Neonative Diploid-Polyploid Hotspots of Paspalum notatum: Identifying Novel Genetic Diversity for Conservation in South America
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chromosome Counts
2.3. Ploidy Analyses by Flow Cytometry
2.4. Georeferenced Maps
3. Results
3.1. Chromosome Number and Ploidy Level
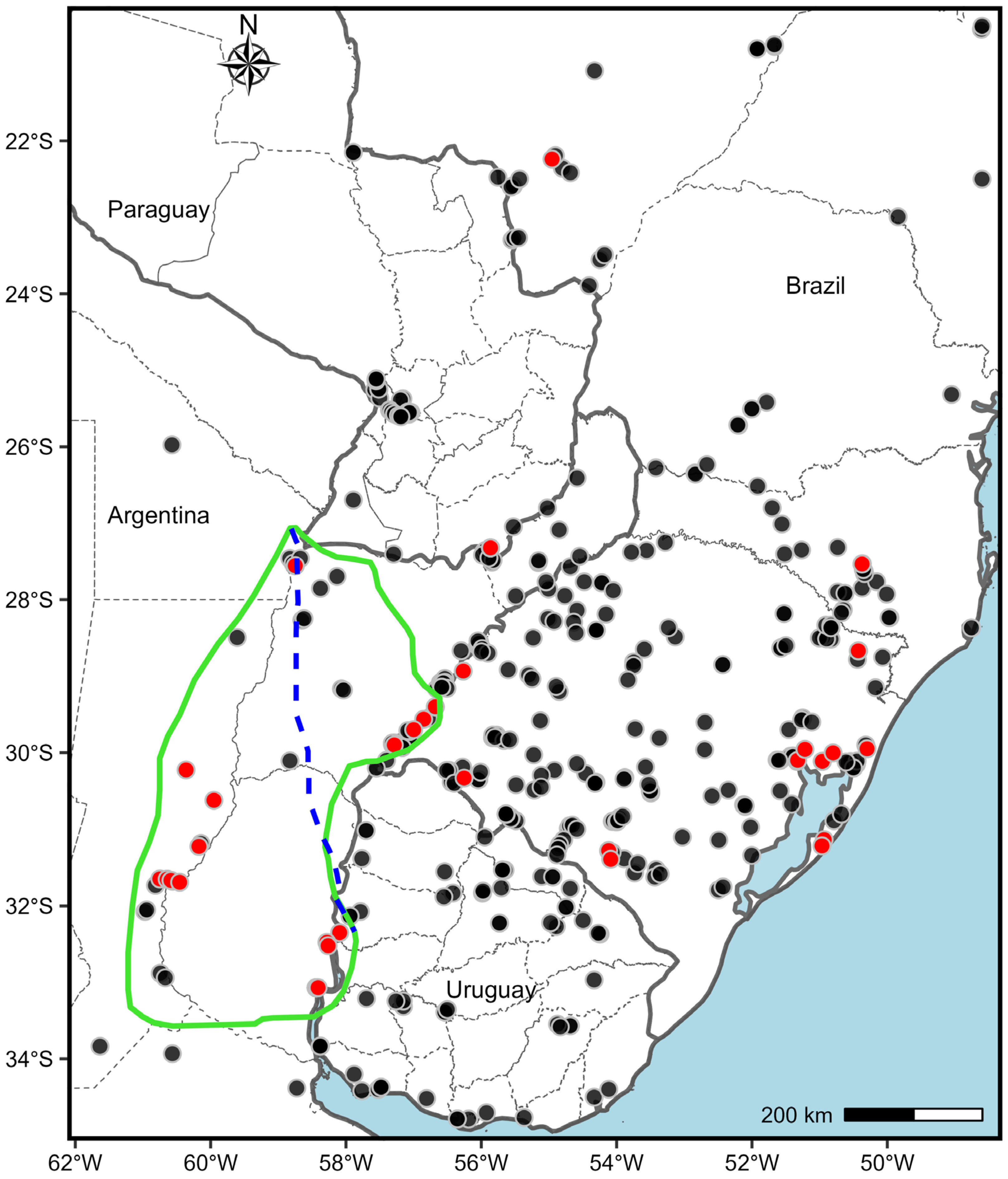
3.2. Cytotype Geographical Distribution
4. Discussion
4.1. Chromosome Numbers and Ploidy Levels in Bahiagrass
4.2. Primary Gene Pool and Origin Center of P. notatum
4.3. Neonative Diploid-Polyploid Centers of P. notatum
4.4. Neopoliploidization in Neonative Centers of P. notatum
4.5. Neonative Centers and Neighbouring Microscale Dynamics
4.6. Further Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2n. | Sporophitic chromosome number |
| x | Basic chromosome number, ploidy |
| n | Gametic chromosome number |
| BIII | 2n + n hybrid |
References
- den Nijs, J.H.C.; Marhold, M.M.K.; Hurka, H. Plant Evolution in disturbed habitat: An introduction. Folia Geobot. 1999, 34, 399–403. [Google Scholar] [CrossRef]
- Essl, F.; Dullinger, S.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Katsanevakis, S.; Kühn, I.; Lenzner, B.; Pauchard, A.; Pyšek, P.; et al. A Conceptual Framework for Range-Expanding Species that Track Human-Induced Environmental Change. BioScience 2019, 69, 908–919. [Google Scholar] [CrossRef]
- Stace, C.A.; Crawley, M.J. Alien Plants; Collins New Naturalist Library, book no. 129; HarperCollins: New York, NY, USA, 2015. [Google Scholar]
- Edelman, N.B.; Mallet, J. Prevalence and adaptive impact of introgression. Annu. Rev. Genet. 2021, 55, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, M.B.; Valls, J.F.M.; Abreu, A.G.; Heiden, G.; Ribeiro-Silva, S.; José, S.C.B.R.; Santos, I.R.I.; Passos, A.M.A.; Burle, M.L. Status of the Ex Situ and In Situ Conservation of Brazilian Crop Wild Relatives of Rice, Potato, Sweet Potato, and Finger Millet: Filling the Gaps of Germplasm Collections. Agronomy 2021, 11, 638. [Google Scholar] [CrossRef]
- Harlan, J.R.; de Wet, J.M.J. Toward a rational classification of cultivated plants. Taxon 1971, 20, 509–517. [Google Scholar] [CrossRef]
- Krapovickas, A.; Gregory, W.C. Taxonomía del género Arachis (Leguminosae). Bonplandia 1994, 8, 1–186. [Google Scholar] [CrossRef]
- Valls, J.F.M. Caracterização do germoplasma de espécies de Paspalum coletado no sul do Brasil. In Anais da Reunião do Grupo Técnico Regional em Melhoramento e Utilização de Recursos Forrageiros; Grupo Campos, EMPASC: Lages, Brazil, 1990; pp. 184–222. [Google Scholar]
- Acuña, C.A.; Martínez, E.J.; Zilli, A.L.; Brugnoli, E.A.; Espinoza, F.; Marcón, F.; Urbani, M.H.; Quarin, C.L. Reproductive systems in Paspalum: Relevance for germplasm collection and conservation, breeding techniques, and adoption of released cultivars. Front. Plant Sci. 2019, 10, 1377. [Google Scholar] [CrossRef]
- Martínez, E.J.; Urbani, M.H.; Quarin, C.L.; Ortiz, J.P.A. Inheritance of apospory in bahiagrass, Paspalum notatum. Hereditas 2001, 135, 19–25. [Google Scholar] [CrossRef]
- Hojsgaard, D.H.; Honfi, A.I.; Rua, G.; Daviña, J.R. Chromosome numbers and ploidy levels of Paspalum species from subtropical South America (Poaceae). Genet. Resour. Crop Evol. 2009, 56, 533–545. [Google Scholar] [CrossRef]
- Honfi, A.I.; Reutemann, A.V.; Schneider, J.S.; Escobar, L.M.; Martínez, E.J.; Daviña, J.R. Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure. Genes 2025, 16, 242. [Google Scholar] [CrossRef]
- Dahmer, N.; Wittmann, M.T.S.; Dall’agnol, M.; Castro, B. Cytogenetic data for Paspalum notatum Flügge accessions. Sci. Agric. 2008, 65, 381–388. [Google Scholar] [CrossRef]
- Espinoza, F.; Daurelio, L.D.; Pessino, S.C.; Valle, E.M.; Quarin, C.L. Genetic characterization of Paspalum notatum accessions by AFLP markers. Plant Syst. Evol. 2006, 258, 147–159. [Google Scholar] [CrossRef]
- Forbes, I.J.R.; Burton, G.W. Cytology of diploids, natural and induced tetraploids, and intra-species hybrids of Bahiagrass, Paspalum notatum Flügge. Crop Sci. 1961, 1, 402–406. [Google Scholar] [CrossRef]
- Souza-Chies, T.; Essi, L.; Rua, G.; Valls, J.F.M.; Miz, R. A preliminary approach to the phylogeny of the genus Paspalum (Poaceae). Genetica 2006, 126, 15–32. [Google Scholar] [CrossRef]
- Pozzobon, M.T.; Valls, J.F.M. Chromosome number in germplasm accessions of Paspalum notatum (Gramineae). Braz. J. Genet. 1997, 20, 29–34. [Google Scholar] [CrossRef]
- D’Aurelio, L.D.; Espinoza, F.; Quarin, C.L.; Pessino, S.C. Genetic diversity in sexual diploid and apomictic tetraploid populations of Paspalum notatum situated in sympatry or allopatry. Plant Syst. Evol. 2004, 244, 189–199. [Google Scholar] [CrossRef]
- Burton, G.W. The method of reproduction in common bahiagrass, Paspalum notatum. J. Am. Soc. Agron. 1948, 40, 443–452. [Google Scholar] [CrossRef]
- Fachinetto, J.M.; Dall’agnol, M.; Schifino-Wittmann, M.T.; Simioni, C.; Ávila, M.R. New wild diploids in Paspalum notatum Flügge (Poaceae): Potential accessions for use in breeding. Crop Breed. Appl. Biotechnol. 2018, 18, 432–436. [Google Scholar] [CrossRef]
- Parodi, L.R. Contribución al estudio de las gramíneas del género Paspalum de la flora uruguaya. Rev. Mus. Plata 1937, 1, 211–250. [Google Scholar]
- Parodi, L.R. La variación de Paspalum notatum Flüggé. Rev. Argent. Agron. 1948, 15, 53–57. [Google Scholar]
- Vega, J.M.; Podio, M.; Orjuela, J.; Siena, L.A.; Pessino, S.C.; Combes, M.C.; Mariac, C.; Albertini, E.; Pupilli, F.; Ortiz, J.P.A.; et al. Chromosome-scale genome assembly and annotation of Paspalum notatum Flüggé var. saurae. Sci. Data 2024, 11, 891. [Google Scholar] [CrossRef]
- Burton, G.W. A search for the origin of Pensacola Bahia grass. Econ. Bot. 1967, 21, 379–382. [Google Scholar] [CrossRef]
- Gould, F.W. Chromosome numbers of Mexican grasses. Can. J. Bot. 1966, 44, 1683–1696. [Google Scholar] [CrossRef]
- Quarin, C.L.; Norrmann, G.A.; Urbani, M.H. Polyploidization in aposporous Paspalum species. Apomixis Newsl. 1989, 2, 44–46. [Google Scholar]
- Tischler, C.R.; Burson, B.L. Evaluating different bahiagrass cytotypes for heat tolerance and leaf epicular wax content. Euphytica 1995, 84, 229–235. [Google Scholar] [CrossRef]
- Martínez, E.J.; Acuña, C.A.; Hojsgaard, D.H.; Quarin, C.L. Segregation for asexual seed production in Paspalum achieved by male gametes of apomictic triploid plants. Ann. Bot. 2007, 100, 1239–1247. [Google Scholar] [CrossRef]
- Barreto, I.L. O gênero Paspalum (Gramineae) no Rio Grande do Sul. Livre Docência Thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil, 1974. [Google Scholar]
- Canto Dorow, T.S. Revisão taxonômica das espécies sul rio-grandenses de Paspalum L. (grupo Notata) Poaceae-Paniceae, com ênfase na análise da variação intraspecifica de P. notatum Flüggé. Master’s Thesis, UFRGS, Porto Alegre, Brazil, 1993; pp. 1–179. [Google Scholar]
- Cidade, F.W.; Dall’Agnol, M.; Bered, F.; de Souza-Chies, T.T. Genetic diversity of the complex Paspalum notatum Flügge (Paniceae: Panicoideae). Genet. Resour. Crop Evol. 2008, 55, 235–246. [Google Scholar] [CrossRef]
- Burton, G.W.; Forbes, I.; Jackson, J. Effect of ploidy on fertility and heterosis in pensacola bahiagrass. Crop Sci. 1970, 10, 63–65. [Google Scholar] [CrossRef]
- Galdeano, F.; Urbani, M.H.; Sartor, M.E.; Honfi, A.I.; Espinoza, F.; Quarin, C.L. Relative DNA content in diploid, polyploid, and multiploid species of Paspalum (Poaceae) with relation to reproductive mode and taxonomy. J. Pl. Res. 2016, 129, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Gates, R.N.; Quarin, C.L.; Pedreira, C.G.S. Bahiagrass. In Warm-Season (C4) Grasses; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; Agronomy Monography 45; ASA, CSSA, SSSA: Madison, WI, USA, 2004; pp. 651–680. [Google Scholar] [CrossRef]
- Burton, G.W. Great diversity for increased yield within narrow grass populations. In Proceedings of the XVI International Grassland Congress, Nice, France, 4–11 October 1989; pp. 301–302. [Google Scholar]
- Blount, A.R.; Acuña, C.A. Bahiagrass. In Genetic Resources, Chromosome Engineering, and Crop Improvement: Forage Crops; Singh, R.J., Ed.; CRC Press: Boca Raton, FL, USA, 2009; Volume 5, pp. 81–101. [Google Scholar] [CrossRef]
- Newman, Y.J.; Vendramini, A.; Blount, A.R. Bahiagrass (Paspalum notatum): Overview and Management; SS-AGR-332; Agronomy Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 2010; pp. 1–9. [Google Scholar] [CrossRef]
- Urbani, M.H.; Acuña, C.A.; Doval, D.W.; Sartor, M.E.; Galdeano, F.; Blount, A.R.; Quarin, C.L. Registration of ‘Boyero UNNE’ bahiagrass. J. Plant Regist. 2017, 11, 26–32. [Google Scholar] [CrossRef]
- Matta, F.P.; Fávero, A.P.; Vigna, B.B.Z.; Cavallari, M.M.; Alves, F.; Oliveira, F.A.; Pereira de Souza, A.; Pozzobon, M.T.; Sousa Azevedo, A.L.; Gusmão, M.R. Characterization of Paspalum genotypes for turfgrass cultivars development. Crop Sci. 2024, 64, 1928–1941. [Google Scholar] [CrossRef]
- Andrade, B.O.; Bonilha, C.L.; Overbeck, G.E.; Vélez-Martin, E.; Rolim, R.G.; Schneider, A.A.; Lucas, D.B.; Garcia, É.N.; Torchelsen, P. Classification of South Brazilian grasslands: Implications for conservation. Appl. Veg. Sci. 2019, 22, 168–184. [Google Scholar] [CrossRef]
- Castañeda-Álvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Maxted, N.; et al. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022. [Google Scholar] [CrossRef]
- Feulgen, R.; Rossenbeck, H. Mikroskopischchemischer Nachweis einer Nucleinsaure vom Typus der Thymonucleisaure und die darauf beruhende elektive Farbung vom Zellkernen in mikroskopischen Praparaten. Z. Phys. Chem. 1924, 135, 203–248. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 29 July 2025).
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Barbosa, M.; Ghosh, A.; Mandel, A. Geodata: Download Geographic Data. R Package Version 0.6-3. 2025. Available online: https://github.com/rspatial/geodata (accessed on 29 July 2025).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 27 July 2025).
- Dunnington, D. Ggspatial: Spatial Data Framework for ggplot2. R Package Version 1.1.9.9000. 2025. Available online: https://github.com/paleolimbot/ggspatial (accessed on 27 July 2025).
- Burton, G.W. Breeding Pensacola bahiagrass, Paspalum notatum: I. Method of reproduction. Agron. J. 1955, 47, 311–314. [Google Scholar] [CrossRef]
- Martínez, E.J.; Espinoza, F.; Quarin, C.L. BIII progeny (2n+n) from apomictic Paspalum notatum obtained through early pollination. J. Hered. 1994, 85, 295–297. [Google Scholar] [CrossRef]
- Saura, F. Cariología de gramíneas en Argentina. Rev. Fac. Agron. Vet. Buenos Aires 1948, 12, 51–67. [Google Scholar]
- Quarin, C.L. Recuentos cromosómicos en gramíneas de Argentina subtropical. Hickenia 1977, 1, 73–78. [Google Scholar]
- Rieseberg, L.H.; Carney, S.A. Plant hybridization. Tansley Review No. 102. New Phytol. 1998, 140, 599–624. [Google Scholar] [CrossRef]
- Herben, T.; Suda, J.; Klimešová, J. Polyploid species rely on vegetative reproduction more than diploids: A re-examination of the old hypothesis. Ann. Bot. 2017, 120, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Harlan, J.R. Anatomy of gene centers. Am. Nat. 1951, 85, 97–103. [Google Scholar] [CrossRef]
- Dahmer-Balbinot, N. Variabilidade citogenética em uma coleção de acessos de Paspalum notatum Flügge. Master’s Thesis, Universidade Federal do Rio Grande do Sulc, Porto Alegre, Brazilia, 2007; pp. 1–71. [Google Scholar]
- Novo, P.E.; Valls, J.F.M.; Galdeano, F.; Honfi, A.I.; Espinoza, F.; Quarin, C.L. Interspecific hybrids between Paspalum plicatulum and P. oteroi: A key tool for forage breeding. Sci. Agric. 2016, 73, 356–362. [Google Scholar] [CrossRef]
- Reyno, R.; Narancio, R.; Speranza, P.; Do Canto, J.; López-Carro, B.; Hernández, P.; Burgueño, J.; Real, D.; Dalla Rizza, M. Molecular and cytogenetic characterization of a collection of bahiagrass (Paspalum notatum Flüggé) native to Uruguay. Genet. Resour. Crop Evol. 2012, 59, 1823–1832. [Google Scholar] [CrossRef]
- Zilli, A.L.; Acuña, C.A.; Schulz, R.R.; Brugnoli, E.A.; Guidalevich, V.; Quarin, C.L.; Martínez, E.J. Widening the gene pool of sexual tetraploid Bahiagrass: Generation and reproductive characterization of a sexual synthetic tetraploid population. Crop Sci. 2018, 58, 762–772. [Google Scholar] [CrossRef]
- Burton, G.W. A cytological study of some species in the genus Paspalum. J. Agric Res. 1940, 3, 193–198. [Google Scholar]
- Fachinetto, J.M.; Schneider, R.; Hubber, K.G.C.; Dall’Agnol, M. Avaliação agronômica e análise da persistência em uma coleção de acessos de Paspalum notatum Flüggé (Poaceae). Agrária 2012, 7, 189–195. [Google Scholar]
- Reutemann, A.V.; Rua, G.H.; Daviña, J.R.; Honfi, A.I. Poaceae. IAPT chromosome data 31/11. Taxon 2019, 68, 1379–1380. [Google Scholar]
- Pagliarini, M.S.; Carraro, L.R.; de Freitas, P.M.; Adamowski, E.D.V.; Rocha Batista, L.A.; Valls, J.F.M. Cytogenetic characterization of Brazilian Paspalum accessions. Hereditas 2001, 135, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Cidade, F.W.; Vigna, B.B.Z.; de Souza, F.H.D.; Valls, J.F.M.; Dall’Agnol, M.; Zucchi, M.I.; Souza-Chies, T.T.; Souza, A.P. Genetic variation in polyploid forage grass: Assessing the molecular genetic variability in the Paspalum genus. BMC Genet. 2013, 14, 50. [Google Scholar] [CrossRef]
- Rebozzio, R.N.; Sartor, M.E.; Quarin, C.L.; Espinoza, F. Residual sexuality and its seasonal variation in natural apomictic Paspalum notatum accessions. Biol. Plant. 2011, 55, 391–395. [Google Scholar] [CrossRef]
- Adamowski, E.V.; Pagliarini, M.S.; Bonato, A.B.M.; Batista, L.A.R.; Valls, J.F.M. Chromosome numbers and meiotic behavior of some Paspalum accessions. Genet. Mol. Biol. 2005, 28, 773–780. [Google Scholar] [CrossRef]
- Honfi, A.I.; Quarin, C.L.; Valls, J.F.M. Estudio Cariológicos en Gramíneas Sudamericanas. Darwiniana 1990, 30, 87–94. [Google Scholar]
- Pozzobon, M.T.; dos Santos Sousa, M.W.; Valls, J.F.M. Chromosome numbers of Paspalum species, IAPT chromosome data 36/4. Taxon 2022, 71, E13–E18. [Google Scholar] [CrossRef]
- Chaparro, C.; Escobar, L.M.; Schneider, J.S.; Eckers, F.; Perichon, M.C.; Daviña, J.R.; Honfi, A.I. Niveles de ploidía de algunas especies de Paspalum L. de Paraguay. Steviana 2023, 15, 25–36. [Google Scholar] [CrossRef]
- Rivarola, A.; Daviña, J.R.; Honfi, A.I. Poaceae, IAPT/IOPB chromosome data 23. Taxon 2016, 55, 1457. [Google Scholar] [CrossRef][Green Version]
- Zilli, A.L.; Brugnoli, E.A.; Marcón, F.; Billa, M.B.; Rios, E.F.; Martínez, E.J.; Acuña, C.A. Heterosis and expressivity of apospory in tetraploid bahiagrass hybrids. Crop Sci. 2015, 55, 1189–1201. [Google Scholar] [CrossRef]
- Suarez, V.C. Paspalum notatum Flüggé (Poaceae): Estudio comparativo del comportamiento cromosómico em meiosis y tamaño y viabilidad de los granos de polen em dos grupos de diferente contenido de ADN. Ph.D. Thesis, Universidad de la República (UDELAR), Montevideo, Uruguay, 2014; 79p. [Google Scholar]
- Davidse, G.; Pohl, R.W. Chromosome numbers of Tropical American grasses (Gramineae). Ann. Missouri Bot. Gard. 1978, 65, 637–649. [Google Scholar] [CrossRef]
- Espinoza, F.; Quarin, C.L. 2n+n hibridization of apomictic Paspalum dilatatum with diploid Paspalum species. Int. J. Plant Sci. 2000, 161, 221–225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pagliarini, M.S.; Takayama, S.Y.; Freitas, P.M.; Carraro, L.R.; Adamowski, E.V.; Batista, L.A.R. Failure of cytokinesis and 2n gamete formation in Brazilian accessions of Paspalum. Euphytica 1999, 108, 129–135. [Google Scholar] [CrossRef]
- Burson, B.L.; Bennett, H.W. Cytogenetics of Paspalum urvillei x P. jurgensii and P. urvillei x P. vaginatum hybrids. Crop Sci. 1972, 12, 105–108. [Google Scholar]
- Burson, B.L.; Lee, H.; Bennett, H.W. Genome relations between tetraploid Paspalum dilatatum and four diploid Paspalum species. Crop Sci. 1973, 13, 739–743. [Google Scholar] [CrossRef]
- Burson, B.L. Genome Relations Among Four Diploid Paspalum Species. Bot. Gaz. 1981, 142, 592–596. [Google Scholar] [CrossRef]
- Rua, G.H.; Speranza, P.R.; Vaio, M.; Arakaki, M. A phylogenetic analysis of the genus Paspalum Poaceae based on cpDNA and morphology. Pl Syst. Evol. 2010, 288, 227–243. [Google Scholar] [CrossRef]
- Catanzaro, M.P.; Bonasora, M.G.; Speranza, P.R.; Medina, N.M.; Valls, J.F.M.; Rua, G.H. Paspalum chilense Poaceae, Paspaleae: A new species from southern South America. Phytotaxa 2015, 197, 245–256. [Google Scholar] [CrossRef]
- Steiner, M.G.; Dall’agnol, M.; Nabinger, C.; Scheffer-Basso, S.M.; Weiler, R.L.; Simioni, C.; Schifino-Wittmann, M.T.; Da Motta, É.A.M. Forage potential of native ecotypes of Paspalum notatum and P. guenoarum. An. Acad. Bras. Ciênc. 2017, 89, 1753–1760. [Google Scholar] [CrossRef]
- Norrmann, G.A.; Quarin, C.L.; Burson, B.L. Cytogenetics and reproductive behavior of different chromosome races in six Paspalum species. J. Hered. 1989, 80, 24–28. [Google Scholar] [CrossRef]
- Quarin, C.L. The nature of apomixis and its origin in Panicoid grasses. Apomixis Newsl. 1992, 5, 8–15. [Google Scholar]
- Savidan, Y. Apomixis: Genetics and breeding. Plant Breed. Rev. 2000, 18, 13–86. [Google Scholar] [CrossRef]
- Stebbins, G.L. Chromosome Evolution in Higher Plants; Addison Wesley Publishing CO: Boston, MA, USA, 1970. [Google Scholar]
- Karunarathne, P.; Schedler, M.; Martínez, E.J.; Honfi, A.I.; Novichkova, A.; Hojsgaard, D.H. Intraspecific ecological niche divergence and reproductive shifts foster cytotype displacement and provide ecological opportunity to polyploids. Ann. Bot. 2018, 121, 1183–1196. [Google Scholar] [CrossRef] [PubMed]
- Karunarathne, P.; Reutemann, A.V.; Schedler, M.; Glücksberg, A.; Martínez, E.J.; Honfi, A.I.; Hojsgaard, D.H. Sexual modulation and the evolutionary implications of a TUG OF WAR between sexual-apomictic reproductive modes in a polyploid grass species. Sci. Rep. 2020, 10, 8319. [Google Scholar] [CrossRef]
- Kearney, M. Hybridization, glaciation and geographical parthenogenesis. Trends Ecol. Evol. 2005, 20, 495–502. [Google Scholar] [CrossRef]
- Hörandl, E. Geographical Parthenogenesis in Alpine and Arctic Plants. Plants 2023, 12, 844. [Google Scholar] [CrossRef]
- Largier, J.L. Considerations in estimating larval dispersal distances from oceanographic data. Ecol. Appl. 2003, 13, 71–89. [Google Scholar] [CrossRef]
- Rogers, H.S.; Beckman, N.G.; Hartig, F.; Johnson, J.S.; Pufal, G.; Shea, K.; Zurell, D.; Bullock, J.M.; Cantrell, R.S.; Loiselle, B.; et al. The total dispersal kernel: A review and future directions. AoB Plants 2019, 11, plz042. [Google Scholar] [CrossRef] [PubMed]
- Reutemann, A.V.; Martínez, E.J.; Schedler, M.; Daviña, J.R.; Hojsgaard, D.H.; Honfi, A.I. Uniparentality: Advantages for range expansion in diploid and diploid-autopolyploid species. Bot. J. Linn. Soc. 2022, 200, 563–585. [Google Scholar] [CrossRef]
- Overbeck, G.E.; Vélez-Martin, E.; Scarano, F.R.; Lewinsohn, T.M.; Fonseca, C.R.; Meyer, S.T.; Müller, S.C.; Ceotto, P.; Dadalt, L.; Durigan, G.; et al. Conservation in Brazil needs to include non-forest ecosystems. Divers. Distrib. 2015, 21, 1455–1460. [Google Scholar] [CrossRef]
- Overbeck, G.E.; Scasta, J.D.; Furquim, F.F.; Boldrini, I.I.; Weir, J.R. The South Brazilian grasslands—A South American tallgrass prairie? Parallels and implications of fire dependency. Perspect. Ecol. Conserv. 2018, 16, 24–30. [Google Scholar]
- Pillar, V.D.; Overbeck, G.E. Nature conservation policies are biased toward forests and neglect grassy ecosystems worldwide. Science 2025, 388, 6747. [Google Scholar] [CrossRef]
- Petermann, J.S.; Buzhdygan, O.Y. Grassland biodiversity. Curr. Biol. 2021, 31, 1195–1201. [Google Scholar] [CrossRef] [PubMed]

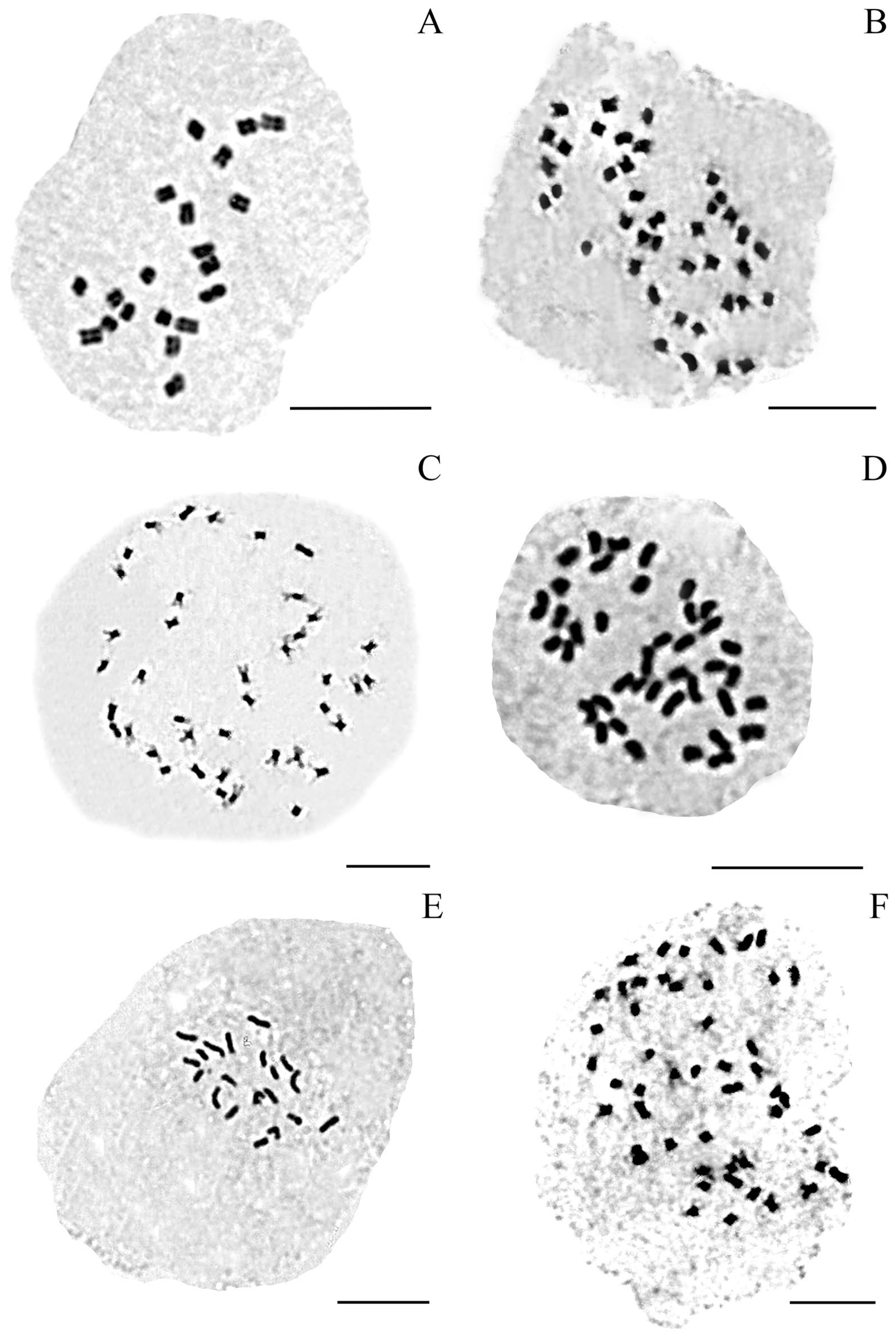
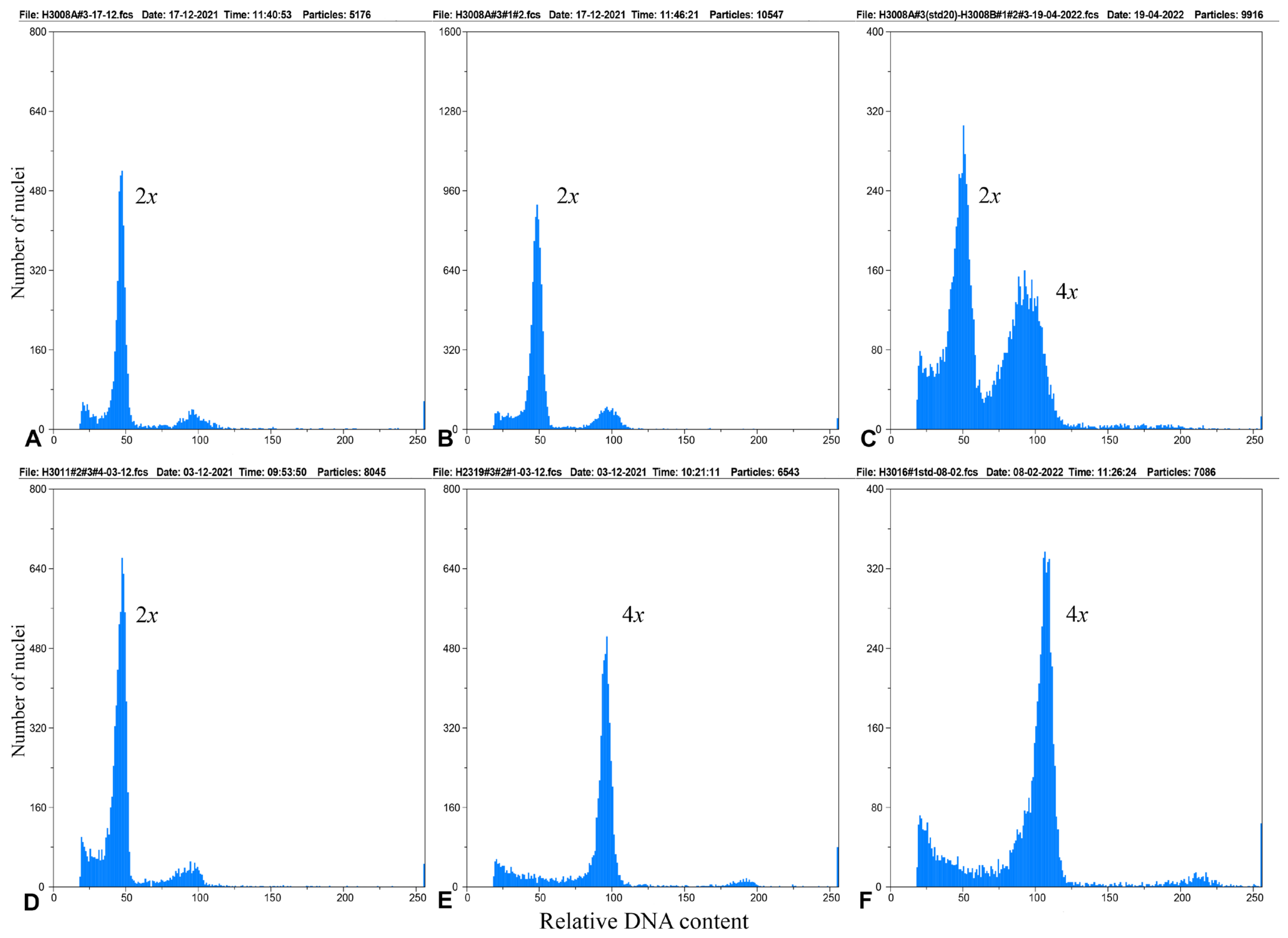

| x | ID (N) | Provenance |
|---|---|---|
| 2x | H1961 #3 to #29 (27) | Argentine. Entre Ríos, Gualeguaychú. |
| 2x | Q 4119 (1) | Argentine. Santa Fe, 7 km S from San Javier, (CTES Herbarium, Quarin unpubl.) |
| 2x | H1740 #1, #3 to #27 (26) | Paraguay. Itapúa, Encarnación. |
| 2x | H3008 #1 to #4 (4) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| 2x | H3010 #1, #2 (2) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| 2x | H3011 #2 to #4 (3) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| x | ID (N) | Provenance |
|---|---|---|
| 4x | H1271 (1) | Argentine. Misiones, Guaraní, Arroyo Yerbas del Paraiso. |
| 4x | H1350 (1) | Argentine. Misiones, Capital, Bañados del Zaimán. |
| 4x | H2137 (1) | Argentine. Corrientes. Paso de los Libres city. Bartolomé Mitre Square. |
| 4x | H2543 #1 to #3 (3) | Argentine. Misiones, Eldorado, Provintial Route 17, 11 km near Pozo Azul. |
| 4x | H2554 #1 to #5 (5) | Argentine. Misiones, Capital, Garupá, near the Train Station. |
| 4x | H2665 (1) | Argentine. Misiones, Concepción de la Sierra, Provintial Routea 2. |
| 4x | H2671 (1) | Argentine. Misiones, Ruta provincial 2, El Palmar del Río. |
| 4x | H2674 #1 to #11 (11) | Argentine. Misiones, Oberá, Panambí, Ruta provincial 2. |
| 4x | H2295 #1 to #3 (3) | Brazil. Parana, Route BR-280, A 57 km from Francisco Beltrão. |
| 4x | H2304 #1,#2,#6,#7 (4) | Brazil. Santa Catarina, BR-116. 1,5 km S da prefeitura of Correia Pinto. [ca. V14411]. |
| 4x | H2309B (1) | Brazil. Santa Catarina, SC-114 de Urupema a Lages, near Lages. [ca. V4508, V12156]. |
| 4x | H2310 #1,#4 (2) | Brazil. Santa Catarina, Correia Pinto, BR-116 y Rua Duque de Caxias y Rua Cándido [ca. V14411]. |
| 4x | H2312 #1 to #5 (5) | Brazil. Santa Catarina, SC-114 de Urupema a Lages, near Lages, [ca. V4508, V12156]. |
| 4x | H2319 #1 (1) | Brazil. Santa Catarina, BR-116 a 50 km al N de Vacaría [ca. V12159]. |
| 4x | H2690 (1) | Brazil. Rio de Janeiro, Jardim Botânico do Rio de Janeiro. [H2690RB #8] |
| 4x | H2691 #2, #4, #6, #8, #9 (5) | Brazil. Rio de Janeiro, Barra de Tijuca, on the sidewalk. |
| 4x | H2693 #1,#3 (2) | Brazil. Rio de Janeiro, Jardim Botânico do Rio de Janeiro. |
| 4x | H3003 #1,#3 (2) | Brazil. Santa Catarina, BR-116 km 210. |
| 4x | H3007 #2 (1) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| 4x | H3008 C #2, #3 (2) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| 4x | H3016 #1, #2 (2) | Brazil. Santa Catarina, BR-116 Correia Pinto. [ca. V14411] |
| 4x | H2528 (1) | Paraguay. Central. National route 2, a 18 km of Caacupé City. |
| 4x | Ch–1 to Ch–3 (3) | Paraguay. Cordillera. Compañía Capilla cué. |
| 4x | Ch–4 to Ch–6 (3) | Paraguay. Cordillera. Compañía Capilla cué, on the side of the road Faustino Rivas. |
| 4x | Ch–7 to Ch–14 (8) | Paraguay. Cordillera. Compañía Capilla cué. |
| 4x | Ch–25 (1) | Paraguay. Central. Luque City, on the side of the road |
| 4x | Ch–26 to Ch–27 (2) | Paraguay. Central. Luque, Central promenade of the Ñú Guazú highway |
| 4x | Ch–29 (1) | Paraguay. Central. Luque city, on the side of the road |
| 4x | Ch–31, Ch–33, Ch 33B to Ch–37 (7) | Paraguay. Paraguarí. On the side of the National route PY1. |
| 4x | Ch–38, Ch–40 (2) | Paraguay. Paraguarí. National route PY1, on the roadside, in front of the Yaguarón Hill. |
| 4x | Ch–41, Ch–44, Ch–45 (3) | Paraguay. Paraguarí. National route PY1, Paraguarí City. |
| 4x | Ch–42 (1) | Paraguay. Paraguarí. National route PY1, on the roadside to access to Paraguarí City. |
| 4x | Ch–43 (1) | Paraguay. Paraguarí. National route PY1, on the roadside to Paraguarí City. |
| 4x | Ch–47 to Ch–61 (15) | Paraguay. Presidente Hayes. Transchaco route, on the roadside. |
| 5x | H3007 #1, #3 (2) | Brazil. Santa Catarina, BR-116, 5 km N Rio Canoas. [ca. V8211] |
| Glossary |
| Neonative: A species or biotype or cytotype which result in a by-product of hybridization between a native species and an alien taxon. |
| Neonative center: Geographical area of origin of neonative taxa. |
| Gene pool: Sum of all the genes, alleles, and karyotypes shared by individuals of a species. |
| Center of origin: Geographical area where the species originated. In species with several intraspecific ploidy levels, the diploid location is an indicator of the evolutionary ancestry starting point for polyploids. In allopolyploid species, the center of origin refers to the site where progenitor species have probably hybridized. |
| Neocenters: New centers of diversity of a species. |
| Diversity center: Geographical area with prominent genetic diversity of a species. |
| Primary center of origin: primordial area with highest morphological and genetic diversity and where is most plausible originated the species. |
| Secondary diversity center: Geographical area where there is great genetic variability of a crop or species, but which is not the main center of origin of that species. |
| Ectopic area: Outside area from the natural geographical distribution of the taxon, biotype, cytotype or species. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar, L.M.; Reutemann, A.V.; Perichon, M.C.; Schneider, J.S.; Sartor, C.A.; Chaparro, C.; Daviña, J.R.; Valls, J.F.M.; Martínez, E.J.; Honfi, A.I. Neonative Diploid-Polyploid Hotspots of Paspalum notatum: Identifying Novel Genetic Diversity for Conservation in South America. Genes 2025, 16, 1098. https://doi.org/10.3390/genes16091098
Escobar LM, Reutemann AV, Perichon MC, Schneider JS, Sartor CA, Chaparro C, Daviña JR, Valls JFM, Martínez EJ, Honfi AI. Neonative Diploid-Polyploid Hotspots of Paspalum notatum: Identifying Novel Genetic Diversity for Conservation in South America. Genes. 2025; 16(9):1098. https://doi.org/10.3390/genes16091098
Chicago/Turabian StyleEscobar, Lucas M., Anna Verena Reutemann, María C. Perichon, Juan S. Schneider, Carolina A. Sartor, Clarisse Chaparro, Julio R. Daviña, José F. M. Valls, Eric J. Martínez, and Ana I. Honfi. 2025. "Neonative Diploid-Polyploid Hotspots of Paspalum notatum: Identifying Novel Genetic Diversity for Conservation in South America" Genes 16, no. 9: 1098. https://doi.org/10.3390/genes16091098
APA StyleEscobar, L. M., Reutemann, A. V., Perichon, M. C., Schneider, J. S., Sartor, C. A., Chaparro, C., Daviña, J. R., Valls, J. F. M., Martínez, E. J., & Honfi, A. I. (2025). Neonative Diploid-Polyploid Hotspots of Paspalum notatum: Identifying Novel Genetic Diversity for Conservation in South America. Genes, 16(9), 1098. https://doi.org/10.3390/genes16091098









