The Phenotype of Physcomitrium patens SMC6 Mutant with Interrupted Hinge Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Cultivation
2.2. Treatments and Sensitivity Assay
2.3. Spontaneous and Induced Mutagenesis
2.4. Mutant Lines
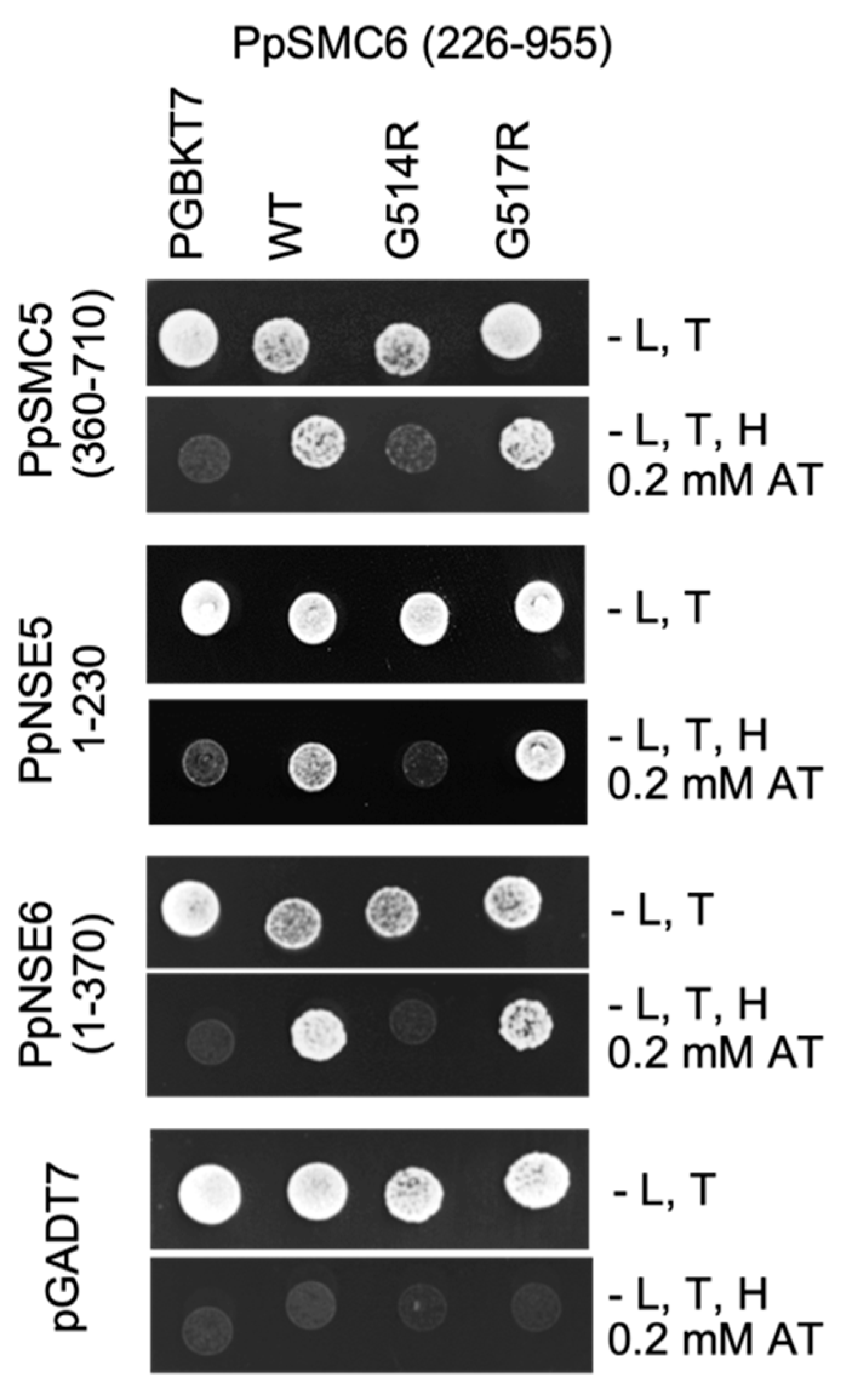
2.5. Yeast Two-Hybrid (Y2H) Constructs and Assays
2.6. DNA Isolation and Analysis of rDNA Copy Numbers
2.7. Morphotype Analysis
2.8. Microscopic Analysis
2.9. Single-Cell Gel Electrophoresis (Comet) Assay
2.10. GT Assay
3. Results
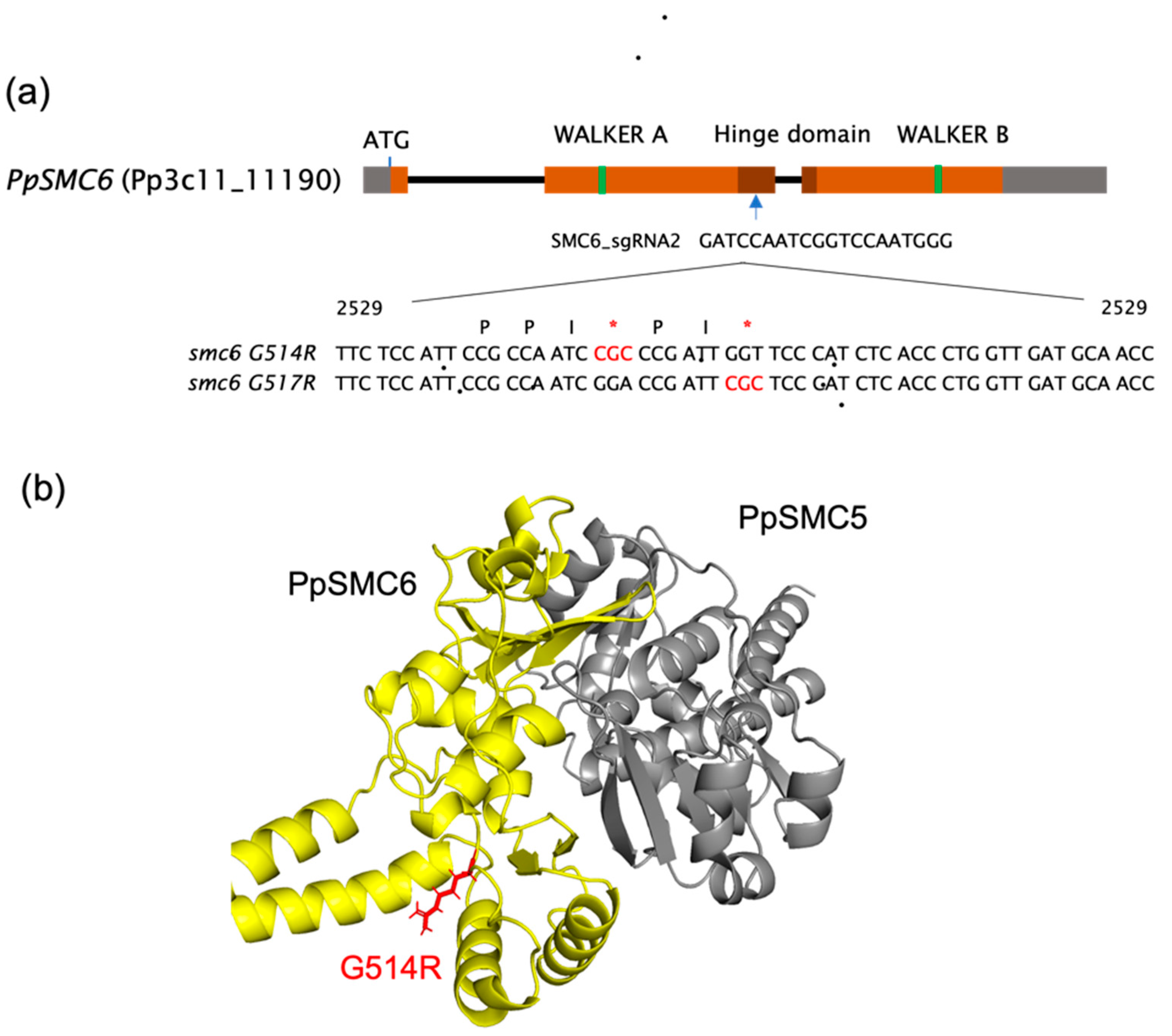
3.1. Generation and Analysis of the P. patens SMC Hinge Mutants
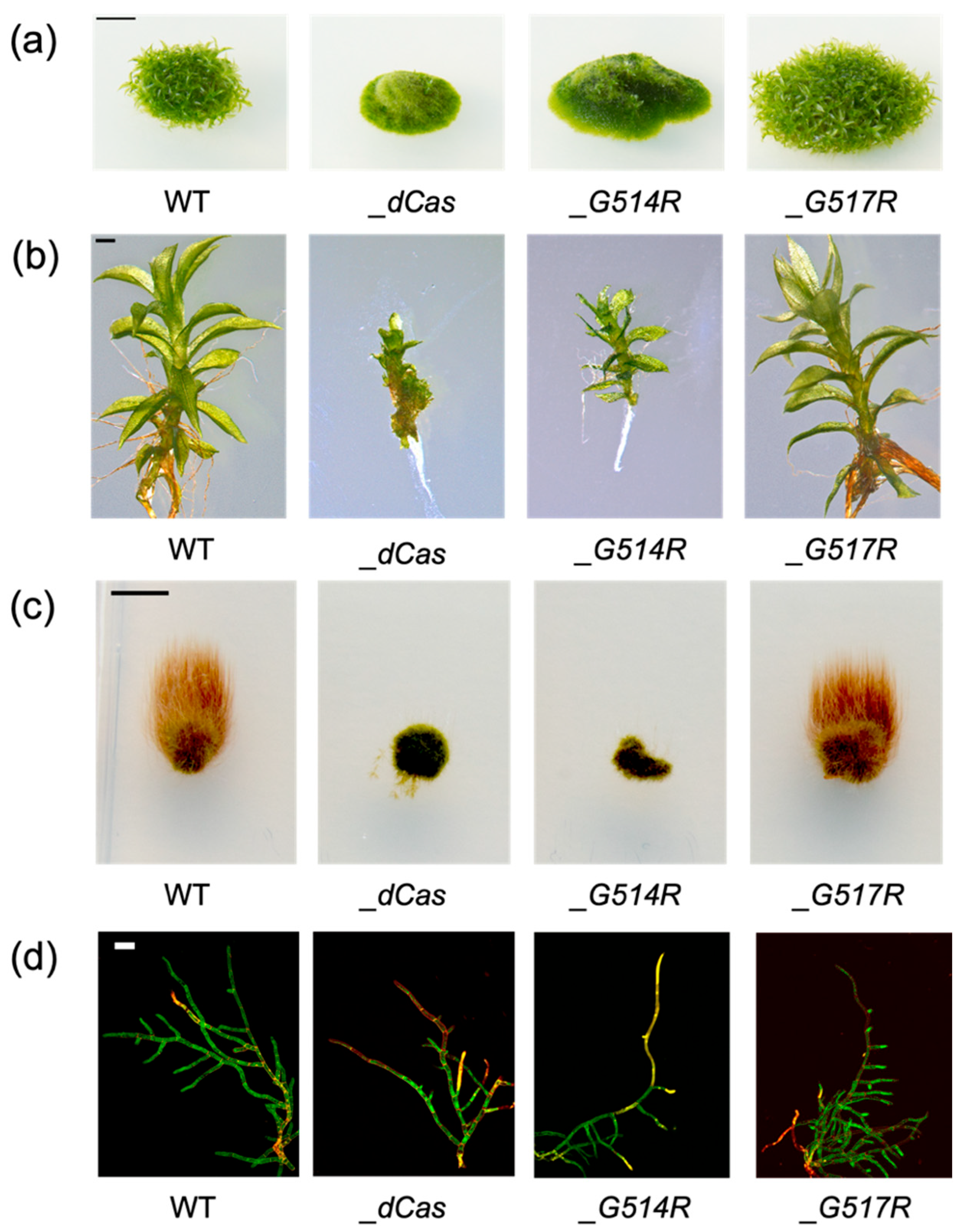
3.2. Morphology of P. patens SMC6-Hinge Mutants
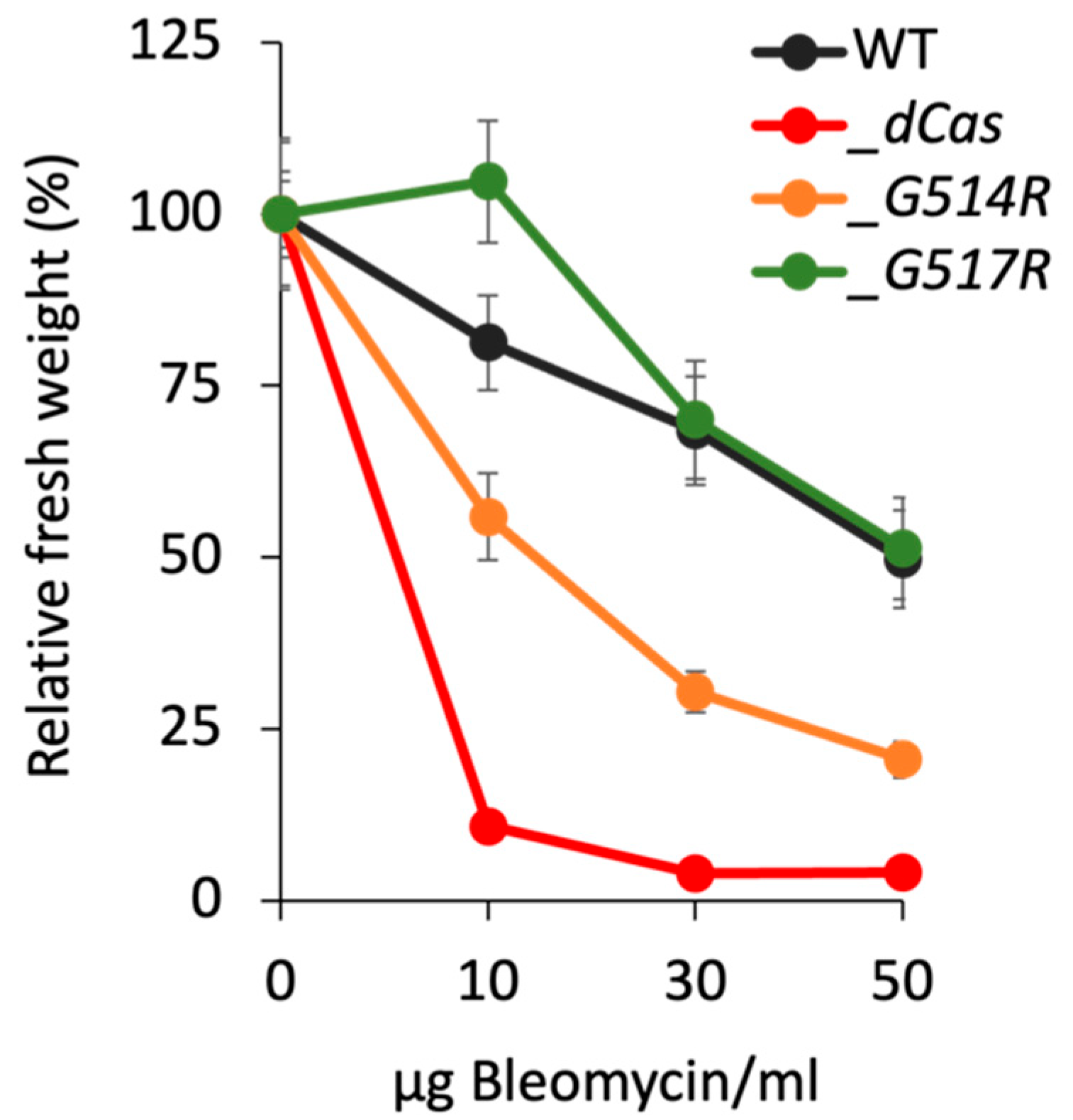
3.3. Growth-Response of P. patens SMC6-Hinge Mutants to Bleomycin
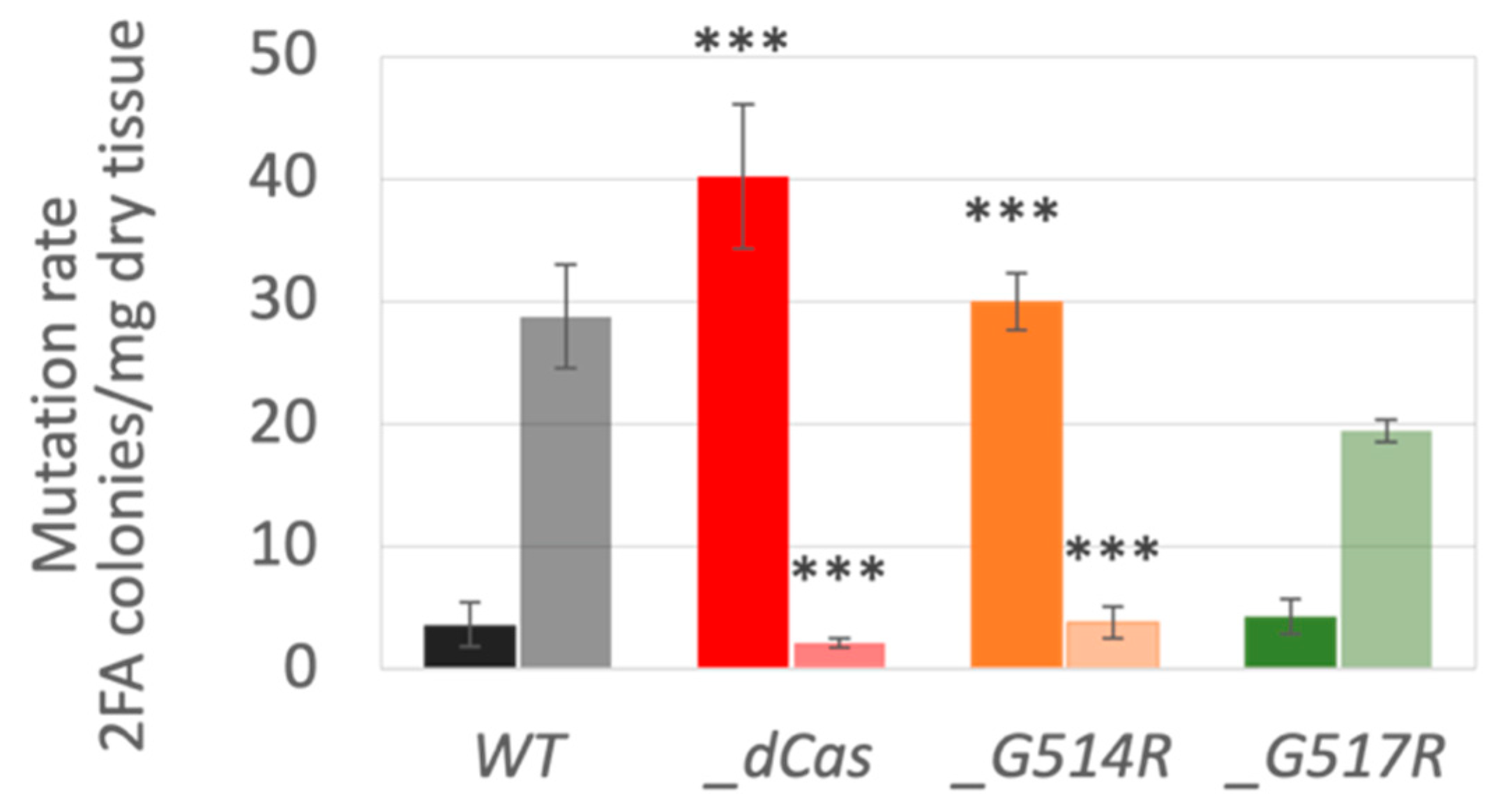
3.4. Spontaneous and Induced Mutagenesis in P. patens SMC6-Hinge Mutants
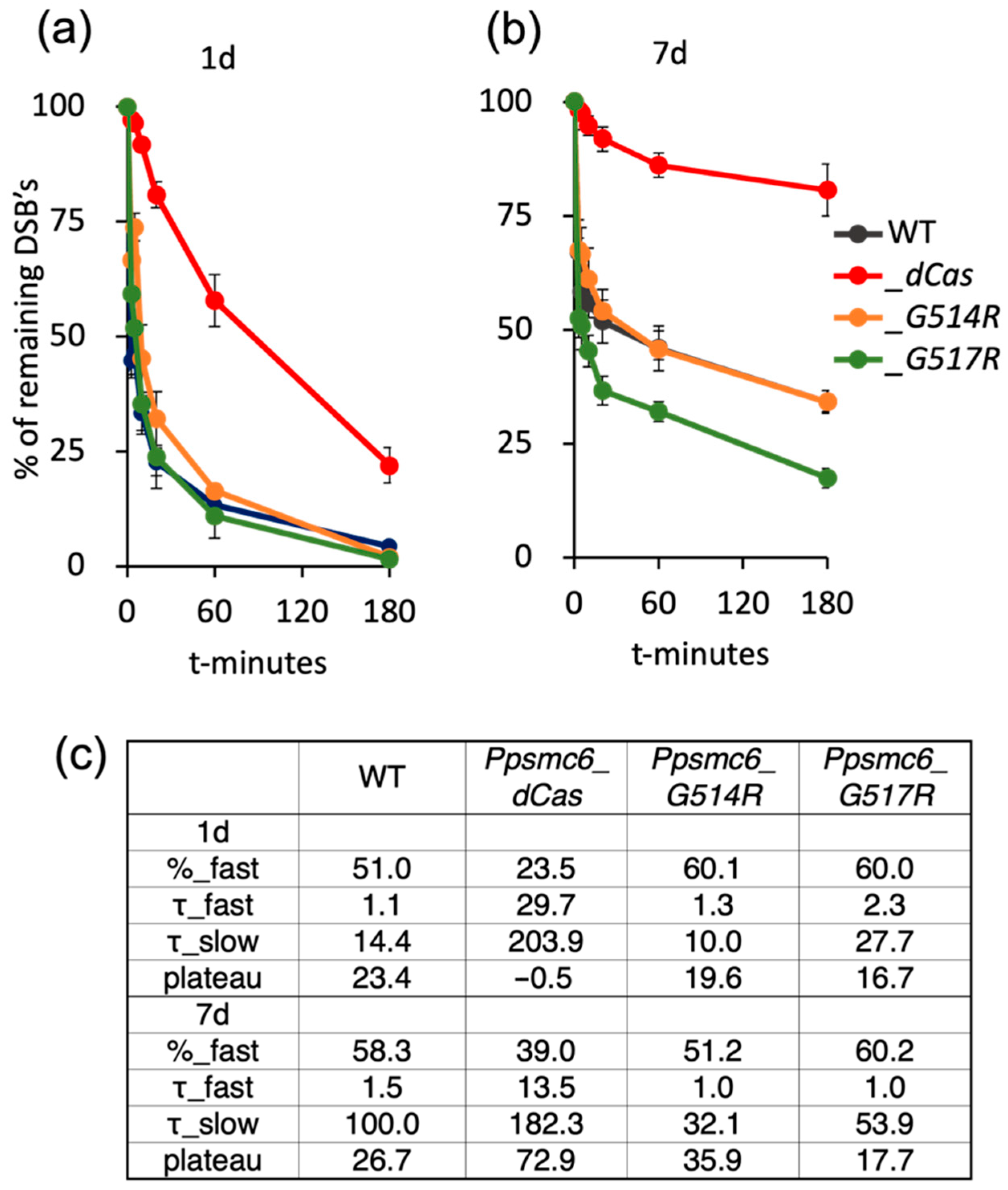
3.5. DSB Repair in P. patens SMC6-Hinge Mutants Is Not Reduced
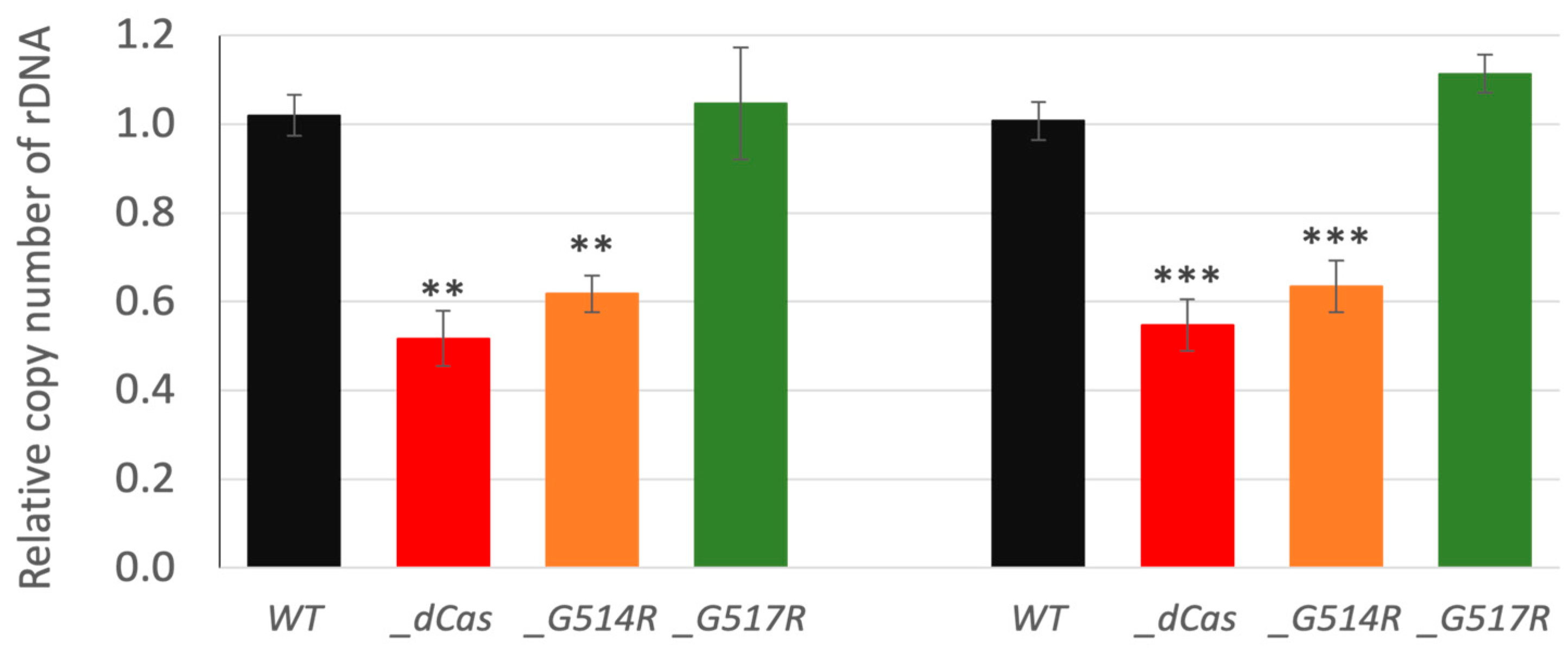
3.6. The Role of P. patens SMC6-Hinge Mutants in Maintenance of Genome Stability
3.7. Absence of GT in P. patens SMC6 G514R Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSB | DNA double strand break |
| dsDNA | double-stranded DNA |
| 2FA | 2-Fluoroadenin |
| GT | Gene Targeting |
| HR | Homologous Recombination |
| NHEJ | Non-Homologous End Joining (DSB repair pathway) |
| NPTII | Neomycin PhosphoTransferase conferring resistance to Kanamycin/G418 |
| PI | Propidium iodide |
| ssDNA | single-stranded DNA |
| τ | Half-life |
| Y2H | Yeast two Hybrid system |
References
- Palecek, J.; Vidot, S.; Feng, M.; Doherty, A.J.; Lehmann, A.R. The Smc5-Smc6 DNA Repair Complex. Bridging of the Smc5-Smc6 Heads by the KLEISIN, Nse4, and Non-Kleisin Subunits. J. Biol. Chem. 2006, 281, 36952–36959. [Google Scholar] [CrossRef] [PubMed]
- Alt, A.; Dang, H.Q.; Wells, O.S.; Polo, L.M.; Smith, M.A.; McGregor, G.A.; Welte, T.; Lehmann, A.R.; Pearl, L.H.; Murray, J.M.; et al. Specialized Interfaces of Smc5/6 Control Hinge Stability and DNA Association. Nat. Commun. 2017, 8, 14011. [Google Scholar] [CrossRef]
- Vondrova, L.; Kolesar, P.; Adamus, M.; Nociar, M.; Oliver, A.W.; Palecek, J.J. A Role of the Nse4 Kleisin and Nse1/Nse3 KITE Subunits in the ATPase Cycle of SMC5/6. Sci. Rep. 2020, 10, 9694. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, J.; Haluska, C.; Zhang, X.; Wang, L.; Liu, G.; Wang, Z.; Jin, D.; Cheng, T.; Wang, H.; et al. Cryo-EM Structures of Smc5/6 in Multiple States Reveal Its Assembly and Functional Mechanisms. Nat. Struct. Mol. Biol. 2024, 31, 1532–1542. [Google Scholar] [CrossRef]
- Shintomi, K.; Hirano, T. How Are Cohesin Rings Opened and Closed? Trends Biochem. Sci. 2007, 32, 154–157. [Google Scholar] [CrossRef]
- Kurze, A.; Michie, K.A.; Dixon, S.E.; Mishra, A.; Itoh, T.; Khalid, S.; Strmecki, L.; Shirahige, K.; Haering, C.H.; Löwe, J.; et al. A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion. EMBO J. 2011, 30, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Griese, J.J.; Witte, G.; Hopfner, K.-P. Structure and DNA Binding Activity of the Mouse Condensin Hinge Domain Highlight Common and Diverse Features of SMC Proteins. Nucleic Acids Res. 2010, 38, 3454–3465. [Google Scholar] [CrossRef]
- Uhlmann, F. SMC Complexes: From DNA to Chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef]
- Hirano, T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell 2016, 164, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, J.; Taylor, E.; Palecek, J.; Fousteri, M.; Andrews, E.A.; Sweeney, S.; Shinagawa, H.; Watts, F.Z.; Lehmann, A.R. Composition and Architecture of the Schizosaccharomyces Pombe Rad18 (Smc5-6) Complex. Mol. Cell. Biol. 2005, 25, 172–184. [Google Scholar] [CrossRef]
- Lelkes, E.; Jemelková, J.; Holá, M.; Štefanovie, B.; Kolesár, P.; Vágnerová, R.; Dvořák Tomaštíková, E.; Pecinka, A.; Angelis, K.J.; Paleček, J.J. Characterization of the Conserved Features of the NSE6 Subunit of the Physcomitrium patens SMC5/6 Complex. Plant J. 2023, 115, 1084–1099. [Google Scholar] [CrossRef]
- Vaculíková, J.; Holá, M.; Králová, B.; Lelkes, E.; Štefanovie, B.; Vágnerová, R.; Angelis, K.J.; Paleček, J.J. NSE5 Subunit Interacts with Distant Regions of the SMC Arms in the Physcomitrium patens SMC5/6 Complex. Plant J. 2024, 119, 1481–1493. [Google Scholar] [CrossRef]
- Holá, M.; Vágnerová, R.; Angelis, K.J. Kleisin NSE4 of the SMC5/6 Complex Is Necessary for DNA Double Strand Break Repair, but Not for Recovery from DNA Damage in Physcomitrella (Physcomitrium patens). Plant Mol. Biol. 2021, 107, 355–364. [Google Scholar] [CrossRef]
- Knight, C.D.; Cove, D.J.; Cumming, A.C.; Quatrano, R.S. Moss Gene Technology. In Molecular Plant Biology: A Practical Approach; Oxford University Press: Oxford, UK, 2002; Chapter 14; Volume 2. [Google Scholar]
- Holá, M.; Kozák, J.; Vágnerová, R.; Angelis, K.J. Genotoxin Induced Mutagenesis in the Model Plant Physcomitrella patens. BioMed Res. Int. 2013, 2013, 535049. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef]
- Cove, D.J.; Perroud, P.-F.; Charron, A.J.; McDaniel, S.F.; Khandelwal, A.; Quatrano, R.S. Somatic Hybridization in the Moss Physcomitrella patens Using PEG-Induced Protoplast Fusion. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot5141. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J.; Schild, A.; Ashton, N.W.; Hartmann, E. Genetic and Physiological Studies of The Effect of Light on The Development of The Moss, Physcomitrella patens. Photochem. Photobiol. 1978, 27, 249–254. [Google Scholar] [CrossRef]
- Trouiller, B.; Schaefer, D.G.; Charlot, F.; Nogue, F. MSH2 Is Essential for the Preservation of Genome Integrity and Prevents Homeologous Recombination in the Moss Physcomitrella patens. Nucleic Acids Res. 2006, 34, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Mallett, D.R.; Chang, M.; Cheng, X.; Bezanilla, M. Efficient and Modular CRISPR-Cas9 Vector System for Physcomitrella patens. Plant Direct 2019, 3, e00168. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.J.; Bednarova, K.; Kozakova, L.; Liao, C.; Guerineau, M.; Colnaghi, R.; Vidot, S.; Marek, J.; Bathula, S.R.; Lehmann, A.R.; et al. Interactions between the Nse3 and Nse4 Components of the SMC5-6 Complex Identify Evolutionarily Conserved Interactions between MAGE and EID Families. PLoS ONE 2011, 6, e17270. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Trouiller, B.; Charlot, F.; Choinard, S.; Schaefer, D.G.; Nogue, F. Comparison of Gene Targeting Efficiencies in Two Mosses Suggests That It Is a Conserved Feature of Bryophyte Transformation. Biotechnol. Lett. 2007, 29, 1591–1598. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Vidali, L. Efficient Polyethylene Glycol (PEG) Mediated Transformation of the Moss Physcomitrella patens. J. Vis. Exp. 2011, e2560. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Jeggo, P.; Lobrich, M. The Influence of Heterochromatin on DNA Double Strand Break Repair: Getting the Strong, Silent Type to Relax. DNA Repair 2010, 9, 1273–1282. [Google Scholar] [CrossRef]
- Peng, X.P.; Lim, S.; Li, S.; Marjavaara, L.; Chabes, A.; Zhao, X. Acute Smc5/6 Depletion Reveals Its Primary Role in rDNA Replication by Restraining Recombination at Fork Pausing Sites. PLoS Genet. 2018, 14, e1007129. [Google Scholar] [CrossRef]
- Hola, M.; Vagnerova, R.; Angelis, K.J. Mutagenesis during Plant Responses to UVB Radiation. Plant Physiol. Biochem. 2015, 93, 29–33. [Google Scholar] [CrossRef]
- Kushnirov, V.V. Rapid and Reliable Protein Extraction from Yeast. Yeast 2000, 16, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelis, K.J.; Holá, M.; Vágnerová, R.; Vaculíková, J.; Paleček, J.J. The Phenotype of Physcomitrium patens SMC6 Mutant with Interrupted Hinge Interactions. Genes 2025, 16, 1091. https://doi.org/10.3390/genes16091091
Angelis KJ, Holá M, Vágnerová R, Vaculíková J, Paleček JJ. The Phenotype of Physcomitrium patens SMC6 Mutant with Interrupted Hinge Interactions. Genes. 2025; 16(9):1091. https://doi.org/10.3390/genes16091091
Chicago/Turabian StyleAngelis, Karel J., Marcela Holá, Radka Vágnerová, Jitka Vaculíková, and Jan J. Paleček. 2025. "The Phenotype of Physcomitrium patens SMC6 Mutant with Interrupted Hinge Interactions" Genes 16, no. 9: 1091. https://doi.org/10.3390/genes16091091
APA StyleAngelis, K. J., Holá, M., Vágnerová, R., Vaculíková, J., & Paleček, J. J. (2025). The Phenotype of Physcomitrium patens SMC6 Mutant with Interrupted Hinge Interactions. Genes, 16(9), 1091. https://doi.org/10.3390/genes16091091






