODY carriers. Diabetologia 2013, 56, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Fevrier, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar]
- Kritis, A.A.; Argyrokastritis, A.; Moschonas, N.K.; Power, S.; Katrakili, N.; Zannis, V.I.; Cereghini, S.; Talianidis, I. Isolation and characterization of a third isoform of human hepatocyte nuclear factor 4. Gene 1996, 173, 275–280. [Google Scholar] [CrossRef]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Kyithar, M.P.; Bacon, S.; Pannu, K.K.; Rizvi, S.R.; Colclough, K.; Ellard, S.; Byrne, M.M. Identification of HNF1A-MODY and HNF4A-MODY in Irish families: Phenotypic characteristics and therapeutic implications. Diabetes Metab. 2011, 37, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Ellard, S.; Colclough, K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum. Mutat. 2006, 27, 854–869. [Google Scholar] [CrossRef] [PubMed]
- Mari, A.; Pacini, G.; Murphy, E.; Ludvik, B.; Nolan, J.J. A model-based method for assessing insulin sensitivity from the oral glucose tolerance test. Diabetes Care 2001, 24, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, C.; Chen, C.; Zhang, Z.; Xiao, H.; Xie, F.; Lei, L.; Chen, Y.; Mao, B.; Jiang, M.; et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2014, 29, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Zhou, H.; Xiao, H.X.; Li, Y.; Zhou, T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Mees, S.T.; Mardin, W.A.; Sielker, S.; Willscher, E.; Senninger, N.; Schleicher, C.; Colombo-Benkmann, M.; Haier, J. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann. Surg. Oncol. 2009, 16, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Boguslawska, J.; Wojcicka, A.; Piekielko-Witkowska, A.; Master, A.; Nauman, A. MiR-224 targets the 3'UTR of type 1 5'-iodothyronine deiodinase possibly contributing to tissue hypothyroidism in renal cancer. PLoS ONE 2011, 6, e24541. [Google Scholar] [CrossRef] [PubMed]
- Kloppel, G.; Drenck, C.R.; Carstensen, A.; Heitz, P.U. The B cell mass at the clinical onset of type I diabetes. Behring Inst. Mitt. 1984, 75, 42–49. [Google Scholar] [PubMed]
- Vesterhus, M.; Haldorsen, I.S.; Raeder, H.; Molven, A.; Njolstad, P.R. Reduced pancreatic volume in hepatocyte nuclear factor 1A-maturity-onset diabetes of the young. J. Clin. Endocrinol. Metab. 2008, 93, 3505–3509. [Google Scholar] [CrossRef] [PubMed]
- Fajans, S.S.; Bell, G.I.; Polonsky, K.S. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 2001, 345, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Siewko, K.; Poplawska-Kita, A.; Telejko, B.; Maciulewski, R.; Zielinska, A.; Nikolajuk, A.; Górska, M.; Szelachowska, M. Prognostic markers for the development of type 1 diabetes in first-degree relatives of diabetic patients. Endokrynol. Pol. 2014, 65, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.M.; Sturis, J.; Menzel, S.; Yamagata, K.; Fajans, S.S.; Dronsfield, M.J.; Bain, S.C.; Hattersley, A.T.; Velho, G.; Froguel, P.; et al. Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes 1996, 45, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Leung, S.W. Identification of microRNA biomarkers in type 2 diabetes: A meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Blondal, T.; Nielsen, S.J.; Baker, A.; Andreasen, D.; Mouritzen, P.; Teilum, M.W.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bacon, S.; Engelbrecht, B.; Schmid, J.; Pfeiffer, S.; Gallagher, R.; McCarthy, A.; Burke, M.; Concannon, C.; Prehn, J.H.M.; Byrne, M.M. MicroRNA-224 is Readily Detectable in Urine of Individuals with Diabetes Mellitus and is a Potential Indicator of Beta-Cell Demise. Genes 2015, 6, 399-416. https://doi.org/10.3390/genes6020399
Bacon S, Engelbrecht B, Schmid J, Pfeiffer S, Gallagher R, McCarthy A, Burke M, Concannon C, Prehn JHM, Byrne MM. MicroRNA-224 is Readily Detectable in Urine of Individuals with Diabetes Mellitus and is a Potential Indicator of Beta-Cell Demise. Genes. 2015; 6(2):399-416. https://doi.org/10.3390/genes6020399
Chicago/Turabian StyleBacon, Siobhán, Britta Engelbrecht, Jasmin Schmid, Shona Pfeiffer, Ross Gallagher, Ailbhe McCarthy, Marie Burke, Caoimhín Concannon, Jochen H. M. Prehn, and Maria M. Byrne. 2015. "MicroRNA-224 is Readily Detectable in Urine of Individuals with Diabetes Mellitus and is a Potential Indicator of Beta-Cell Demise" Genes 6, no. 2: 399-416. https://doi.org/10.3390/genes6020399
APA StyleBacon, S., Engelbrecht, B., Schmid, J., Pfeiffer, S., Gallagher, R., McCarthy, A., Burke, M., Concannon, C., Prehn, J. H. M., & Byrne, M. M. (2015). MicroRNA-224 is Readily Detectable in Urine of Individuals with Diabetes Mellitus and is a Potential Indicator of Beta-Cell Demise. Genes, 6(2), 399-416. https://doi.org/10.3390/genes6020399






