MicroRNA Regulation in the Freeze-Tolerant Heart of Dryophytes versicolor
Abstract
1. Introduction
2. Results
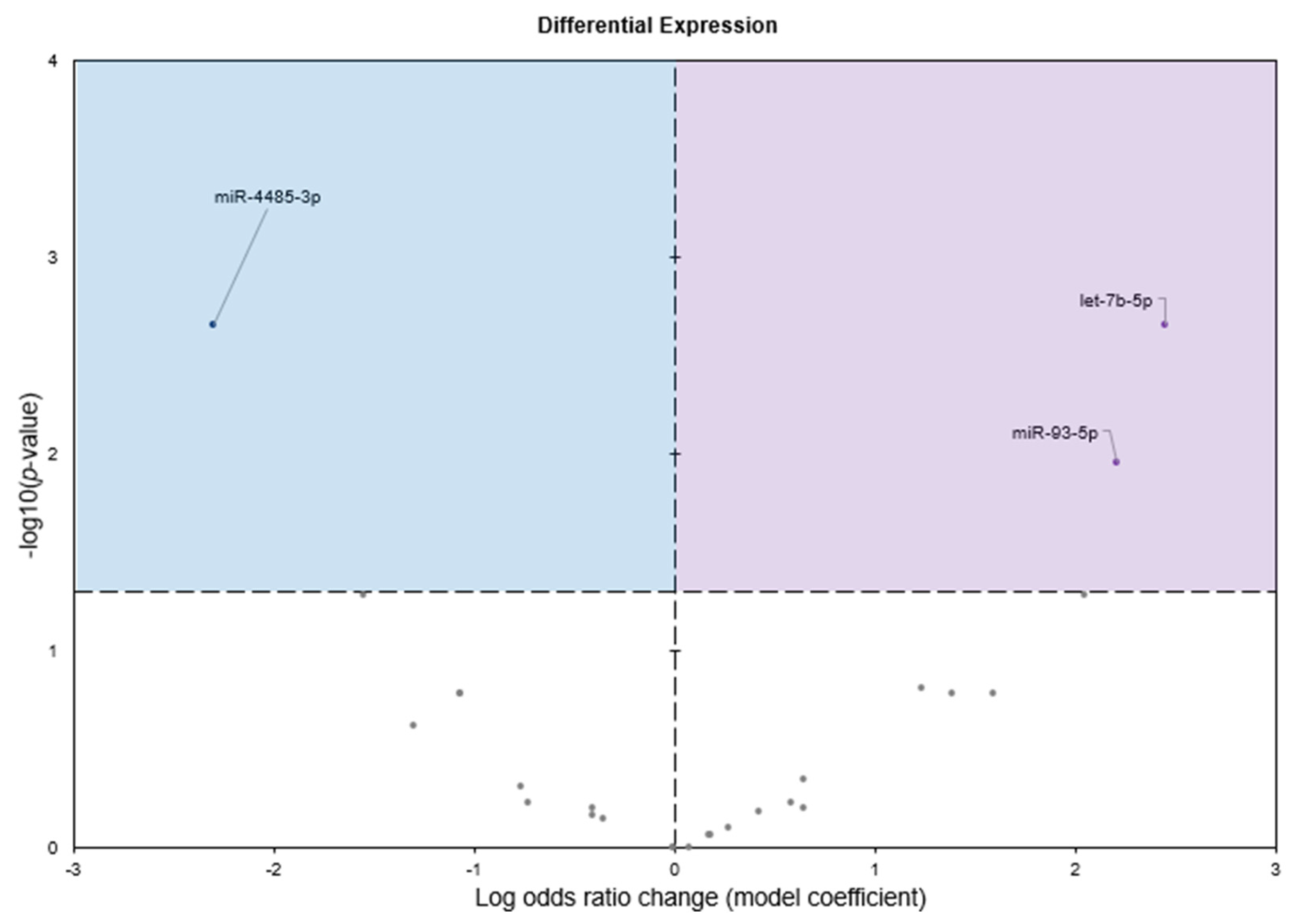
2.1. Small RNA Sequencing Summary
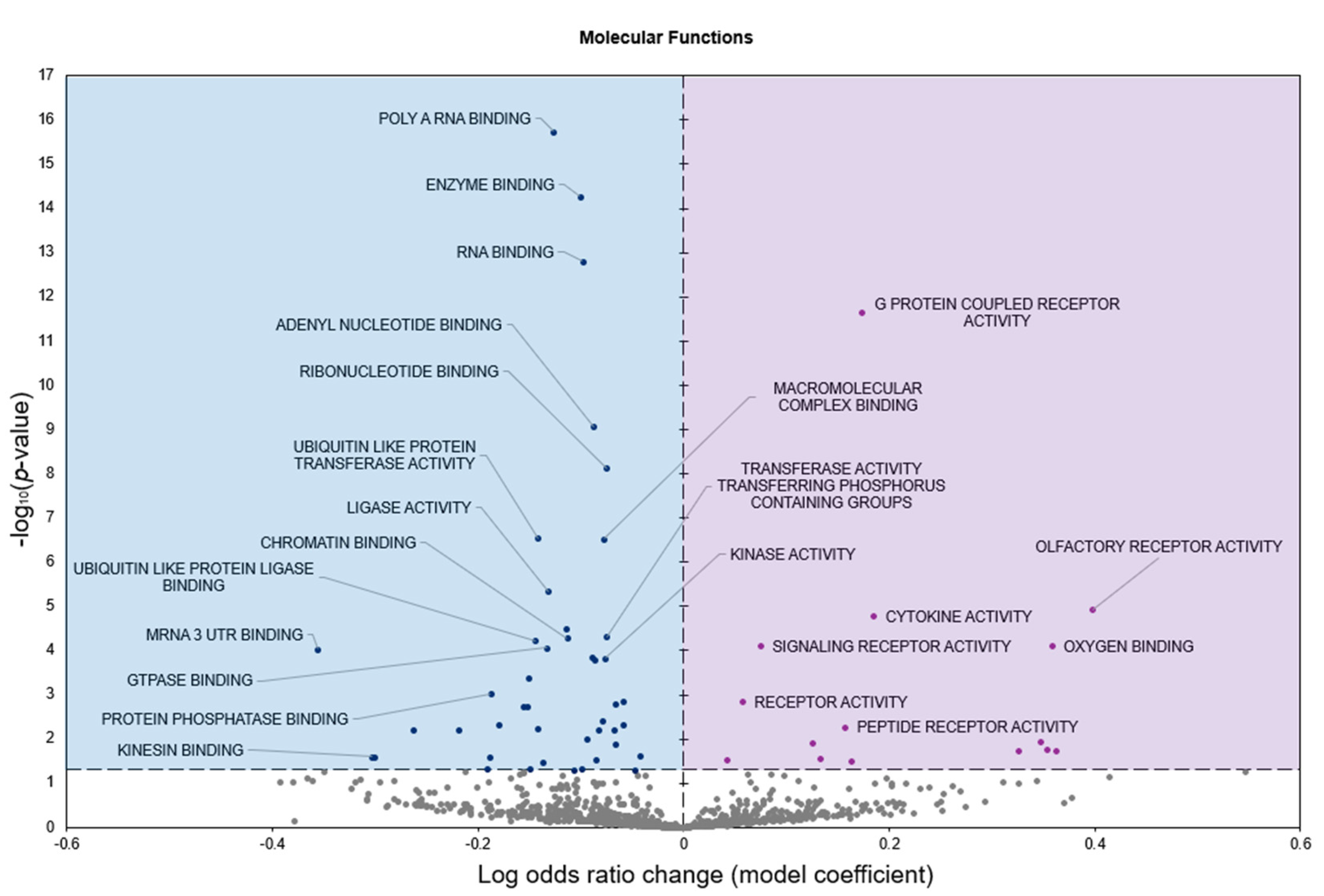
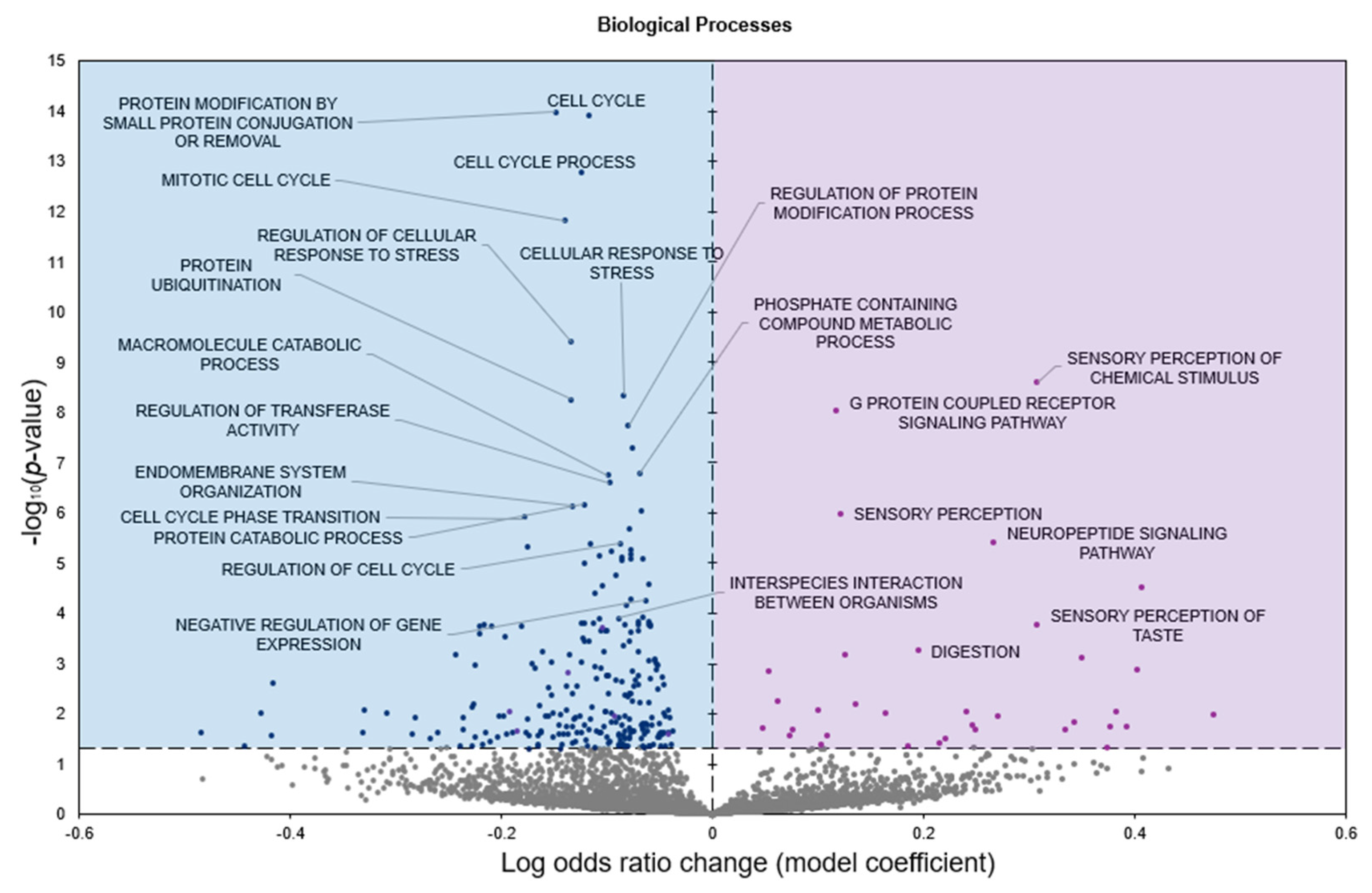
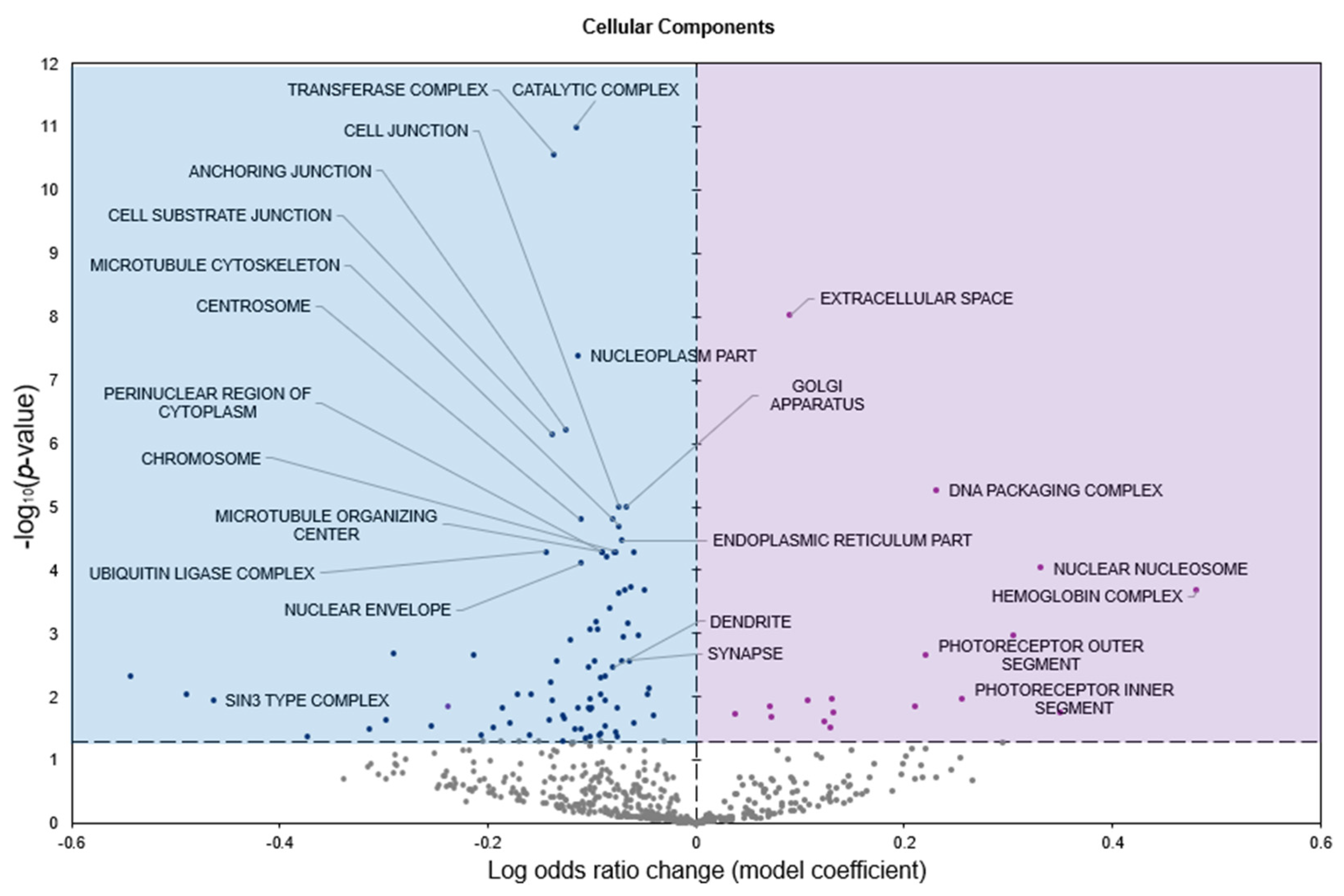
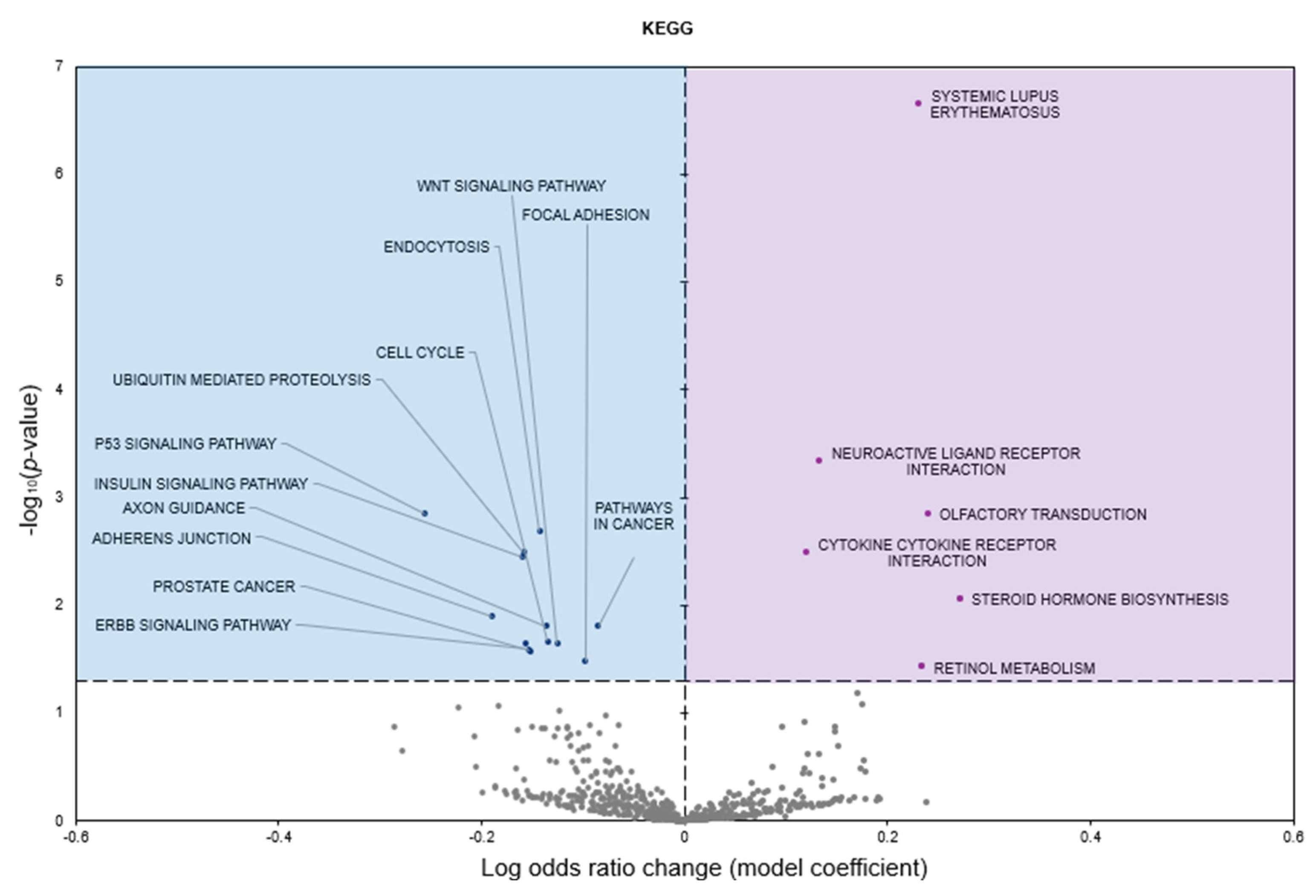
2.2. Gene Set Analyses
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Small RNA Sequencing
4.3. Read Processing
4.4. Differential Expression Analysis
4.5. Gene Set Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | MicroRNA |
| MRD | Metabolic Rate Depression |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Rehman, S.; Storey, K.B. Dynamics of Epigenetic Regulation in Dryophytes Versicolor Skeletal Muscle: Lysine Methylation and Acetylation Involvement in Metabolic Rate Depression. J. Therm. Biol. 2024, 122, 103865. [Google Scholar] [CrossRef] [PubMed]
- Ingelson-Filpula, W.A.; Storey, K.B. MicroRNA Biogenesis Proteins Follow Tissue-Dependent Expression during Freezing in Dryophytes Versicolor. J. Comp. Physiol. B 2022, 192, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Molecular Physiology of Freeze Tolerance in Vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Freeze Tolerance: Constraining Forces, Adaptive Mechanisms. Can. J. Zool. 1988, 66, 1122–1127. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression in Animals: Transcriptional and Translational Controls. Biol. Rev. Soc. 2004, 79, 207–233. [Google Scholar] [CrossRef]
- Breedon, S.A.; Varma, A.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Mejías, C.; Nespolo, R.F.; Storey, K.B. Torpor-Responsive microRNAs in the Heart of the Monito Del Monte, Dromiciops gliroides. BioFactors 2023, 49, 1061–1073. [Google Scholar] [CrossRef]
- Ingelson-Filpula, W.A.; Hadj-Moussa, H.; Storey, K.B. MicroRNA Transcriptomics in Liver of the Freeze-Tolerant Gray Tree Frog (Dryophytes versicolor) Indicates Suppression of Energy-Expensive Pathways. Cell Biochem. Funct. 2023, 41, 309–320. [Google Scholar] [CrossRef]
- Ingelson-Filpula, W.A.; Breedon, S.A.; Storey, K.B. MicroRNA, Myostatin, and Metabolic Rate Depression: Skeletal Muscle Atrophy Resistance in Hibernating Myotis Lucifugus. Cells 2024, 13, 2074. [Google Scholar] [CrossRef]
- Attaie, Y.; Storey, K.B. MicroRNA-Mediated Regulation in Anoxic Lithobates sylvaticus Liver. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 55, 101487. [Google Scholar] [CrossRef]
- Varma, A.; Breedon, S.A.; Storey, K.B. Sub-Zero microRNA Expression in the Liver of the Frozen Hatchling Painted Turtle, Chrysemys picta marginata. Sci. Total Environ. 2023, 857, 159304. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, W.; Yang, Y.; Zhang, Z. miRNA-93-5p Promotes Gemcitabine Resistance in Pancreatic Cancer Cells by Targeting the PTEN-Mediated PI3K/Akt Signaling Pathway. Ann. Clin. Lab. Sci. 2021, 51, 310–320. [Google Scholar] [PubMed]
- Wang, H.; Su, X.; Zhang, Q.-Q.; Zhang, Y.-Y.; Chu, Z.-Y.; Zhang, J.-L.; Ren, Q. MicroRNA-93-5p Participates in Type 2 Diabetic Retinopathy through Targeting Sirt1. Int. Ophthalmol. 2021, 41, 3837–3848. [Google Scholar] [CrossRef] [PubMed]
- Len, J.S.; Koh, W.S.D.; Tan, S.-X. The Roles of Reactive Oxygen Species and Antioxidants in Cryopreservation. Biosci. Rep. 2019, 39, BSR20191601. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pan, W.; Shen, Y.; Chen, Z.; Zhang, L.; Zhang, Y.; Luo, Q.; Ying, X. IGF1/IGF1R and microRNA Let-7e down-Regulate Each Other and Modulate Proliferation and Migration of Colorectal Cancer Cells. Cell Cycle 2018, 17, 1212–1219. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, W.; Liu, Y.; Guo, A.; Yang, D. Let-7b Acts as a Tumor Suppressor in Osteosarcoma via Targeting IGF1R. Oncol. Lett. 2019, 17, 1646–1654. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of Let-7 and Its Target Oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR Recruits Let-7/RISC to Repress c-Myc Expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, S.; Yang, W.; Du, F. Let-7b-5p Suppresses the Proliferation and Migration of Pulmonary Artery Smooth Muscle Cells via down-Regulating IGF1. Clinics 2022, 77, 100051. [Google Scholar] [CrossRef]
- Xu, H.; Liu, C.; Zhang, Y.; Guo, X.; Liu, Z.; Luo, Z.; Chang, Y.; Liu, S.; Sun, Z.; Wang, X. Let-7b-5p Regulates Proliferation and Apoptosis in Multiple Myeloma by Targeting IGF1R. Acta Biochim. Biophys. Sin. 2014, 46, 965–972. [Google Scholar] [CrossRef]
- Fu, X.; Mao, X.; Wang, Y.; Ding, X.; Li, Y. Let-7c-5p Inhibits Cell Proliferation and Induces Cell Apoptosis by Targeting ERCC6 in Breast Cancer. Oncol. Rep. 2017, 38, 1851–1856. [Google Scholar] [CrossRef]
- Sripada, L.; Singh, K.; Lipatova, A.V.; Singh, A.; Prajapati, P.; Tomar, D.; Bhatelia, K.; Roy, M.; Singh, R.; Godbole, M.M.; et al. Hsa-miR-4485 Regulates Mitochondrial Functions and Inhibits the Tumorigenicity of Breast Cancer Cells. J. Mol. Med. 2017, 95, 641–651. [Google Scholar] [CrossRef]
- Rehman, S.; Parent, M.; Storey, K.B. Histone Arginine Methylation as a Regulator of Gene Expression in the Dehydrating African Clawed Frog (Xenopus laevis). Genes 2024, 15, 1156. [Google Scholar] [CrossRef]
- Bloskie, T.; Storey, K.B. Histone H3 and H4 Modifications Point to Transcriptional Suppression as a Component of Winter Freeze Tolerance in the Gall Fly Eurosta solidaginis. Int. J. Mol. Sci. 2023, 24, 10153. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi-Takihara, K.; Kishimoto, T. Cytokines and Their Receptors in Cardiovascular Diseases—Role of Gp130 Signalling Pathway in Cardiac Myocyte Growth and Maintenance. Int. J. Exp. Pathol. 2000, 81, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Epelman, S.; Liu, P.P.; Mann, D.L. Role of Innate and Adaptive Immunity in Cardiac Injury and Repair. Nat. Rev. Immunol. 2015, 15, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Molecular Biology of Freezing Tolerance. Compr. Physiol. 2013, 3, 1283–1308. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Mitochondria, Metabolic Control and microRNA: Advances in Understanding Amphibian Freeze Tolerance. BioFactors 2020, 46, 220–228. [Google Scholar] [CrossRef]
- Tamura, M.; Gu, J.; Matsumoto, K.; Aota, S.; Parsons, R.; Yamada, K.M. Inhibition of Cell Migration, Spreading, and Focal Adhesions by Tumor Suppressor PTEN. Science 1998, 280, 1614–1617. [Google Scholar] [CrossRef]
- Dieni, C.A.; Bouffard, M.C.; Storey, K.B. Glycogen Synthase Kinase-3: Cryoprotection and Glycogen Metabolism in the Freeze-Tolerant Wood Frog. J. Exp. Biol. 2012, 215, 543–551. [Google Scholar] [CrossRef]
- Costanzo, J.P.; Reynolds, A.M.; do Amaral, M.C.F.; Rosendale, A.J.; Lee, R.E. Cryoprotectants and Extreme Freeze Tolerance in a Subarctic Population of the Wood Frog. PLoS ONE 2015, 10, e0117234. [Google Scholar] [CrossRef]
- Sugden, P.H.; Fuller, S.J. Regulation of Protein Turnover in Skeletal and Cardiac Muscle. Biochem. J. 1991, 273, 21–37. [Google Scholar] [CrossRef]
- Roepstorff, K.; Grøvdal, L.; Grandal, M.; Lerdrup, M.; van Deurs, B. Endocytic Downregulation of ErbB Receptors: Mechanisms and Relevance in Cancer. Histochem. Cell Biol. 2008, 129, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Storey, K.B. Small RNA and Freeze Survival: The Cryoprotective Functions of MicroRNA in the Frozen Muscle Tissue of the Grey Tree Frog. Metabolites 2024, 14, 387. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-W.; Biggar, K.K.; Luu, B.E.; Szereszewski, K.E.; Storey, K.B. Analysis of microRNA Expression during the Torpor-Arousal Cycle of a Mammalian Hibernator, the 13-Lined Ground Squirrel. Physiol. Genom. 2016, 48, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hadj-Moussa, H.; Storey, K.B. Current Progress of High-Throughput microRNA Differential Expression Analysis and Random Forest Gene Selection for Model and Non-Model Systems: An R Implementation. J. Integr. Bioinform. 2016, 13, 306. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a Genome-Centric Resource for Non-Coding RNA Families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Naranjo, M.; Breedon, S.A.; Storey, K.B. Cardiac microRNA Expression Profile in Response to Estivation. Biochimie 2023, 210, 22–34. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. RBiomirGS: An All-in-One miRNA Gene Set Analysis Solution Featuring Target mRNA Mapping and Expression Profile Integration. PeerJ 2018, 6, e4262. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, S.; Breedon, S.A.; Rhzali, I.; Storey, K.B. MicroRNA Regulation in the Freeze-Tolerant Heart of Dryophytes versicolor. Genes 2025, 16, 997. https://doi.org/10.3390/genes16090997
Rehman S, Breedon SA, Rhzali I, Storey KB. MicroRNA Regulation in the Freeze-Tolerant Heart of Dryophytes versicolor. Genes. 2025; 16(9):997. https://doi.org/10.3390/genes16090997
Chicago/Turabian StyleRehman, Saif, Sarah A. Breedon, Imane Rhzali, and Kenneth B. Storey. 2025. "MicroRNA Regulation in the Freeze-Tolerant Heart of Dryophytes versicolor" Genes 16, no. 9: 997. https://doi.org/10.3390/genes16090997
APA StyleRehman, S., Breedon, S. A., Rhzali, I., & Storey, K. B. (2025). MicroRNA Regulation in the Freeze-Tolerant Heart of Dryophytes versicolor. Genes, 16(9), 997. https://doi.org/10.3390/genes16090997






