Sex-Specific Polygenic Risk Scores and Replication in a Model-Free Analysis of Schizophrenia Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Sex-Specific Heterogeneity
2.3. Polygenic Risk Scores
2.4. Prediction
2.5. Replication
3. Results and Discussion
3.1. Variant by Variant Analysis
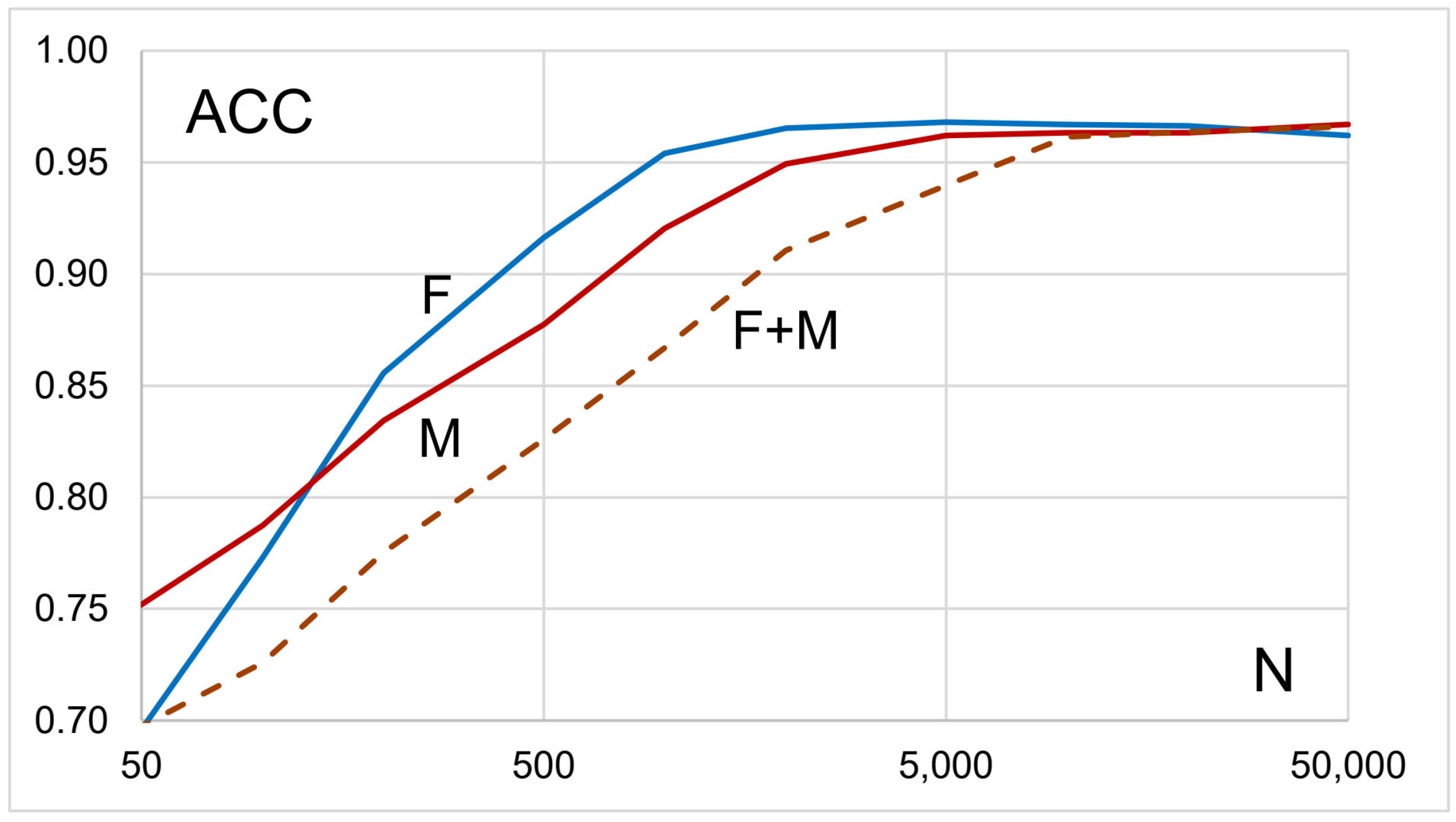
3.2. Polygenic Risk Scores
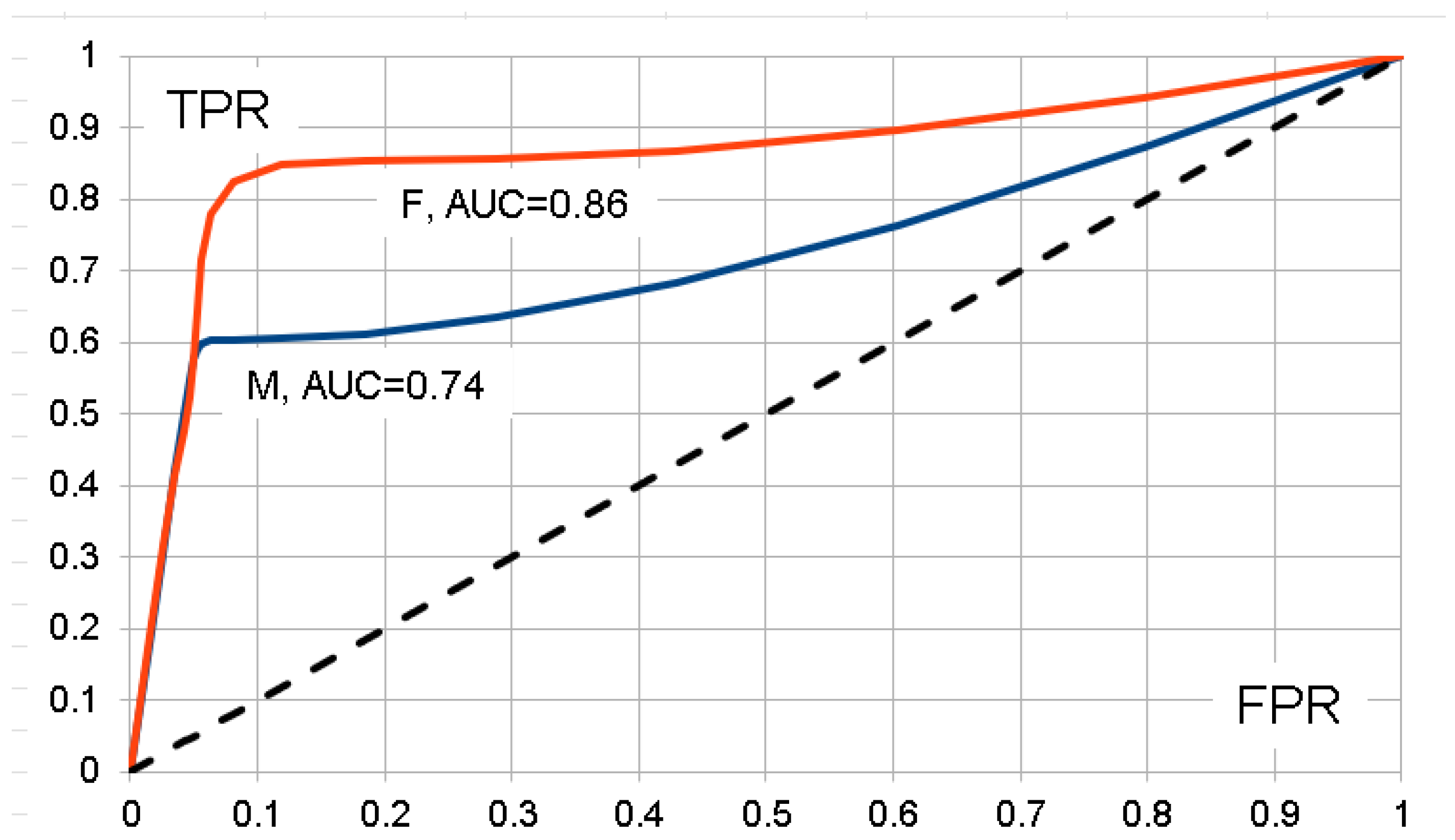
3.3. Replication
- The CUL9 gene was recently shown to be one of five genes causally related to schizophrenia [28].
- In older literature, the PTPRZ1 gene was genetically associated with schizophrenia, and PTPRZ1-transgenic mice exhibited molecular and cellular changes implicated in the pathogenesis of schizophrenia [29].
- In a bivariate meta-analysis, MED19 and other genes were found to be associated with schizophrenia [30].
- Darier disease, an autosomal dominant skin disorder, is caused by mutations in the ATP2A2 gene [31]. In a population-based study, relatives of individuals with Darier disease had a significantly higher risk of having bipolar disorder than relatives of matched individuals from the general population, suggesting that genetic variability within the ATP2A2 gene also confers susceptibility for bipolar disorder [31], but no such association was found with schizophrenia. Our results now strongly demonstrate that the ATP2A2 gene is in fact associated with schizophrenia.
3.4. Limitations and Future Research Directions
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PRS | Polygenic risk score |
| OR | Odds ratio |
| PPV | Positive predictive value |
| NPV | Negative predictive value |
| ACC | Prediction accuracy |
| TPR | True prediction ratio |
| FPR | False prediction ratio |
| ROC | Receiver operating characteristic |
| AUC | Area under the ROC curve |
References
- Zoghbi, A.W.; Dhindsa, R.S.; Goldberg, T.E.; Mehralizade, A.; Motelow, J.E.; Wang, X.; Alkelai, A.; Harms, M.B.; Lieberman, J.A.; Markx, S.; et al. High-impact rare genetic variants in severe schizophrenia. Proc. Natl. Acad. Sci. USA 2021, 118, e2112560118. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; Legge, S.E.; Rees, E.; Walters, J.T.R.; O’Donovan, M.C. Genomic findings in schizophrenia and their implications. Mol. Psychiatry 2023, 28, 3638–3647. [Google Scholar] [CrossRef] [PubMed]
- Ott, J.; Sun, D. Multilocus association analysis under polygenic models. Int. J. Data Min. Bioinform. 2012, 6, 482–489. [Google Scholar] [CrossRef] [PubMed]
- MacLean, C.J.; Ploughman, L.M.; Diehl, S.R.; Kendler, K.S. A new test for linkage in the presence of locus heterogeneity. Am. J. Hum. Genet. 1992, 50, 1259–1266. [Google Scholar]
- Hoh, J.; Ott, J. Scan statistics in genome-wide scan for complex trait loci. In Scan Statistics: Methods and Applications; Glaz, J., Pozdnyakov, V., Wallenstein, S., Eds.; Birkhäuser: Boston, MA, USA, 2009; pp. 195–202. [Google Scholar]
- Hoh, J.; Wille, A.; Ott, J. Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res. 2001, 11, 2115–2119. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Privé, F.; Arbel, J.; Vilhjálmsson, B.J. LDpred2: Better, faster, stronger. Bioinformatics 2021, 36, 5424–5431. [Google Scholar] [CrossRef]
- Privé, F.; Arbel, J.; Aschard, H.; Vilhjálmsson, B.J. Identifying and correcting for misspecifications in GWAS summary statistics and polygenic scores. Hum. Genet. Genom. Adv. 2022, 3, 100136. [Google Scholar] [CrossRef]
- Ge, T.; Chen, C.-Y.; Ni, Y.; Feng, Y.-C.A.; Smoller, J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019, 10, 1776. [Google Scholar] [CrossRef]
- Lloyd-Jones, L.R.; Zeng, J.; Sidorenko, J.; Yengo, L.; Moser, G.; Kemper, K.E.; Wang, H.; Zheng, Z.; Magi, R.; Esko, T.; et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019, 10, 5086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Privé, F.; Vilhjálmsson, B.; Speed, D. Improved genetic prediction of complex traits from individual-level data or summary statistics. Nat. Commun. 2021, 12, 4192. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience 2019, 8, giz082. [Google Scholar] [CrossRef]
- Zabad, S.; Gravel, S.; Li, Y. Fast and accurate Bayesian polygenic risk modeling with variational inference. Am. J. Hum. Genet. 2023, 110, 741–761. [Google Scholar] [CrossRef]
- Martinelli, A.; Leone, S.; Baronio, C.M.; Archetti, D.; Redolfi, A.; Adorni, A.; Caselani, E.; D’Addazio, M.; Di Forti, M.; Laffranchini, L.; et al. Sex differences in schizophrenia spectrum disorders: Insights from the DiAPAson study using a data-driven approach. Soc. Psychiatry Psychiatr. Epidemiol. 2025, 60, 1983–1997. [Google Scholar] [CrossRef]
- Koch, E.; Nyberg, L.; Lundquist, A.; Pudas, S.; Adolfsson, R.; Kauppi, K. Sex-specific effects of polygenic risk for schizophrenia on lifespan cognitive functioning in healthy individuals. Transl. Psychiatry 2021, 11, 520. [Google Scholar] [CrossRef]
- Mas-Bermejo, P.; Papiol, S.; Torrecilla, P.; Lavín, V.; Kwapil, T.R.; Barrantes-Vidal, N.; Rosa, A. Sex-specific association between schizophrenia polygenic risk and subclinical schizophrenia-related traits. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2025, 136, 111161. [Google Scholar] [CrossRef]
- Jiang, J.-C.; Singh, K.; Nitin, R.; Davis, L.K.; Wray, N.R.; Shah, S. Sex-specific association between genetic risk of psychiatric disorders and cardiovascular diseases. Circ. Genom. Precis. Med. 2024, 17, e004685. [Google Scholar] [CrossRef]
- Privé, F.; Vilhjálmsson, B.J.; Aschard, H.; Blum, M.G.B. Making the most of clumping and thresholding for polygenic scores. Am. J. Hum. Genet. 2019, 105, 1213–1221. [Google Scholar] [CrossRef]
- Yarkoni, T.; Westfall, J. Choosing prediction over explanation in psychology: Lessons from machine learning. Perspect. Psychol. Sci. 2017, 12, 1100–1122. [Google Scholar] [CrossRef]
- Lo, A.; Chernoff, H.; Zheng, T.; Lo, S.H. Why significant variables aren’t automatically good predictors. Proc. Natl. Acad. Sci. USA 2015, 112, 13892–13897. [Google Scholar] [CrossRef]
- Plomin, R.; von Stumm, S. Polygenic scores: Prediction versus explanation. Mol. Psychiatry 2022, 27, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Jarauta, E.; Bea-Sanz, A.M.; Marco-Benedi, V.; Lamiquiz-Moneo, I. Genetics of hypercholesterolemia: Comparison between familial hypercholesterolemia and hypercholesterolemia nonrelated to LDL receptor. Front. Genet. 2020, 11, 554931. [Google Scholar] [CrossRef] [PubMed]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesth. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Agresti, A. An Introduction to Categorical Data Analysis, 3rd ed.; Wiley: Hoboken, NJ, USA, 2019; p. xiii. 375p. [Google Scholar]
- Ning, J.; Wang, X.; Ott, J. Digenic analysis confirms known and uncovers novel schizophrenia risk genes. Med. Res. Arch. 2024, 12. [Google Scholar] [CrossRef]
- Nakamura, T.; Takata, A. The molecular pathology of schizophrenia: An overview of existing knowledge and new directions for future research. Mol. Psychiatry 2023, 28, 1868–1889. [Google Scholar] [CrossRef]
- Takahashi, N.; Sakurai, T.; Bozdagi-Gunal, O.; Dorr, N.P.; Moy, J.; Krug, L.; Gama-Sosa, M.; Elder, G.A.; Koch, R.J.; Walker, R.H.; et al. Increased expression of receptor phosphotyrosine phosphatase-β/ζ is associated with molecular, cellular, behavioral and cognitive schizophrenia phenotypes. Transl. Psychiatry 2011, 1, e8. [Google Scholar] [CrossRef]
- Greco, L.A.; Reay, W.R.; Dayas, C.V.; Cairns, M.J. Pairwise genetic meta-analyses between schizophrenia and substance dependence phenotypes reveals novel association signals with pharmacological significance. Transl. Psychiatry 2022, 12, 403. [Google Scholar] [CrossRef]
- Cederlöf, M.; Bergen, S.E.; Långström, N.; Larsson, H.; Boman, M.; Craddock, N.; Östberg, P.; Lundström, S.; Sjölander, A.; Nordlind, K.; et al. The association between Darier disease, bipolar disorder, and schizophrenia revisited: A population-based family study. Bipolar Disord. 2015, 17, 340–344. [Google Scholar] [CrossRef]
- Turgut, A.O.; Koca, D. The effects of case/control ratio and sample size on genome-wide association studies: A simulation study. Vet. Med. Sci. 2024, 10, e1444. [Google Scholar] [CrossRef]
- Carlisle, S.M.; Qin, H.; Hendrickson, R.C.; Muwanguzi, J.E.; Lefkowitz, E.J.; Kennedy, R.E.; Yan, Z.; Yacoubian, T.A.; Benveniste, E.N.; West, A.B.; et al. Sex-based differences in the activation of peripheral blood monocytes in early Parkinson disease. Npj Park. Dis. 2021, 7, 36. [Google Scholar] [CrossRef]
- Koch, E.; Kauppi, K.; Chen, C.-H. Candidates for drug repurposing to address the cognitive symptoms in schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2023, 120, 110637. [Google Scholar] [CrossRef] [PubMed]
- Hukic, D.S.; Lavebratt, C.; Olsson, E.; Östenson, C.-G.; Eriksson, S.V.; Erlinge, D.; Schalling, M.; Ösby, U. Troponin T levels associated with genetic variants in NOTCH2 and MTNR1B in women with psychosis. Psychiatry Res. 2017, 250, 217–220. [Google Scholar] [CrossRef]
- Saha, E.; Ben Guebila, M.; Fanfani, V.; Fischer, J.; Shutta, K.H.; Mandros, P.; DeMeo, D.L.; Quackenbush, J.; Lopes-Ramos, C.M. Gene regulatory networks reveal sex difference in lung adenocarcinoma. Biol. Sex Differ. 2024, 15, 62. [Google Scholar] [CrossRef]
- Hjorthøj, C.; Compton, W.; Starzer, M.; Nordholm, D.; Einstein, E.; Erlangsen, A.; Nordentoft, M.; Volkow, N.D.; Han, B. Association between cannabis use disorder and schizophrenia stronger in young males than in females. Psychol. Med. 2023, 53, 7322–7328. [Google Scholar] [CrossRef]
| Known Phenotype | Prediction of “Case” | Prediction of “Control” |
|---|---|---|
| case | a | b |
| control | c | d |
| N | n, F | %, F | n, M | %, M | n, F + M | %, F + M |
|---|---|---|---|---|---|---|
| 100 | 207 | 22.2% | 446 | 20.6% | 851 | 27.5% |
| 200 | 131 | 14.1% | 371 | 17.1% | 702 | 22.7% |
| 500 | 65 | 7.0% | 260 | 12.0% | 534 | 17.2% |
| 1000 | 37 | 4.0% | 167 | 7.7% | 382 | 12.3% |
| 2000 | 26 | 2.8% | 91 | 4.2% | 323 | 10.4% |
| 5000 | 34 | 3.6% | 72 | 3.3% | 126 | 4.1% |
| 10,000 | 30 | 3.2% | 84 | 3.9% | 110 | 3.6% |
| 20,000 | 29 | 3.1% | 83 | 3.8% | 121 | 3.9% |
| 50,000 | 35 | 3.8% | 71 | 3.3% | 105 | 3.4% |
| chr | Variant | bp | Gene | Variant Type |
|---|---|---|---|---|
| 5 | rs2250599 | 14,861,105 | ANKH | intron, genic upstream transcript |
| 6 | rs16896326 | 43,185,671 | CUL9 | intron, genic upstream transcript |
| 6 | rs11962528 | 67,901,631 | LOC105377845 | intron |
| 7 | rs1196509 | 121,975,547 | PTPRZ1 | intron |
| 10 | rs978554 | 99,742,899 | CUTC | intron |
| 11 | rs10896638 | 57,703,469 | MED19 | 500B downstream transcript |
| 12 | rs9540 | 110,351,050 | ATP2A2 | 3’UTR, benign |
| 14 | rs2293792 | 73,249,825 | PAPLN | intron, genic upstream transcript |
| 15 | rs16948431 | 64,957,012 | ANKDD1A | intron |
| 17 | rs7209186 | 69,327,056 | ABCA5 | 5’UTR, genic upstream transcript |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ott, A.; Ott, J. Sex-Specific Polygenic Risk Scores and Replication in a Model-Free Analysis of Schizophrenia Data. Genes 2025, 16, 1080. https://doi.org/10.3390/genes16091080
Ott A, Ott J. Sex-Specific Polygenic Risk Scores and Replication in a Model-Free Analysis of Schizophrenia Data. Genes. 2025; 16(9):1080. https://doi.org/10.3390/genes16091080
Chicago/Turabian StyleOtt, Anna, and Jurg Ott. 2025. "Sex-Specific Polygenic Risk Scores and Replication in a Model-Free Analysis of Schizophrenia Data" Genes 16, no. 9: 1080. https://doi.org/10.3390/genes16091080
APA StyleOtt, A., & Ott, J. (2025). Sex-Specific Polygenic Risk Scores and Replication in a Model-Free Analysis of Schizophrenia Data. Genes, 16(9), 1080. https://doi.org/10.3390/genes16091080







