Insights into Cross-Kingdom Plant Pathogenic Bacteria
Abstract
: Plant and human pathogens have evolved disease factors to successfully exploit their respective hosts. Phytopathogens utilize specific determinants that help to breach reinforced cell walls and manipulate plant physiology to facilitate the disease process, while human pathogens use determinants for exploiting mammalian physiology and overcoming highly developed adaptive immune responses. Emerging research, however, has highlighted the ability of seemingly dedicated human pathogens to cause plant disease, and specialized plant pathogens to cause human disease. Such microbes represent interesting systems for studying the evolution of cross-kingdom pathogenicity, and the benefits and tradeoffs of exploiting multiple hosts with drastically different morphologies and physiologies. This review will explore cross-kingdom pathogenicity, where plants and humans are common hosts. We illustrate that while cross-kingdom pathogenicity appears to be maintained, the directionality of host association (plant to human, or human to plant) is difficult to determine. Cross-kingdom human pathogens, and their potential plant reservoirs, have important implications for the emergence of infectious diseases.1. Introduction
Plant pathogenic bacteria are responsible for major economical losses in agricultural industries worldwide, prompting massive research efforts to understand their ecology, pathology and epidemiology. Studies of many agriculturally relevant pathogens like Pseudomonas syringae, Xylella fastidiosa, Erwinia amylovora, and Xanthomonas campestris [1-4] have revealed an extensive specialization to plant systems, including a wealth of plant-specific pathogenicity determinants such as type III secretion systems, plant hormone analogs, and enzymes that target plant-specific cell wall components [1,5-7]. Despite this, we often neglect the fact that the interactions of phytopathogenic bacteria are not confined to plants, and may include other organisms in the environment. Recent studies have begun to show that many plant pathogens have the capacity to colonize other hosts outside of the plant kingdom, including insects, animals, and humans [8,9].
Human pathogens are studied almost exclusively for their detrimental impact on human health. Pathogens such as Escherichia coli, Listeria monocytogenes, and Campylobacter jejuni possess a diverse set of genetic factors that enable pathogenicity, ranging from specialized secretion systems to toxins and adhesins, all of which are involved in manipulating or circumventing the human immune system [10-12]. We often consider these human pathogenic bacteria to be dedicated animal pathogens, causing disease and epidemics; yet, the constant interaction of human carriers with their environment predisposes these pathogens to alternative niches that include non-animal hosts. Not surprisingly, we have begun to discover that some animal pathogens, which are known to cause serious human diseases, also have plant pathogenic capabilities. The epidemiology and disease strategies of these pathogens, whether considered primarily a plant or human pathogen, are of particular interest from the perspectives of both the biology and evolution of cross-kingdom pathogenesis.
The disease strategies used by cross-kingdom pathogens to infect unrelated hosts are of particular interest since plant and animal hosts have distinctive physical barriers and defense responses. While specialists may have evolved dedicated factors for overcoming the physical barriers and innate defenses in a certain host, a cross-kingdom pathogen would require a diverse library of genes and disease strategies to overcome the specific defense obstacles of each of its hosts. This would either necessitate a suite of pathogenicity factors to enable attachment, disease development, and dispersal for each host, or a universal disease strategy in which the same suite of pathogenicity factors is used for all hosts. Signatures of host adaptation will be apparent in these determinants, which may include incremental mutations and genetic rearrangements, along with the acquisition of novel genetic elements that may contribute to virulence or host specificity [13]. The identification of these genetic determinants in cross-kingdom pathogens with both human- and plant-pathogenic potential can provide a better understanding of the evolution of phytopathogenicity, as well as the role of plants as potential reservoirs for clinically relevant bacteria.
This review will examine several bacterial pathogens that exhibit cross-kingdom pathogenicity, where both humans and plants are potential hosts (Table 1). We begin by exploring the pathogenicity of Pantoea and Burkholderia, which are commonly regarded as plant pathogens, but have been shown to cause human disease. We then examine the disease strategies of the human pathogens Salmonella, Serratia, Enterobacter and Enterococcus, which have been shown to have plant-pathogenic potential. We highlight what is known about the disease determinants and strategies for each pathogen in each of its hosts, and identify those examples where specific determinants are used in multiple hosts. Finally, we address the evolutionary means by which plant pathogens may evolve to be pathogenic to humans, as well as the possible routes of microbes that can lead to the evolution and maintenance of cross-kingdom pathogenicity.
2. Plant Pathogens that Infect Humans
Plant pathogens have evolved a repertoire of pathogenicity factors that are used to invade plant host cells, facilitate disease development, and eventually promote pathogen dispersal [14]. Surprisingly, some of these plant pathogens cause disease in humans, and are frequently isolated from human infections in the nosocomial environment. The genus Pantoea, for example, currently includes seven species (Pantoea agglomerans, Pantoea ananatis, Pantoea citrea, Pantoea dispersa, Pantoea punctata, Pantoea stewartii, and Pantoea terra) [15-21] that are known to cause plant disease. P. agglomerans causes crown and root gall disease of gypsophila and beet [15,20], P. ananatis causes bacterial blight and dieback of Eucalyptus [16], brown stalk rot of maize [22], and stem necrosis of rice [23], and P. citrea is the causal agent of pink disease in pineapple [21]. The type III secretion system (T3SS) appears to be an essential plant pathogenicity factor for many species of Pantoea, enabling efficient colonization and onset of disease through the injection of bacterial effector proteins that disrupt defense signalling in host cells [24]. Pantoea agglomerans pvs. gypsophilae and betae are known for using a T3SS for causing galls on their host plants [25]. While P. agglomerans pv. betae can induce gall formation on gypsophila and beet plants, P. agglomerans pv. gypsophilae is only capable of causing gall formation on gypsophila. Host range is determined in part by the type III effector protein, PthG, which is recognized by the beet plant resulting in a defense response, but functions as a virulence factor in gypsophila [25]. Studies with transgenic Nicotiana tabacum plants expressing PthG suggest that the effector may interfere in the plant auxin signalling pathways, resulting in higher observed auxin and ethylene levels, and subsequent blockage of root and shoot development [25]. The manipulation of plant-specific hormones is likely directly responsible for callus/gall formation, and highlights the specialization of this pathogen to plant systems.
Despite this specialization to plants, species of Pantoea have also been discovered to be pathogenic to humans. Now classified as an opportunistic human pathogen, P. agglomerans was implicated in US and Canadian outbreaks of septicaemia caused by contaminated closures on infusion fluid bottles [26]. P. agglomerans has since been associated in the contamination of intravenous fluid, parenteral nutrition, blood products, propofol, and transference tubes, causing illness and even death [27-31]. P. agglomerans has also been obtained from joint fluids of patients with synovitis, osteomyelitis, and arthritis [17], where infection often occurs following injuries with wood slivers, plant thorns, or wooden splinters [17,32-34]. This is also true for P. ananatis, P. septica, and P. dispersa, which are known for causing disease in onion and sugar cane plants, but have been implicated in multiple cases of bacteremia and septicaemia [35-37]. Phylogenetic studies examining the relationship between plant and clinical isolates have shown that they are indistinguishable, making their potential pathogenicity in plant and animal hosts unclear [38].
Much like Pantoea, species of Burkholderia are also recognized phytopathogens, but are ubiquitous in the terrestrial and aquatic environment. Burkholderia species, including Burkholderia cepacia, Burkholderia cenocepacia, Burkholderia ambifaria, and Burkholderia pyrrocinia were initially identified as inhabitants of agricultural soil, with some, like Burkholderia glathei being found in fossil soils in Germany [39]. Given their ubiquity in the terrestrial environment, it is not surprising that some strains, like Burkholderia plantarii and Burkholderia gladioli have been shown to be pathogenic to onion, rice, gladiolus and iris [39], and possess key plant pathogenicity factors. Burkholderia glumae, the most important bacterial pathogen of rice in Japan, Korea and Taiwan, was shown to carry a plant T3SS, which is essential for its ability to cause plant disease [40]. An analysis of the proteins regulated by HrpB, which activates the expression of T3SS genes, identified 34 secreted extracellular proteins, 21 of which had putative HrpB-binding sequences in their upstream regulatory regions [41]. Another set of 16 proteins appeared to be secreted via a type II protein secretion system (T2SS), and may play a role in plant pathogenesis [41]. Mutants lacking either the T2SS or the T3SS produced the toxin, toxoflavin, but were less virulent to rice panicles, indicating the importance of these systems to disease [41]. In fact, Burkholderia pseudomallei possesses multiple T3SS gene clusters, one of which, T3SS2, showed high similarity to the T3SSs of phytopathogenic Xanthomonas spp. and Ralstonia solanacearum [42], and in addition, arabinose-negative strains of B. pseudomallei are more virulent in tomato plants, and tend to carry a T3SS, suggesting that these two phenotypes are linked to virulence in plants [43]. Strains of some species, like B. cepacia, are specialized to plant roots and have been found in the maize rhizosphere [44], while others are pathogenic to onion, and cause severe rots using a cocktail of plant-specific enzymes following biofilm formation [45].
Interestingly, B. cepacia that causes onion rot can also cause life-threatening pulmonary infections in individuals with chronic granulomatous disease, or cystic fibrosis (CF) [46]. Between 1980 and 1990, B. cepacia was associated with a number of cases of “cepacia syndrome” in CF treatment centres and social gatherings, where CF patients became infected by other B. cepacia-carrying individuals, with many of the infections being fatal [39,47-50]. Recently, fatal infections were also observed in non-CF patients in reanimation wards in Europe and North America [51]. Investigation into the ability of various strains of B. cepacia to penetrate airway barriers revealed that all three of the B. cepacia strains tested circumvented the mechanical barriers of mucus and ciliary transport to penetrate the airway epithelium [52]. Different strains of B. cepacia use distinct invasion pathways and virulence determinants, which may account for differences in the virulence of strains [52].
Several disease determinants for different species of Burkholderia in human hosts have been determined. For example, B. pseudomallei, which infects tomato plants, contains a complete pqsA-pqsE operon, which is highly similar to the genes responsible for the synthesis of the virulence-associated signalling molecule 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) found in Pseudomonas aeruginosa [53]. Introduction of the B. pseudomallei hhqA and hhqE genes into P. aeruginosa pqsA and pqsE mutants restored PQS production and virulence. Likewise, the presence of a capsular polysaccharide has been implicated in the pathogenicity of Burkholderia toward humans. Parental strains of B. mallei with the capsule were highly virulent in hamsters and mice, while capsule mutants were avirulent in both animal models [54].
| Pathogen | Plant host/Niche | Plant pathogenicity factors 1 | Human disease/Condition | Human pathogenicity factors 1 |
|---|---|---|---|---|
| Enterobacter cloacae 2 | Macadamia, dragon fruit, orchids, papaya | Unknown | Respiratory/skin/urinary infection, septicaemia | cytotoxin, ompX |
| Enterococcus faecalis 2 | Arabidopsis thaliana | fsrB, sprE | Urinary/abdominal/cutaneous infections, septicaemia | Hemolysin, salB, esp, fsrB, sprE |
| Burkholderia ambifaria | Soil, maize roots | Dioxygenase | Unknown | Unknown |
| Burkholderia cenocepacia 2 | Soil, maize roots, onion | T4SS | Septicaemia | Unknown |
| Burkholderia cepacia 2 | Soil, rice, maize, wheat, onion | Unknown | Septicaemia | Unknown |
| Burkholderia gladioli 2 | Onion, gladiolus, iris, rice | Unknown | Septicaemia | Unknown |
| Burkholderia glathei | Soil | Unknown | Unknown | Unknown |
| Burkholderia glumae 2 | Rice | toxR, T3SS | Chronic granulomatous disease | Unknown |
| Burkholderia mallei 2 | Soil | Unknown | Melioidosis/Glanders | Capsular polysaccharide |
| Burkholderia plantarii 2 | Rice, gladiolus, iris | rpoS, quorum sensing | Melioidosis | Rhamnolipids |
| Burkholderia pseudomallei 2 | Tomato | T3SS | Melioidosis/Glanders | pqsA–pqsE operon |
| Burkholderia pyrrocinia 2 | Soil | Unknown | Melioidosis/Glanders | Unknown |
| Pantoea agglomerans 2 | Crown/root gall | pthG, T3SS | Arthritis/septicaemia | T3SS |
| Pantoea ananatis 2 | Eucalyptus, maize, rice | T3SS | Septicaemia | Unknown |
| Pantoea citrea 2 | Pineapple | T3SS | Septicaemia | Unknown |
| Pantoea dispersa 2 | Seeds | T3SS | Septicaemia | Unknown |
| Pantoea punctata | Japanese mandarin oranges | T3SS | Unknown | Unknown |
| Pantoea septica | Unknown | Unknown | Septicaemia | Unknown |
| Pantoea stewartii | Maize | T3SS | Unknown | Unknown |
| Pantoea terrea | Japanese mandarin oranges | Unknown | Unknown | Unknown |
| Salmonella enterica 2 | Tomato, Arabidopsis thaliana | agfA, agfB, FilI | Gastroenteritis/typhoid fever | Flagellum (fhD), T3SS |
| Serratia marcescens 2 | Squash, pumpkin | Fimbrial genes, biofilm, oxyR | Septicaemia, urinary tract infection | LPS, iron uptake, hemolysin, protease |
1T3SS: type III secretion system; T4SS: type IV secretion system;2Bacterial pathogens capable of both plant and human disease.
3. Human Pathogens that Can Infect Plants
Our human-centric view of disease often leads to sweeping assumptions that human pathogenic bacteria are devoted to human hosts. Species of Salmonella, Serratia, Enterobacter, and Enterococcus are commonly considered problematic human pathogens that are frequently found in the nosocomial environment, and which cause food poisoning, general infections, and septicaemia [55-58] (Table 1). Relatively recent studies, however, have begun to uncover that these human-pathogenic bacterial species are also capable of colonizing and causing disease in a wide variety of plant hosts. Notably, many of these studies have been conducted under laboratory conditions, providing evidence for the phytopathogenic potential of these bacterial species; but, the incidence of plant disease caused by many of these human pathogens in the natural environment remains unknown.
Salmonella is the causal agent of many human, animal, and bird diseases worldwide, including gastroenteritis and typhoid fever, and results in approximately 1.4 million human illnesses and 600 deaths annually in the United States [56]. Successful host infection by Salmonella appears dependent on many pathogenicity determinants, including two T3SSs and a large suite of secreted effectors that function to mediate both intercellular and intracellular survival in the host [59]. Other virulence factors include the flhD gene, which regulates the production of the flagellum, is essential for the full invasive potential of the bacterium [60]. Virulence factors like flavohemoglobin protect against nitric oxides, and mediate bacterial survival in macrophages after phagocytosis [61].
Despite its clear adaptation to surviving in human hosts, Salmonella has also been isolated from the phyllosphere of tomato crops [62]. S. enterica levels on wild tomato (Solanum pimpinellifolium) were lower than on domesticated tomato cultivars (Solanum lycopersicum), with S. enterica preferentially colonizing type 1 trichomes. Plants irrigated with contaminated water had larger S. enterica populations than plants grown from seeds planted in infected soil; however, both routes of contamination resulted in detectable S. enterica populations in the phyllosphere, suggesting that S. enterica has the ability to associate with plants and has adapted to survive in the phyllosphere. agfA and agfB were identified as being involved in enabling S. enterica to associate with plants [63]. agfA mutants were unaffected in their ability to attach or colonize alfalfa sprouts, whereas agfB mutants showed reduced colonization ability. This suggests that agfB alone plays a role in plant attachment [63].
Salmonella is not only capable of colonizing the phyllosphere; it has been shown to infect the plant Arabidopsis under laboratory conditions, and cause death of plant organs [64,65]. Inoculation of Salmonella in Arabidopsis via shoot or root tissues resulted in chlorosis, wilting, and eventually death of the infected tissues within seven days [64]. The specific virulence factors involved are not yet known; however, its plant pathogenicity, much like its human pathogenicity appears dependent, in part, on the flagellum. FliI, a protein needed for flagellar assembly has been shown to be essential for plant pathogenesis, possibly through its involvement in a specialized protein export pathway [66]. The FliI protein is similar to the HrpB6 protein of the rice pathogen Xanthomonas, which is a key component of the T3SS, and mediates interactions between Xanthomonas and its host [66]. Secretion of proteins may therefore be integral for both human and plant association for Salmonella, although it is still unknown whether the same proteins can be used for pathogenesis in both hosts.
Serratia marcescens is a human pathogen commonly found in the respiratory and urinary tracts of humans, and is responsible for approximately 1.4% of nosocomial infections in the United States [57]. Through transposon mutagenesis, genes involved in lipopolysaccharide (LPS) biosynthesis, iron uptake, and hemolysin production were discovered to be essential for bacterial virulence in the host Caenorhabditis elegans [67]. Further studies have shown that purified protease proteins of S. marcescens administered to the lung tissue of guinea pigs and mice produced pneumonia-like symptoms and haemorrhaging, which was similar to those animals with acute Serratia pneumonia [68]. Yet, despite its animal virulence factors, Serratia has also been found to be a common phytopathogen. S. marcescens is recognized as a phloem-resident pathogen that causes cucurbit yellow vine disease of pumpkin (Cucurbita moschata L.) and squash (Cucurbita pepo L.), which is marked by wilting, phloem discoloration, and yellowing of foliage [69-71]. Recent studies have shown that S. marcescens produces a biofilm along the sides of the phloem vessels, blocking the transport of nutrients and eventually causing the plant to wilt and die [72,73]. A genetic screen to identify genes that modulate biofilm formation in S. marcescens revealed the involvement of fimbrial genes, as well as an oxyR homolog—a conserved bacterial transcription factor having a primary role in the oxidative stress response [74]. The involvement of these plant disease genes in human pathogenicity has not been determined.
Another bacterium that has also shown cross-kingdom pathogenesis is the Gram-negative bacterium Enterobacter cloacae, an important nosocomial pathogen responsible for bacteremia, lower respiratory tract infections, skin and soft-tissue infections, as well as urinary tract infections [55]. Recently, an E. cloacae infection of the bloodstream was traced back to contaminated human albumin [75]. E. cloacae synthesizes a Shiga-like toxin II-related cytotoxin, which was implicated in an infant case of haemolytic-uremic syndrome [76]. Studies have shown that the concentration of the outer membrane protein (OmpX) produced by E. cloacae during infection influenced the ability of the bacterium to invade rabbit intestinal tissue [77]. Overproduction of OmpX led to a 10-fold increase in the invasiveness of E. cloacae in rabbit intestinal enterocytes in situ, whereas the mutant and wildtype strains were unable to effectively invade the same tissue [77]. Interestingly, OmpX shares high amino acid similarity to the virulence proteins PagC and Rck of Salmonella typhimurium as well as the virulence-associated Ail protein of Yersinia enterocolitica. Although there is evidence that E. cloacae has evolved to colonize the human host, it has also been identified as the causal agent of grey kernel disease of macadamia (Macadamia integrifolia) [78]. The onset of grey kernel disease affects not only the quality of the kernels produced by the tree, but results in grey discoloration and a foul odour [78]. E. cloacae also causes bacterial soft rot disease in dragon fruit (Hylocereus spp.) [18], bacterial leaf rot in Odontioda orchids [79], and is also responsible for internal yellowing disease in papaya [78]. Again, the specificity of the virulence factors used in each of these hosts is not known.
Cross-kingdom pathogenesis is not limited to Gram-negative bacteria. Enterococci are part of the normal intestinal flora of humans and animals, but are also important pathogens responsible for serious infections, especially in immunocompromised patients [58]. With increasing antibiotic resistance, enterococci are recognized as nosocomial pathogens that can be challenging to treat. The genus Enterococcus includes more than 17 species, but only a few can cause local or systemic clinical infections including urinary tract and abdominal infections, wound infections, bacteremia, and endocarditis [80]. Clinical isolates of Enterococcus faecalis have been found to produce hemolysin—a virulence factor that leads to the lysis of red blood cells [81]. Hemolytic strains exhibit multiple drug resistance more frequently than non-hemolytic strains, while strains isolated from fecal specimens of healthy individuals display a low (17%) incidence of hemolysin production [81]. The salB gene of E. faecalis has also been shown to be a virulence factor, since it increases bacterial adherence to extracellular matrix proteins and promotes biofilm formation during infection [82]. The surface protein Esp was shown to contribute to the colonization and persistence of the bacterium in the urinary tract [83]. E. faecalis has also been shown to exhibit pathogenicity toward insects in addition to human hosts, where the extracellular gelatinase of E. faecalis was found to destroy the host defence system through the degradation of inducible antimicrobial peptides in insect hemolymph and in human serum [84]. Similarly, a putative quorum-sensing system gene (fsrB) and a serine protease (sprE) were shown to play an important role in mammalian and nematode models of infection [58].
E. faecalis is not only capable of infecting mammalian and nematode hosts, but also the plant Arabidopsis thaliana, causing plant death seven days after inoculation under laboratory conditions [58]. The manifestation of disease initially begins once the bacterium has successfully attached itself to the leaf surface, where upon entry of the leaf tissue through the stomata or wounds, E. faecalis multiplies and colonizes the intercellular spaces of the plant host and causes rotting and disruption of the plant cell wall and membrane structures. The phytopathogenicity of E. faecalis appears to involve some of the same genetic determinants involved in animal pathogenesis, including the quorum sensing system gene (fsrB) and a serine protease (sprE) [58]. The quorum-sensing mutant (ΔfsrB) was attenuated in virulence in the A. thaliana root model, since fewer bacteria were able to attach to the root surfaces, and biofilm formation was greatly reduced. Similarly, the serine protease mutant (ΔsprE) no longer caused plant death, suggesting that this gene plays an important role in E. faecalis plant pathogenesis [58]. E. faecalis clearly uses a general disease strategy that allows it to use the same virulence factors for exploiting two different hosts.
4. Evolutionary Models
Cross-kingdom pathogenicity represents an intriguing balance between the costs and benefits of bacterial generalization versus specialization. While there are benefits to specializing on one host, and evolving to exploit its subtle vulnerabilities, the availability of this host would limit the success of the pathogen, thereby favouring host diversification. Conversely, a broad host range would ensure host availability for the pathogen, but at the significant cost of having to utilize a universal, and likely suboptimal infection strategy for exploiting that host. When the alternative host is from a different kingdom, the dynamics become even more intriguing, since the disease strategy is likely to be quite different. The evolution of cross-kingdom pathogenicity and its origins can be described using several simplistic models. In the context of the examples described here, one model describes the evolution of a successful animal pathogen that acquires the ability to cause disease in a plant host (human to plant). If we presuppose that animal pathogens are readily acquired from the environment and are already adapted to animal hosts [85], there are several ways in which these pathogens may acquire plant-pathogenic potential. Deposition of animal pathogens back into the environment, such that they are introduced near or onto plants recurrently may facilitate the exchange of genetic information with other organisms in the environment, and the gain of a selective advantage that enables persistence. One primary route to this general environment is through the natural strategies used by bacteria to ensure dissemination from their infected host. For humans, symptoms such as diarrhea that result from an infection provide the bacteria with an escape route from the host into the environment [86] (Figure 1). Released bacteria move from human wastes to natural watersheds, with subsequent reuse of these sources for irrigation of commercially relevant crops increasing bacterial titres in the phyllosphere [64,65]. But, the movement of human pathogens to the general environment may also occur through indirect routes. The Pharoah ant (Monomorium pharaonis) is able to transport human pathogenic bacteria including Salmonella, Staphylococcus, and Streptococcus within a nosocomial setting [87], and there have been reports of the common house fly, Musca domestica, carrying Pseudomonas aeruginosa, Enterococcus faecalis and Staphylococcus aureus [88] (Figure 1). These insects function as transports for human pathogenic bacteria, moving them from the clinical setting to the general environment [9], and likely onto plants. Acclimation to a plant host would occur over extended periods of time, and would be accelerated with repeated cycling of pathogens from humans to the general environment.
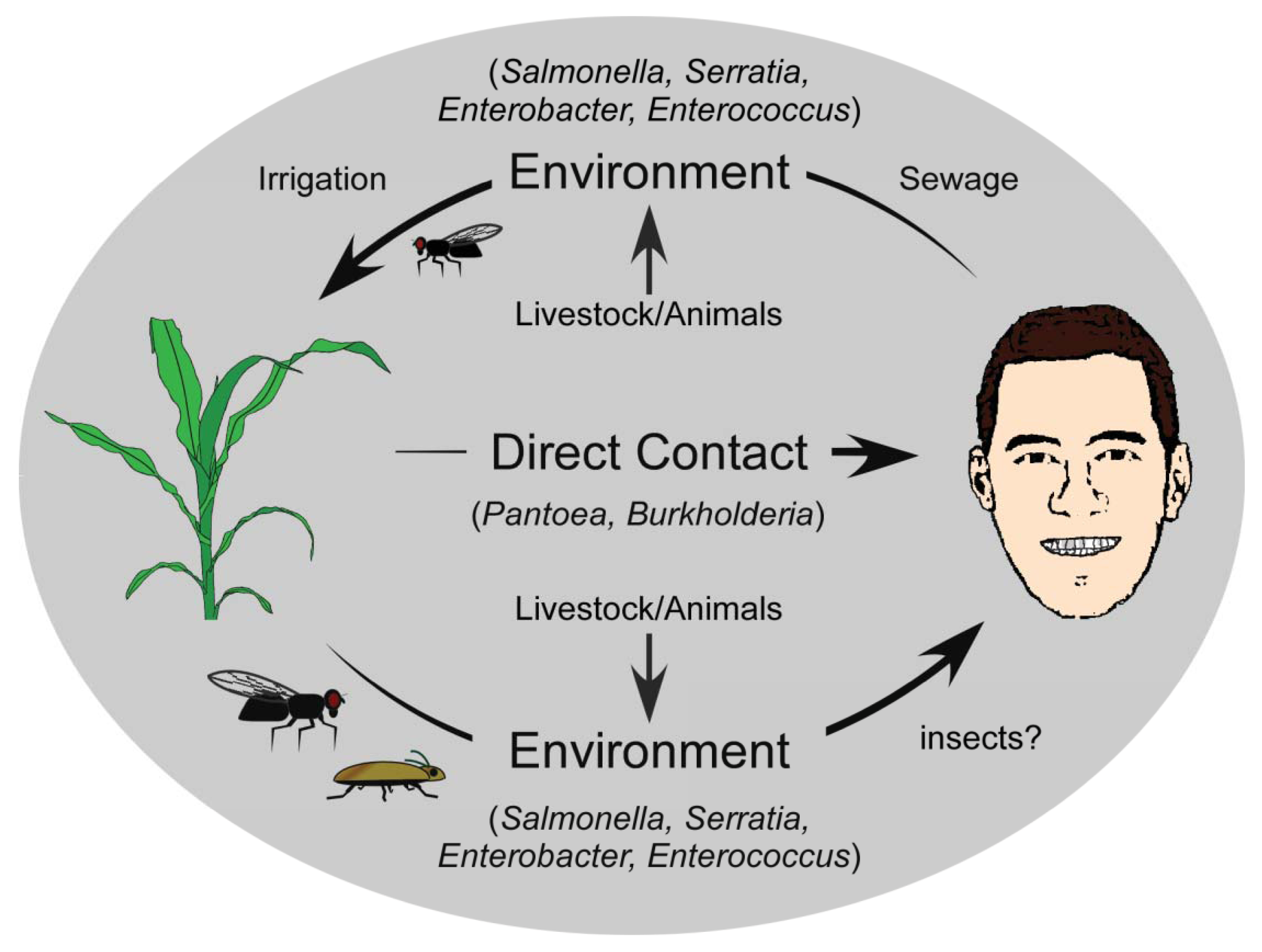
Under this same model, the evolution of cross-kingdom pathogenicity can also be viewed as a secondary trait that arose following the acquisition of key factors by bacteria, which acted either as anti-feeding deterrents against potential animal predators, or functioned to ensure survival and persistence in the host and/or general environment [91]. These determinants may have targeted key defense pathways of animal predators, which would have included the innate immune system—the most basal and common defense pathway of many eukaryotes. This would have allowed pathogens carrying such determinants to exploit both humans/animals and plants by targeting a common pathway, enabling pathogen replication and dispersal [92]. Still, such pathogens would have had to rely on incidental host association and/or contact mediated by external processes, as the microbe would not have specific strategies for host entry or association. The maintenance of cross-kingdom pathogenicity would require only the selective pressure that initially resulted in the evolution of the virulence or defense factor.
A second model that can account for the evolution of the cross-kingdom pathogens has plant pathogenicity as the ancestral state [89], with established plant pathogens having evolved the ability to exploit animals (including humans) as alternative hosts. The simplest mode of transmission is direct contact with an epiphytically colonized or diseased plant by humans, whether by ingestion, scrapes or abrasions, or contact with the skin or other mucosal membranes, providing the bacteria with a direct route to a potential host. Several plant pathogens that cause opportunistic human infections are associated with commercially relevant crop plants. Species of Pantoea, for example, are found quite commonly on plants, including beets [15,20], maize [22], rice [23], and pineapple [21], and human infections occur frequently following abrasions caused by rose thorns and splinters, suggesting that these plant-associated strains can lead to human infection directly (Figure 1) [17,32-34]. Similarly, Burkholderia infects various plant species including onion, rice, sorghum and velvet beans [39,47,90], and may also have a direct route to humans. Indirect transfer to humans or even to the nosocomial environment may be mediated by other organisms like ants and flies, which have been implicated as carriers of many bacterial species [87,88], and may provide environmental isolates with a direct route to other environments and hosts.
Although there is evidence to support these models of evolution, it is difficult to establish evolutionary directionality, or the underlying selective pressures that lead to the evolution of cross-kingdom pathogenicity. The ability of human pathogens to exploit plants as alternative hosts is of particular significance from the perspective of host jumps and host-specific adaptation, since plants would ultimately serve as an extensive reservoir for clinically relevant bacteria. Some plant endophytes, which are organisms that live inside of plants often asymptomatically, have been shown to exhibit human pathogenic potential. Species like Morganella morganii, Klebsiella pneumoniae, Pantoea agglomerans and Streptomyces sp., have clinical relevance, but have been more closely investigated for their association with plants [93,94]. M. morganii is a phosphate solubilising bacterium [95], K. pneumoniae a nitrogen-fixing endophyte [96], P. agglomerans a gall-forming phytopathogen, and Streptomyces sp. a plant endophyte and plant pathogen [97-99]. Many of these animal-pathogenic endophytes have been shown to be latent plant pathogens, where after establishing a symbiotic mutualistic relationship with their plant host, the bacterium infects and causes severe disease within the plant host, possibly due to environmental changes [100]. It has also been observed that an organism that is an endophyte in one plant species may be pathogenic in a different plant species [101]. Yet, many human pathogens have been found to occur naturally in the rhizosphere [93], and it has been suggested that the mechanisms necessary for surviving in the rhizosphere are similar to those necessary for causing human infection [102]. Contributing to the maintenance of these crosskingdom pathogens may be anthropogenic factors. Certain species with human and plant pathogenic potential, such as Pantoea and Burkholderia cepacia are also commonly used as biocontrol agents against Erwinia amylovora and Rhizoctonia solani, respectively [103,104]. The extensive use of these bacteria as biocontrol agents on commercially relevant crops can lead to broad dispersal in the environment, increasing their populations dramatically and providing them with greater opportunity for interaction with other microbes. Horizontal gene transfer with other plant and human pathogenic bacteria in the environment can lead to extensive exchange of host-specific virulence factors or niche- specific determinants, creating rapidly-evolving populations that may exhibit oscillations between host-associative and free-living states.
5. Conclusions
Advances in molecular genetics coupled with the exploration of the pathogenic potential of seemingly dedicated pathogens is beginning to reveal that many phytopathogenic bacteria are capable of exploiting human hosts, and many human pathogens are capable of exploiting plant hosts. The constant cycling of pathogens from the general environment to human environments may help to maintain cross-kingdom pathogenicity, with plants serving as intermediate hosts or reservoirs for human pathogens. The ability of these cross-kingdom pathogens to maintain their population levels in a variety of environments likely increases their pan genome and evolutionary potential, ultimately making these pathogens significant from the perspective of emerging and remerging infectious diseases.
Acknowledgments
We would like to thank Taylor Duda, April Sefton, Lucas Robinson, and Kollin Schmalenberg for their helpful suggestions and comments. John Stavrinides is supported by funding from the NSERC Discovery Grants Program, the Canada Foundation for Innovation, and the Faculty of Science at the University of Regina.
References
- Hopkins, D.L. Xylella Fastidiosa: Xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 1989, 27, 271–290. [Google Scholar]
- Whalen, M.C.; Innes, R.W.; Bent, A.F.; Staskawicz, B.J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 1991, 3, 49–59. [Google Scholar]
- Jock, S.; Donat, V.; López, M.M.; Bazzi, C.; Geider, K. Following spread of fire blight in Western, Central and Southern Europe by molecular differentiation of Erwinia amylovora strains with PFGE analysis. Environ. Microbiol. 2002, 4, 106–114. [Google Scholar]
- Dow, J.M.; Crossman, L.; Findlay, K.; He, Y.-Q.; Feng, J.-X.; Tang, J.-L. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10995–11000. [Google Scholar]
- Osbourn, A.E.; Barber, C.E.; Daniels, M.J. Identification of plant-induced genes of the bacterial pathogen Xanthomonas campestris pathovar campestris using a promoter-probe plasmid. EMBO J. 1987, 6, 23–28. [Google Scholar]
- Wei, Z.M.; Laby, R.J.; Zumoff, C.H.; Bauer, D.W.; He, S.Y.; Collmer, A.; Beer, S.V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 1992, 257, 85–88. [Google Scholar]
- Guttman, D.S.; Vinatzer, B.A.; Sarkar, S.F.; Ranall, M.V.; Kettler, G.; Greenberg, J.T. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 2002, 295, 1722–1726. [Google Scholar]
- Chantanao, A.; Jensen, H.J. Saprozoic nematodes as carriers and disseminators of plant pathogenic bacteria. J. Nematol. 1969, 1, 216–218. [Google Scholar]
- Nadarasah, G.; Stavrinides, J. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol. Rev. 2011, 35, 555–575. [Google Scholar]
- Enders, U.; Karch, H.; Toyka, K.V.; Michels, M.; Zielasek, J.; Pette, M.; Heesemann, J.; Hartung, H.P. The spectrum of immune responses to Campylobacter jejuni and glycoconjugates in Guillain-Barre syndrome and in other neuroimmunological disorders. Ann. Neurol. 1993, 34, 136–144. [Google Scholar]
- Jarvis, K.G.; Giron, J.A.; Jerse, A.E.; McDaniel, T.K.; Donnenberg, M.S.; Kaper, J.B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 1995, 92, 7996–8000. [Google Scholar]
- Lenz, L.L.; Mohammadi, S.; Geissler, A.; Portnoy, D.A. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 12432–12437. [Google Scholar]
- Woolhouse, M.E.; Taylor, L.H.; Haydon, D.T. Population biology of multihost pathogens. Science 2001, 292, 1109–1112. [Google Scholar]
- Panstruga, R.; Dodds, P.N. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 2009, 324, 748–750. [Google Scholar]
- Cooksey, D.A. Galls of Gypsophila paniculata caused by Erwinia herbicola. Plant Dis. 1986, 70, 464–468. [Google Scholar]
- Coutinho, T.A.; Preisig, O.; Mergaert, J.; Cnockaert, M.C.; Riedel, K.H.; Swings, J.; Wingfield, M.J. Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis. 2002, 86, 20–25. [Google Scholar]
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar]
- Masyahit, M.; Sijam, K.; Awang, Y.; Ghazali, M. First report on bacterial soft rot disease on dragon fruit (Hylocereus spp.) caused by Enterobacter cloacae in peninsular Malaysia. Int. J. Agric. Biol. 2009, 11, 659–666. [Google Scholar]
- Koutsoudis, M.D.; Tsaltas, D.; Minogue, T.D.; von Bodman, S.B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 2006, 103, 5983–5988. [Google Scholar]
- Burr, T.J.; Katz, B.H.; Abawi, G.S.; Crosier, D.C. Comparison of tumorigenic strains of Erwinia herbicola isolated from table beet with E.h. gypsophilae. Plant Dis. 1991, 75, 855–858. [Google Scholar]
- Marín-Cevada, V.; Vargas, V.H.; Juárez, M.; López, V.G.; Zagada, G.; Hernández, S.; Cruz, A.; Caballero-Mellado, J.; López-Reyes, L.; Jiménez-Salgado, T.; et al. Presence of Pantoea citrea, causal agent of pink disease, in pineapple fields in Mexico. Plant Pathol. 2006, 55, 294. [Google Scholar]
- Goszczynska, T.; Botha, W.J.; Venter, S.N.; Coutinho, T.A. Isolation and identification of the casual agent of brown stalk rot, a new disease of maize in South Africa. Plant Dis. 2007, 91, 711–718. [Google Scholar]
- Cother, E.J.; Reinke, R.; McKenzie, C.; Lanoiselet, V.M.; Noble, D.H. An unusual stem necrosis of rice caused by Pantoea ananas and the first record of this pathogen on rice in Australia. Austr. Plant Pathol. 2004, 33, 495–503. [Google Scholar]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar]
- Weinthal, D.; Yablonski, S.; Singer, S.; Barash, I.; Manulis-Sasson, S.; Gaba, V. The type III effector PthG of Pantoea agglomerans pv. gypsophilae modifies host plant responses to auxin, cytokinin and light. Eur. J. Plant Pathol. 2010, 128, 289–302. [Google Scholar]
- Maki, D.G.; Rhame, F.S.; Mackel, D.C.; Bennett, J.V. Nationwide epidemic of septicemia caused by contaminated intravenous products. I. Epidemiologic and clinical features. Am. J. Med. 1976, 60, 471–485. [Google Scholar]
- Matsaniotis, N.S.; Syriopoulou, V.P.; Theodoridou, M.C.; Tzanetou, K.G.; Mostrou, G.I. Enterobacter sepsis in infants and children due to contaminated intravenous fluids. Infect. Control 1984, 5, 471–477. [Google Scholar]
- Alvarez, F.E.; Rogge, K.J.; Tarrand, J.; Lichtiger, B. Bacterial contamination of cellular blood components. A retrospective review at a large cancer center. Ann. Clin. Lab. Sci. 1995, 25, 283–290. [Google Scholar]
- Bennett, S.N.; McNeil, M.M.; Bland, L.A.; Arduino, M.J.; Villarino, M.E.; Perrotta, D.M.; Burwen, D.R.; Welbel, S.F.; Pegues, D.A.; Stroud, L.; et al. Postoperative infections traced to contamination of an intravenous anesthetic, propofol. N. Engl. J. Med. 1995, 333, 147–154. [Google Scholar]
- Habsah, H.; Zeehaida, M.; van Rostenberghe, H.; Noraida, R.; Wan Pauzi, W.I.; Fatimah, I.; Rosliza, A.R.; Nik Sharimah, N.Y.; Maimunah, H. An outbreak of Pantoea spp. in a neonatal intensive care unit secondary to contaminated parenteral nutrition. J. Hosp. Infect. 2005, 61, 213–218. [Google Scholar]
- Bicudo, E.L.; Macedo, V.O.; Carrara, M.A.; Castro, F.F.; Rage, R.I. Nosocomial outbreak of Pantoea agglomerans in a pediatric urgent care center. Braz. J. Infect. Dis. 2007, 11, 281–284. [Google Scholar]
- Flatauer, F.E.; Khan, M.A. Septic arthritis caused by Enterobacter agglomerans. Arch. Intern. Med. 1978, 138, 788. [Google Scholar]
- de Champs, C.; Le Seaux, S.; Dubost, J.J.; Boisgard, S.; Sauvezie, B.; Sirot, J. Isolation of Pantoea agglomerans in two cases of septic mono-arthritis after plant thorn and wood sliver injuries. J. Clin. Microbiol. 2000, 38, 460–461. [Google Scholar]
- Kratz, A.; Greenberg, D.; Barki, Y.; Cohen, E.; Lifshitz, M. Pantoea agglomerans as a cause of septic arthritis after palm tree thorn injury; case report and literature review. Arch. Dis. Child. 2003, 88, 542–544. [Google Scholar]
- Schmid, H.; Schubert, S.; Weber, C.; Bogner, J.R. Isolation of a Pantoea dispersa-like strain from a 71-year-old woman with acute myeloid leukemia and multiple myeloma. Infection 2003, 31, 66–67. [Google Scholar]
- de Baere, T.; Verhelst, R.; Labit, C.; Verschraegen, G.; Wauters, G.; Claeys, G.; Vaneechoutte, M. Bacteremic infection with Pantoea ananatis. J. Clin. Microbiol. 2004, 42, 4393–4395. [Google Scholar]
- Brady, C.L.; Cleenwerck, I.; Venter, S.N.; Engelbeen, K.; de Vos, P.; Coutinho, T.A. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2430–2440. [Google Scholar]
- Volksch, B.; Thon, S.; Jacobsen, I.D.; Gube, M. Polyphasic study of plant- and clinic-associated Pantoea agglomerans strains reveals indistinguishable virulence potential. Infect. Genet. Evol. 2009, 9, 1381–1391. [Google Scholar]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar]
- Maeda, Y.; Kiba, A.; Ohnishi, K.; Hikichi, Y. Implications of amino acid substitutions in GyrA at position 83 in terms of oxolinic acid resistance in field isolates of Burkholderia glumae, a causal agent of bacterial seedling rot and grain rot of rice. Appl. Environ. Microbiol. 2004, 70, 5613–5620. [Google Scholar]
- Kang, Y.; Kim, J.; Kim, S.; Kim, H.; Lim, J.Y.; Kim, M.; Kwak, J.; Moon, J.S.; Hwang, I. Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics 2008, 8, 106–121. [Google Scholar]
- Stevens, M.P.; Haque, A.; Atkins, T.; Hill, J.; Wood, M.W.; Easton, A.; Nelson, M.; Underwood-Fowler, C.; Titball, R.W.; Bancroft, G.J.; et al. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 2004, 150, 2669–2676. [Google Scholar]
- Winstanley, C.; Hart, C.A. Presence of type III secretion genes in Burkholderia pseudomallei correlates with Ara(-) phenotypes. J. Clin. Microbiol. 2000, 38, 883–885. [Google Scholar]
- di Cello, F.; Bevivino, A.; Chiarini, L.; Fani, R.; Paffetti, D.; Tabacchioni, S.; Dalmastri, C. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 1997, 63, 4485–4493. [Google Scholar]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar]
- Govan, J.R.; Hughes, J.E.; Vandamme, P. Burkholderia cepacia: Medical, taxonomic and ecological issues. J. Med. Microbiol. 1996, 45, 395–407. [Google Scholar]
- Ballard, R.W.; Palleroni, N.J.; Doudoroff, M.; Stanier, R.Y.; Mandel, M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J. Gen. Microbiol. 1970, 60, 199–214. [Google Scholar]
- Govan, J.R.; Brown, P.H.; Maddison, J.; Doherty, C.J.; Nelson, J.W.; Dodd, M.; Greening, A.P.; Webb, A.K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 1993, 342, 15–19. [Google Scholar]
- Holmes, A.; Nolan, R.; Taylor, R.; Finley, R.; Riley, M.; Jiang, R.Z.; Steinbach, S.; Goldstein, R. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J. Inflect. Dis. 1999, 179, 1197–1205. [Google Scholar]
- LiPuma, J.J. Burkholderia cepacia. Management issues and new insights. Clin. Chest Med. 1998, 19, 473–486. [Google Scholar]
- Graves, M.; Robin, T.; Chipman, A.M.; Wong, J.; Khashe, S.; Janda, J.M. Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clin. Infect. Dis. 1997, 25, 838–842. [Google Scholar]
- Schwab, U.; Leigh, M.; Ribeiro, C.; Yankaskas, J.; Burns, K.; Gilligan, P.; Sokol, P.; Boucher, R. Patterns of epithelial cell invasion by different species of the Burkholderia cepacia complex in well-differentiated human airway epithelia. Infect. Immun. 2002, 70, 4547–4555. [Google Scholar]
- Diggle, S.P.; Lumjiaktase, P.; Dipilato, F.; Winzer, K.; Kunakorn, M.; Barrett, D.A.; Chhabra, S.R.; Camara, M.; Williams, P. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 2006, 13, 701–710. [Google Scholar]
- DeShazer, D.; Waag, D.M.; Fritz, D.L.; Woods, D.E. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 2001, 30, 253–269. [Google Scholar]
- Fluit, A.C.; Schmitz, F.J.; Verhoef, J. Frequency of isolation of pathogens from bloodstream, nosocomial pneumonia, skin and soft tissue, and urinary tract infections occurring in European patients. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 188–191. [Google Scholar]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar]
- Jha, A.K.; Bais, H.P.; Vivanco, J.M. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect. Immun. 2005, 73, 464–475. [Google Scholar]
- Coburn, B.; Li, Y.; Owen, D.; Vallance, B.A.; Finlay, B.B. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 2005, 73, 3219–3227. [Google Scholar]
- Schmitt, C.K.; Ikeda, J.S.; Darnell, S.C.; Watson, P.R.; Bispham, J.; Wallis, T.S.; Weinstein, D.L.; Metcalf, E.S.; O'Brien, A.D. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Inflect. Immun. 2001, 69, 5619–5625. [Google Scholar]
- Stevanin, T.M.; Poole, R.K.; Demoncheaux, E.A.; Read, R.C. Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 2002, 70, 4399–4405. [Google Scholar]
- Barak, J.D.; Kramer, L.C.; Hao, L.Y. Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 2011, 77, 498–504. [Google Scholar]
- Barak, J.D.; Gorski, L.; Naraghi-Arani, P.; Charkowski, A.O. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005, 71, 5685–5691. [Google Scholar]
- Schikora, A.; Carreri, A.; Charpentier, E.; Hirt, H. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS ONE 2008, 3, e2279. [Google Scholar]
- Cooley, M.B.; Miller, W.G.; Mandrell, R.E. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 2003, 69, 4915–4926. [Google Scholar]
- Dreyfus, G.; Williams, A.W.; Kawagishi, I.; Macnab, R.M. Genetic and biochemical analysis of Salmonella typhimurium FliI, a flagellar protein related to the catalytic subunit of the FoF1 ATPase and to virulence proteins of mammalian and plant pathogens. J. Bacteriol. 1993, 175, 3131–3138. [Google Scholar]
- Kurz, C.L.; Chauvet, S.; Andres, E.; Aurouze, M.; Vallet, I.; Michel, G.P.; Uh, M.; Celli, J.; Filloux, A.; de Bentzmann, S.; et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003, 22, 1451–1460. [Google Scholar]
- Lyerly, D.; Gray, L.; Kreger, A. Characterization of rabbit corneal damage produced by Serratia keratitis and by a Serratia protease. Infect. Immun. 1981, 33, 927–932. [Google Scholar]
- Rascoe, J.; Berg, M.; Melcher, U.; Mitchell, F.L.; Bruton, B.D.; Pair, S.D.; Fletcher, J. Identification, phylogenetic analysis, and biological characterization of Serratia marcescens strains causing cucurbit yellow vine disease. Phytopathology 2003, 93, 1233–1239. [Google Scholar]
- Bruton, D.B.; Mitchell, F.; Fletcher, J.; Pair, S.D.; Wayadande, A.; Melcher, U.; Brady, J.; Bextine, B.; Popham, T.W. Serratia marcescens, a phloem-colonizing, squash bug-transmitted bacterium: Casual agent of cucurbit yellow vine disease. Plant Dis. 1998, 87, 512–520. [Google Scholar]
- Bruton, D.B.; Brady, J.; Mitchell, F.; Bextine, B.; Wayadande, A.; Pair, S.D.; Fletcher, J.; Melcher, U. Yellow vine of cucurbits: Pathogenicity of Serratia marcescens and transmission by Anasa tristis. Phytopathology 2001, 91, S11. [Google Scholar]
- Labbate, M.; Queek, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698. [Google Scholar]
- Labbate, M.; Zhu, H.; Thung, L.; Bandara, R.; Larsen, M.R.; Willcox, M.D.P.; Givskov, M.; Rice, S.A.; Kjelleberg, S. Quorum sensing regulation of adhesion in Serratia marcescens MG1 is surface dependent. J. Bacteriol. 2007, 189, 2702–2711. [Google Scholar]
- Shanks, R.M.Q.; Stella, N.A.; Kalivoda, E.J.; Doe, M.R.; O'Dee, D.A.; Lathrop, K.L.; Guo, F.L.; Nau, G.J. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 2007, 189, 7262–7272. [Google Scholar]
- Wang, S.A.; Tokars, J.I.; Bianchine, P.J.; Carson, L.A.; Arduino, M.J.; Smith, A.L.; Hansen, N.C.; Fitzgerald, E.A.; Epstein, J.S.; Jarvis, W.R. Enterobacter cloacae bloodstream infections traced to contaminated human albumin. Clin. Infect. Dis. 2000, 30, 35–40. [Google Scholar]
- Paton, A.W.; Paton, J.C. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J. Clin. Microbiol. 1996, 34, 463–465. [Google Scholar]
- de Kort, G.; Bolton, A.; Martin, G.; Stephen, J.; van de Klundert, J.A. Invasion of rabbit ileal tissue by Enterobacter cloacae varies with the concentration of OmpX in the outer membrane. Inflect. Immun. 1994, 62, 4722–4726. [Google Scholar]
- Nishijima, K.A.; Wall, M.M.; Siderhurst, M.S. Demonstrating pathogenicity of Enterobacter cloacae on macadamia and identifying associated volatiles of gray kernel of macadamia in Hawaii. Plant Dis. 2007, 91, 1221–1228. [Google Scholar]
- Takahashi, Y.; Takahashi, K.; Watanabe, K.; Kawano, T. Bacterial black spot caused by Burkholderia andropogonis on Odontoglossum and intergeneric hybrid orchids. J. Gen. Plant. Pathol. 2004, 70, 284–287. [Google Scholar]
- Murray, B.E. Diversity among the multidrug-resistant enterococci. Emerg. Infect. Dis. 1998, 4, 46–65. [Google Scholar]
- Ike, Y.; Hashimoto, H.; Clewell, D.B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 1984, 45, 528–530. [Google Scholar]
- Mohamed, J.A.; Huang, D.B. Biofilm formation by enterococci. J. Med. Microbiol. 2007, 56, 1581–1588. [Google Scholar]
- Shankar, N.; Lockatell, C.V.; Baghdayan, A.S.; Drachenberg, C.; Gilmore, M.S.; Johnson, D.E. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 2001, 69, 4366–4372. [Google Scholar]
- Park, S.Y.; Kim, K.M.; Lee, J.H.; Seo, S.J.; Lee, I.H. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect. Immun. 2007, 75, 1861–1869. [Google Scholar]
- Pearce-Duvet, J.M. The origin of human pathogens: Evaluating the role of agriculture and domestic animals in the evolution of human disease. Biol. Rev. Camb. Philos. Soc. 2006, 81, 369–382. [Google Scholar]
- Muller, H.E. Occurrence and pathogenic role of Morganella-Proteus-Providencia group bacteria in human feces. J. Clin. Microbiol. 1986, 23, 404–405. [Google Scholar]
- Beatson, S. Pharaoh's ants as pathogen vectors in hospitals. Lancet 1972, 299, 425–427. [Google Scholar]
- Fotedar, R.; Banerjee, U.; Singh, S.; Shriniwas; Verma, A.K. The housefly (Musca domestica) as a carrier of pathogenic microorganisms in a hospital environment. J. Hosp. Infect. 1992, 20, 209–215. [Google Scholar]
- Stavrinides, J. Origin and Evolution of Phytopathogenic Bacteria. In Plant Pathogenic Bacteria: Genomics and Molecular Biology; Jackson, R.W., Ed.; Academic Press: Norfolk, UK, 2009. [Google Scholar]
- Palleroni, N. The Genus Pseudomonas; The Williams & Wilkins Co.: Baltimore, MD, USA, 1984; Volume 1. [Google Scholar]
- van Baarlen, P.; van Belkum, A.; Summerbell, R.C.; Crous, P.W.; Thomma, B.P.H.J. Molecular mechanisms of pathogenicity: How do pathogenic microorganisms develop cross-kingdom host jumps? FEMS Microbiol. Rev. 2007, 31, 239–277. [Google Scholar]
- Medzhitov, R.; Janeway, C.A. Innate immunity: Impact on the adaptive immune response. Curr. Opin. Immunol. 1997, 9, 4–9. [Google Scholar]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar]
- Andreote, F.D.; Rossetto, P.B.; Souza, L.C.; Marcon, J.; Maccheroni, W., Jr.; Azevedo, J.L.; Araujo, W.L. Endophytic population of Pantoea agglomerans in citrus plants and development of a cloning vector for endophytes. J. Basic Microbiol. 2008, 48, 338–346. [Google Scholar]
- Thaller, M.C.; Berlutti, F.; Schippa, S.; Lombardi, G.; Rossolini, G.M. Characterization and sequence of PhoC, the principal phosphate-irrepressible acid phosphatase of Morganella morganii. Microbiology 1994, 140, 1341–1350. [Google Scholar]
- Iniguez, A.L.; Dong, Y.; Triplett, E.W. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol. Plant Microbe Interact. 2004, 17, 1078–1085. [Google Scholar]
- Coombs, J.T.; Franco, C.M. Visualization of an endophytic Streptomyces species in wheat seed. Appl. Environ. Microbiol. 2003, 69, 4260–4262. [Google Scholar]
- Loria, R.; Bignell, D.; Moll, S.; Huguet-Tapia, J.; Joshi, M.; Johnson, E.; Seipke, R.; Gibson, D. Thaxtomin biosynthesis: The path to plant pathogenicity in the genus Streptomyces. Antonie Van Leeuwenhoek 2008, 94, 3–10. [Google Scholar]
- Rose, C.E., III; Brown, J.M.; Fisher, J.F. Brain abscess caused by Streptomyces infection following penetration trauma: Case report and results of susceptibility analysis of 92 isolates of Streptomyces species submitted to the CDC from 2000 to 2004. J. Clin. Microbiol. 2008, 46, 821–823. [Google Scholar]
- Lund, B.; Wyatt, G. The effect of oxygen and carbon dioxide concentrations on bacterial soft rot of potatoes. I. King Edward potatoes inoculated with Erwinia carotovora var. atroseptica. Potato Res. 1972, 15, 174–179. [Google Scholar]
- van Peer, R.; Punte, H.L.; de Weger, L.A.; Schippers, B. Characterization of root surface and endorhizosphere Pseudomonads in relation to their colonization of roots. Appl. Environ. Microbiol. 1990, 56, 2462–2470. [Google Scholar]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar]
- Parke, J.L.; Gurian-Sherman, D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 2001, 39, 225–258. [Google Scholar]
- Wright, S.A.I.; Zumoff, C.H.; Schneider, L.; Beer, S.V. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 2001, 67, 284–292. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kirzinger, M.W.B.; Nadarasah, G.; Stavrinides, J. Insights into Cross-Kingdom Plant Pathogenic Bacteria. Genes 2011, 2, 980-997. https://doi.org/10.3390/genes2040980
Kirzinger MWB, Nadarasah G, Stavrinides J. Insights into Cross-Kingdom Plant Pathogenic Bacteria. Genes. 2011; 2(4):980-997. https://doi.org/10.3390/genes2040980
Chicago/Turabian StyleKirzinger, Morgan W.B., Geetanchaly Nadarasah, and John Stavrinides. 2011. "Insights into Cross-Kingdom Plant Pathogenic Bacteria" Genes 2, no. 4: 980-997. https://doi.org/10.3390/genes2040980
APA StyleKirzinger, M. W. B., Nadarasah, G., & Stavrinides, J. (2011). Insights into Cross-Kingdom Plant Pathogenic Bacteria. Genes, 2(4), 980-997. https://doi.org/10.3390/genes2040980




