The Genetic Structure of Cape Verdean Population Revealed by Y-Chromosome STRs
Abstract
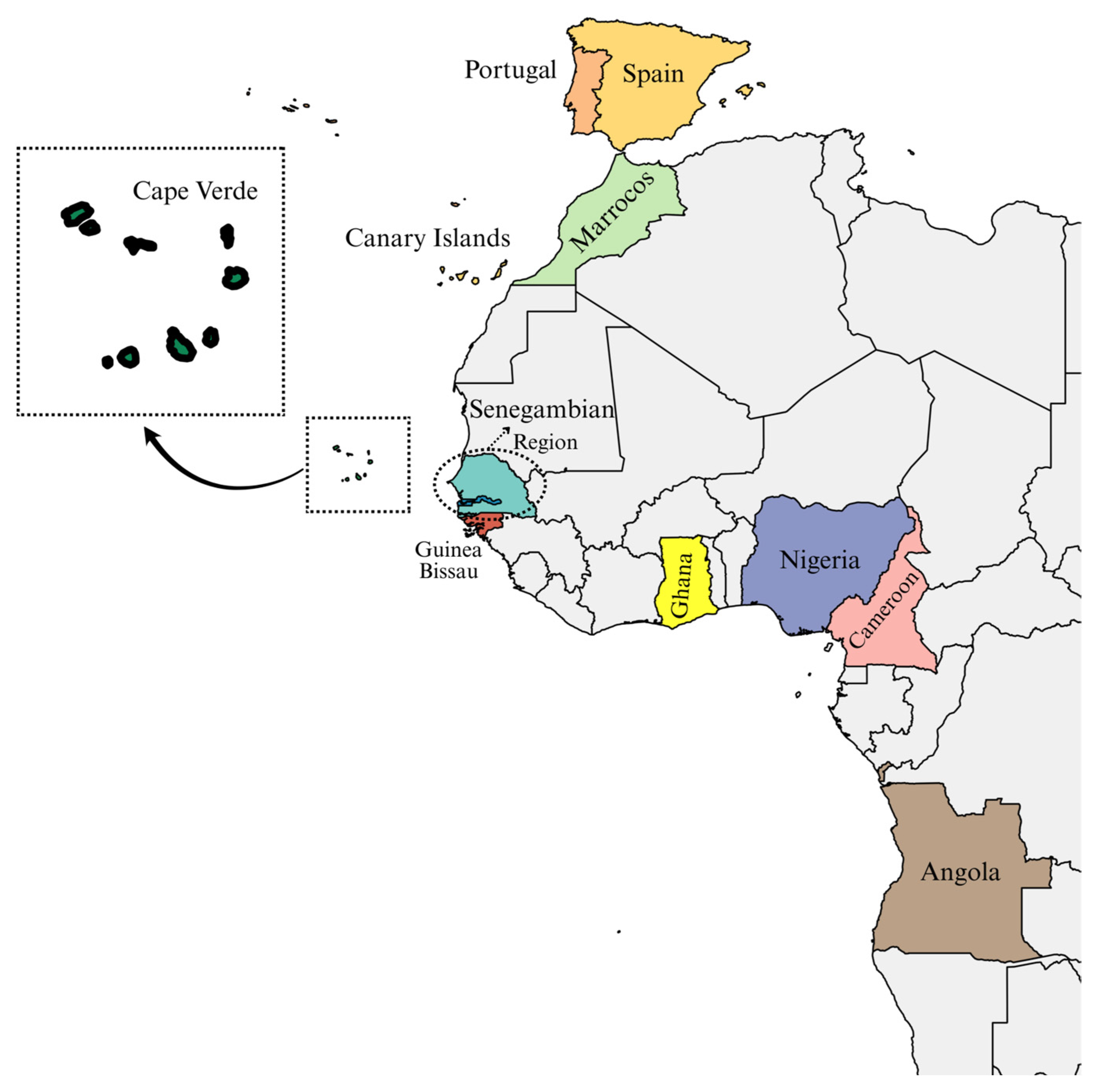
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Y-STR Fragment Analysis
2.3. Statistical Analyses
3. Results
3.1. Analysis of Haplotype and Allele Diversity (and Forensic Parameters)
3.2. Haplogroup Distribution
3.3. Genetic Distance Analysis Based on RST Distances
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CE | Capillary Electrophoresis |
| DC | Discrimination Capacity |
| GD | Gene Diversity |
| HD | Haplotype Diversity |
| HMP | Haplotype Match Probability |
| INMLCF, I.P. | Instituto Nacional de Medicina Legal e Ciências Forenses, Instituto Público |
| MDS | Multidimensional Scaling |
| NGS | Next-Generation Sequencing |
| OL | Off-Ladder |
| PCR | Polymerase Chain Reaction |
| PIC | Polymorphism Information Content |
| PVE | Proportion of Variance Explained |
| QS | Quality Sensor |
| RM Y-STRs | Rapidly Mutating Y-Chromosomal Short Tandem Repeats |
| STR | Short Tandem Repeat |
| YHRD | Y Chromosome Haplotype Reference Database |
| Y-STR | Y-Chromosomal Short Tandem Repeat |
References
- Gonçalves, R.; Fernandes, A.T.; Brehm, A. Cabo Verde islands: Different maternal and paternal heritage testifies the nature of its first settlers. Int. Congr. Ser. 2004, 1261, 372–373. [Google Scholar] [CrossRef]
- Fernandes, A.T.; Velosa, R.; Jesus, J.; Carracedo, A.; Brehm, A. Genetic differentiation of the Cabo Verde archipelago population analysed by STR polymorphisms. Ann. Hum. Genet. 2003, 67, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Laurent, R.; Szpiech, Z.A.; da Costa, S.S.; Thouzeau, V.; Fortes-Lima, C.A.; Dessarps-Freichey, F.; Lémée, L.; Utgé, J.; Rosenberg, N.A.; Baptista, M.; et al. A genetic and linguistic analysis of the admixture histories of the islands of Cabo Verde. eLife 2023, 12, e79827. [Google Scholar] [CrossRef]
- Parra, E.J.; Ribeiro, J.C.; Caeiro, J.L.; Riveiro, A. Genetic structure of the population of Cabo Verde (west Africa): Evidence of substantial European admixture. Am. J. Phys. Anthr. 1995, 97, 381–389. [Google Scholar] [CrossRef]
- Brehm, A.; Pereira, L.; Bandelt, H.J.; Prata, M.J.; Amorim, A. Mitochondrial portrait of the Cabo Verde archipelago: The Senegambian outpost of Atlantic slave trade. Ann. Hum. Genet. 2002, 66, 49–60. [Google Scholar] [CrossRef]
- Beleza, S.; Campos, J.; Lopes, J.; Araújo, I.I.; Hoppfer Almada, A.; Correia e Silva, A.; Parra, E.J.; Rocha, J. The admixture structure and genetic variation of the archipelago of Cape Verde and its implications for admixture mapping studies. PLoS ONE 2012, 7, e51103. [Google Scholar] [CrossRef]
- Gonçalves, R.; Rosa, A.; Freitas, A.; Fernandes, A.; Kivisild, T.; Villems, R.; Brehm, A. Y-chromosome lineages in Cabo Verde Islands witness the diverse geographic origin of its first male settlers. Hum. Genet. 2003, 113, 467–472. [Google Scholar] [CrossRef]
- Resende, A.; Amorim, A.; da Silva, C.V.; Ribeiro, T.; Porto, M.J.; Costa Santos, J.; Afonso Costa, H. Study of genetic markers of CODIS and ESS systems in a population of individuals from Cabo Verde living in Lisboa. Int. J. Leg. Med. 2017, 131, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Amorim, A.; Marques-Santos, R.; Vieira-Silva, C.; Afonso-Costa, H.; Espinheira, R.; Ferreira-Gomes, P.; Costa-Santos, J. Genetic portrait of a native population of Cabo Verde living in Lisboa. Forensic Sci. Int. Genet. 2012, 6, e166–e169. [Google Scholar] [CrossRef]
- Gabinete de Estratégia e Estudos. População estrangeira com estatuto legal de residente em Portugal—Cabo Verde. 2023. Available online: https://www.gee.gov.pt/pt/lista-publicacoes/estatisticas-de-imigrantes-em-portugal-por-nacionalidade/paises/Cabo%20Verde/3947-populacao-estrangeira-com-estatuto-legal-de-residente-em-portugal-cabo-verde/file (accessed on 5 May 2025).
- AIMA I.P.—DPEE—Direção de Planeamento, Estudos e Estatistica. Relatório de Migrações e Asilo 2023; AIMA: London, UK, 2024.
- Kayser, M. Forensic use of Y-chromosome DNA: A general overview. Hum. Genet. 2017, 136, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, K.N.; Goedbloed, M.; Fang, R.; Schaap, O.; Lao, O.; Wollstein, A.; Choi, Y.; van Duijn, K.; Vermeulen, M.; Brauer, S.; et al. Mutability of Y-chromosomal microsatellites: Rates, characteristics, molecular bases, and forensic implications. Am. J. Hum. Genet. 2010, 87, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, K.N.; Keerl, V.; Wollstein, A.; Choi, Y.; Zuniga, S.B.; Ralf, A.; Vermeulen, M.; de Knijff, P.; Kayser, M. A new future of forensic Y-chromosome analysis: Rapidly mutating Y-STRs for differentiating male relatives and paternal lineages. Forensic Sci. Int. Genet. 2012, 6, 208–218. [Google Scholar] [CrossRef]
- Y-Chromosome Haplotype Reference Database (YHRD). Available online: https://yhrd.org (accessed on 5 May 2025).
- Roewer, L. The Y-Short Tandem Repeat Haplotype Reference Database (YHRD) and Male Population Stratification in Europe—Impact on Forensic Genetics. Forensic Sci. Rev. 2003, 15, 165–172. [Google Scholar] [PubMed]
- QIAGEN. Investigator® Argus Y-28 QS Handbook; QIAGEN: Hilden, Germany, 2022; Available online: https://www.qiagen.com/in/resources/download.aspx?id=cf7ae42a-72bc-4d3d-812d-684d0625a179&lang=en (accessed on 5 May 2025).
- Steffen, C.R.; Huszar, T.I.; Borsuk, L.A.; Vallone, P.M.; Gettings, K.B. A multi-dimensional evaluation of the ‘NIST 1032′ sample set across four forensic Y-STR multiplexes. Forensic Sci. Int. Genet. 2022, 57, 102655. [Google Scholar] [CrossRef]
- Haarkötter, C.; Isabel Medina-Lozano, M.; Vinueza-Espinosa, D.C.; Saiz, M.; Gálvez, X.; Carlos Álvarez, J.; Antonio Lorente, J. Evaluating the efficacy of three Y-STRs commercial kits in degraded skeletal remains. Sci. Justice 2024, 64, 543–548. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F. Genetic drift and estimation of effective population size. Genetics 1981, 98, 625–640. [Google Scholar] [CrossRef]
- Gouy, A.; Zieger, M. STRAF—A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 2017, 30, 148–151. [Google Scholar] [CrossRef]
- Cetkovic Gentula, M.; Nevski, A. NEVGEN Y-DNA Haplogroup Predictor. Available online: https://www.nevgen.org (accessed on 16 June 2025).
- Carvalho, M.; Brito, P.; Bento, A.M.; Gomes, V.; Antunes, H.; Costa, H.A.; Lopes, V.; Serra, A.; Balsa, F.; Andrade, L.; et al. Paternal and maternal lineages in Guinea-Bissau population. Forensic Sci. Int. Genet. 2011, 5, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Kofi, A.E.; Hakim, H.M.; Khan, H.O.; Ismail, S.A.; Ghansah, A.; David, A.A.; Mat, N.F.C.; Chambers, G.K.; Edinur, H.A. Population data of 23 Y chromosome STR loci for the five major human subpopulations of Ghana. Int. J. Leg. Med. 2020, 134, 1313–1315. [Google Scholar] [CrossRef]
- Purps, J.; Siegert, S.; Willuweit, S.; Nagy, M.; Alves, C.; Salazar, R.; Angustia, S.M.; Santos, L.H.; Anslinger, K.; Bayer, B.; et al. A global analysis of Y-chromosomal haplotype diversity for 23 STR loci. Forensic Sci. Int. Genet. 2014, 12, 12–23. [Google Scholar] [CrossRef]
- Della Rocca, C.; Cannone, F.; D’Atanasio, E.; Bonito, M.; Anagnostou, P.; Russo, G.; Barni, F.; Alladio, E.; Destro-Bisol, G.; Trombetta, B.; et al. Ethnic fragmentation and degree of urbanization strongly affect the discrimination power of Y-STR haplotypes in central Sahel. Forensic Sci. Int. Genet. 2020, 49, 102374. [Google Scholar] [CrossRef]
- Palha, T.; Gusmão, L.; Ribeiro-Rodrigues, E.; Guerreiro, J.F.; Ribeiro-Dos-Santos, A.; Santos, S. Disclosing the genetic structure of Brazil through analysis of male lineages with highly discriminating haplotypes. PLoS ONE 2012, 7, e40007. [Google Scholar] [CrossRef]
- Pontes, M.L.; Cainé, L.; Abrantes, D.; Lima, G.; Pinheiro, M.F. Allele frequencies and population data for 17 Y-STR loci (AmpFℓSTR® Y-filer™) in a Northern Portuguese population sample. Forensic Sci. Int. 2007, 170, 62–67. [Google Scholar] [CrossRef]
- Aboukhalid, R.; Bouabdellah, M.; Abbassi, M.; Bentayebi, K.; Elmzibri, M.; Squalli, D.; Amzazi, S. Haplotype frequencies for 17 Y-STR loci (AmpFlSTRY-filer) in a Moroccan population sample. Forensic Sci. Int. Genet. 2010, 4, e73–e74. [Google Scholar] [CrossRef]
- Gusmão, L.; Butler, J.M.; Carracedo, A.; Gill, P.; Kayser, M.; Mayr, W.R.; Morling, N.; Prinz, M.; Roewer, L.; Tyler-Smith, C.; et al. DNA Commission of the International Society of Forensic Genetics (ISFG): An update of the recommendations on the use of Y-STRs in forensic analysis. Int. J. Leg. Med. 2006, 120, 191–200. [Google Scholar] [CrossRef]
- Rosa, A.; Ornelas, C.; Brehm, A.; Villems, R. Population data on 11 Y-chromosome STRs from Guiné-Bissau. Forensic Sci. Int. 2006, 157, 210–217. [Google Scholar] [CrossRef]
- Costa, R.; Fadoni, J.; Amorim, A.; Cainé, L. Y-STR Databases-Application in Sexual Crimes. Genes 2025, 16, 484. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef]

| Haplogroup (H) | Sub-Haplogroup (SH) | Frequency SH (%) | Frequency H (%) | Ancestry |
|---|---|---|---|---|
| E | E1a | 3% | 45% | African |
| E1b1a | 28% | |||
| E1b1b | 14% | |||
| E3a | 1% | |||
| L | L1a | 1% | 2% | Asian |
| L1b | 1% | |||
| I | I2a2a | 1% | 4% | European |
| I2a2b | 3% | |||
| R | R1b | 44% | 44% | |
| G | G2a2b1 | 1% | 1% | Middle Eastern |
| J | J1a2a1a2 | 2% | 4% | |
| J2b2a | 3% | |||
| T | - | 1% | 1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, R.; Fadoni, J.; Amorim, A.; Cainé, L. The Genetic Structure of Cape Verdean Population Revealed by Y-Chromosome STRs. Genes 2025, 16, 999. https://doi.org/10.3390/genes16090999
Costa R, Fadoni J, Amorim A, Cainé L. The Genetic Structure of Cape Verdean Population Revealed by Y-Chromosome STRs. Genes. 2025; 16(9):999. https://doi.org/10.3390/genes16090999
Chicago/Turabian StyleCosta, Rita, Jennifer Fadoni, António Amorim, and Laura Cainé. 2025. "The Genetic Structure of Cape Verdean Population Revealed by Y-Chromosome STRs" Genes 16, no. 9: 999. https://doi.org/10.3390/genes16090999
APA StyleCosta, R., Fadoni, J., Amorim, A., & Cainé, L. (2025). The Genetic Structure of Cape Verdean Population Revealed by Y-Chromosome STRs. Genes, 16(9), 999. https://doi.org/10.3390/genes16090999









