Abstract
Background/Objectives: Blood flows through the body and reaches all tissues, contributing to homeostasis and physiological functions. Providing information and understanding on how the transcriptome of whole blood behaves in response to physiological or pathological stimuli is critical. Methods: We collected blood from four healthy individuals and performed long-read RNA sequencing (lrRNA-seq) for the precise identification and expression quantification of RNA variants. Moreover, we compared two genome references: the Genome Reference Consortium Human Build 38 (GRCh38) and the Telomere-to-Telomere (T2T) assembly of the CHM13 cell line (T2T-CHM13). Results: With GRCh38, we could identify an average of about 46,000 genes, 1.3-fold more genes than T2T-CHM13. Similarly, we identified about 185,000 isoforms with GRCh38 and 140,000 with T2T-CHM13, finding similar differences for full splice match (FSM) and incomplete splice match (ISM) transcript isoforms. There were about 90,000 novel isoforms for GRCh38 and 70,000 for T2T-CHM13, 47% and 50% of the total number of identified isoforms, respectively. Differences in isoform numbers between GRCh38 and T2T-CHM13 were identified for the subcategories “Genic Genomic”, “Intergenic”, and “Genic Intron”. Using GRCh38, we generally identified a higher number of non-coding isoforms, as well as a higher number of isoforms aligning within intron and intergenic regions. Nonetheless, GRCh38 might incur false positive results, and T2T-CHM13 is likely more accurate for genome sequences in the repetitive regions. Conclusions: LrRNA-seq is a valid method for the identification of novel isoforms in blood, and this study is a first step toward the creation of a comprehensive database of the structure and expression of transcript isoforms for optimized predictive medicine.
1. Introduction
Blood flows through the body and reaches all peripheral tissues. It contributes to body homeostasis and maintains physiological functions. We reported previously that, in the long-term gene expression profiles of whole blood of patients undergoing regular medical health check-ups, a group of genes was stably expressed over time. These genes were mainly involved in immune system pathways, including antigen cell presentation and interferon-related signaling; meanwhile, in another group of genes, we observed altered gene expression related to altered cellular machinery signaling [1].
This longitudinal examination showed that the whole blood gene expression signature can reveal unmanifested physiological changes. Moreover, it is now possible to predict the presence or absence of disease from RNA information in the blood. However, the accuracy of disease prediction is questionable because the basis of RNA information in the blood is incomplete. We focused on the long-read RNA-seq method, which provides more accurate isoform RNA information than short-read RNA-seq [2]. Moreover, since many isoforms are still undiscovered, lrRNA-seq is a powerful method to identify new isoforms [3,4]. The large number of variants contribute to the intricacy of the transcriptome, influenced by mechanisms such as RNA editing, alternative splicing, and alternative polyadenylation. RNA isoforms with unique structural properties, functionality, and stability are generated [5,6].
The identification, characterization, and quantification of these variants can reveal how full blood gene expression behaves in physiological settings and under different pathological conditions [3]. Based on this background, we collected full blood from four individuals who underwent medical health checkups, and analyzed their overall RNA variant composition and expression using PacBio lrRNA-seq. Because lrRNA-seq can technically read RNA sequences larger than 1 kb, it can detect RNA information with high accuracy. However, the ability of RNA-seq to efficiently identify and quantify gene expression depends on the comprehensiveness of the selected genomic reference. In this regard, we focused on the comparison between two genome references, the Genome Reference Consortium Human Build 38 (GRCh38) [7] and the Telomere-to-Telomere (T2T) assembly of the CHM13 cell line (T2T-CHM13) [8,9]. GRCh38 has been widely used [10], is well annotated, and is used as the standard tool for many existing data [11,12]. The T2T-CHM13 reference provides more accurate genome sequences, including repetitive regions, such as telomeres and centromeres [13], newly added and corrected 238 Mb of sequence, and 1956 new genes [8,14]. In principle, GRCh38 should be chosen for comparison with existing datasets, while T2T-CHM13 should be chosen when focusing on the analysis of new genes and repetitive regions [14,15,16].
Where possible, it would be best to analyze using both references and compare the results. At present, the appropriateness of the reference may vary depending on the research purpose and analysis target. T2T-CHM13 is likely to become more widespread in the future, and a transition to T2T-CHM13 is expected in the long term [14,17]. Regardless of the reference chosen, it is important to clearly record the version of the used reference genome and consider it when interpreting results and comparing them with other studies. Few studies used whole blood for whole transcriptome analysis using long-read RNA-seq, and only two papers used GRCh38 as a reference [18,19]. Whole blood was also used as the source of RNA for another two studies, but in these cases, the target genes of RNA in whole blood had already been determined, and only a limited portion of the RNA was analyzed [20,21]. In addition, previous long-read RNA-seq annotations have used either the GRCh38 reference, or, in a limited number of studies, a reference that integrates both GRCh38 and T2T-CHM13 genome assemblies [14,22,23]; however, these studies used preestablished cell lines or deposited datasets for analysis, and none examined whole blood samples.
Against this background, this is the first comparative study using long-read RNA-seq from whole peripheral blood and data processed using GRCh38 and T2T-CHM13 genomic references for the whole transcriptome. Our future objective is to identify the RNA isoforms present in healthy human adult blood samples to create a database that can show transcript isoform variations when whole human blood is used as a source. This preliminary study is a necessary step toward building such a platform for optimized predictive medicine.
2. Materials and Methods
2.1. Study Design
In this study, we withdrew by standard phlebotomy the peripheral whole blood from 4 participants (males: n = 2; females: n = 2. 1_M36, 2_M_105, 3_M127, 4_M166), who visited the Public Central Hospital of Matto Ishikawa for medical examinations in 2016, as previously described [1,24]. The participants, who were considered healthy and had no apparent disease, were informed about the study, and their consent was obtained. Clinical parameters were recorded for all 4 individuals (Supplementary Table S1). The study protocol information and ethical approvals are described in the “Institutional Review Board Statement” section.
2.2. Whole Blood RNA Isolation and lrRNA-Seq Library Creation
Patient whole blood was collected in 2016 and stored in PAXgene Blood RNA Tubes, IVD (762165, QIAGEN, Hilden, Germany). Total RNA was extracted from stored blood according to the PAXgene Blood RNA Kit (762164, QIAGEN) protocol. A total RNA quality check was performed according to the Agilent RNA 6000 Nano Kit (5067-1511, Agilent Technologies, Inc., Santa Clara, CA, USA) protocol and analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). After confirming that the RIN value of RNA was ≥7, cDNA was synthesized and amplified using the Iso-Seq Express 2.0 kit (103-071-500, PacBio, Menlo Park, CA, USA). The prepared cDNA was then ligated with an SMRT adapter at the 5′ and 3′ ends using the SMRTbell prep kit 3.0 (102-182-700, PacBio). After library preparation, the cDNA libraries were sequenced on a Sequel IIe system (PacBio). The numbers of total reads for samples 1_M36, 2_M_105, 3_M127, and 4_M166 were, respectively: 4,456,301; 4,544,423; 4,288,374; and 4,245,914.
2.3. Sequence Analysis Processing
Raw PacBio Iso-seq data (BAM files) were aligned to the GRCh38 or T2T-CHM13 genome (GCA_009914755.4 [T2T-CHM13v2.0]) using pbmm2 v10.0 (Release Version 1.10.0 · PacificBiosciences/pbmm2 · GitHub). Isoform analysis was performed with Isoseq v4.0.0 (https://github.com/PacificBiosciences/IsoSeq, accessed on 13 October 2023) and classification of full-length isoforms with Pigeon v1.0.0 (https://isoseq.how/classification/pigeon.html, accessed on 12 December 2022) from the aligned data was performed. Sequence analysis processing was performed at the Research and Development Center for Precision Medicine, Tsukuba University. SQANTI3 (ver. 5.0, GitHub—ConesaLab/SQANTI3: Tool for the Quality Control of Long-Read Defined Transcriptomes) was used by the analysis pipeline of Bioengineering Lab. Co. (Sagamihara, Japan). The isoforms were annotated with gene transfer format (GTF) files downloaded from the UCSC Genome Browser [10]: for HG38, https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.ncbiRefSeq.gtf.gz (accessed on 29 February 2024); for T2T, https://hgdownload.soe.ucsc.edu/goldenPath/hs1/bigZips/genes/hs1.ncbiRefSeq.gtf.gz (accessed on 29 February 2024). Both GRCh38 and T2T-CHM13 genome references can be downloaded from the NCBI. Users should note that genomic coordinates and annotations differ between the two references, and mapping and annotation should be performed consistently to ensure reproducibility [14]. The mRNA transcript isoforms were filtered for a count value of at least 2 unique molecules per million of reads. All plots were created by using Origin, Version 2021 (OriginLab Corporation, Northampton, MA, USA), except if stated otherwise.
3. Results
3.1. Known and Novel Isoforms Identified in Whole Blood
The clinical parameters for all four individuals (Supplementary Table S1) did not show major health concerns, and they were considered disease-free. Full blood was used for the identification and quantification of known and novel isoforms by long-read RNA sequencing. The differences and similarities among the four samples were depicted by PCA analysis using either GRCh38 or T2T-CHM13 (Supplementary Figure S1). The samples were approximately equidistant and evenly distributed across the PCA plot, with no distinct clusters, indicating that each sample exhibited unique transcriptomic variability. The SQANTI classification categorizes the transcript isoforms based on splice junctions (SJs) [25,26], briefly (Figure 1A): isoforms matching precisely the reference isoforms are identified as full splice match (FSM), while isoforms labeled as incomplete splice match (ISM) have matching consecutive splice junctions, as well as missing splice junctions at the 5′, 3′, or both ends [26]. New isoforms of already known genes are categorized as Novel in Catalog (NIC), containing known splice junctions or new splice junctions from known donors and acceptors: Novel Not in Catalog (NNC) with new donors and/or acceptors [25]. Genic genomic isoforms partially overlap exons and an intron in an annotated gene, genic intron isoforms are located within an intron, Antisense isoforms overlap with coding sequences on the opposite strand of the annotated gene, Fusion isoforms bridge two annotated loci, and Intergenic isoforms do not overlap with the genomic interval spanned by an annotated gene (Figure 1A).
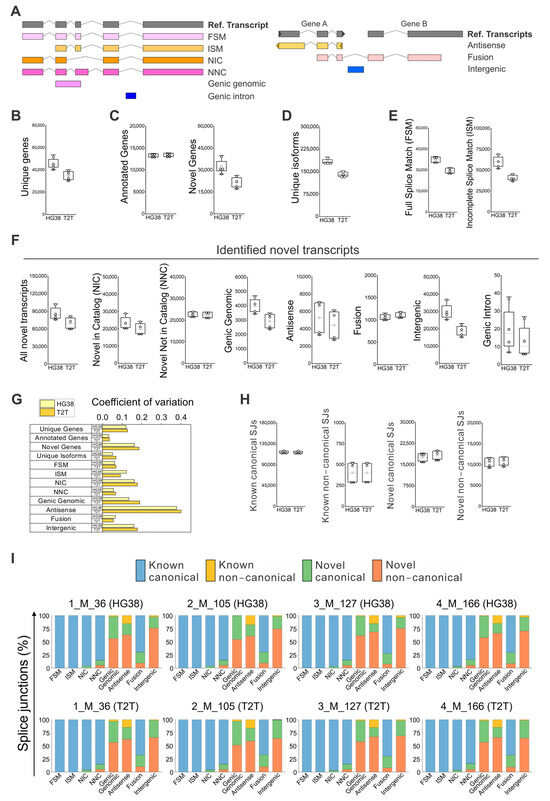
Figure 1.
Identification of known and novel genes and isoforms in peripheral blood using GRCh38 (HG38) and T2T-CHM13 (T2T) genome references. (A) SQANTI classification for isoforms [25,26], briefly: Full Splice Match (FSM), Incomplete Splice Match (ISM), Novel in Catalog (NIC), Novel Not in Catalog (NNC), Genic genomic, Genic intron, Antisense, Fusion, and Intergenic. Iso-Seq data analyzed with GRCh38 and T2T-CHM13 references for the identification of: (B) all unique genes, (C) annotated and novel genes within all unique genes, (D) identified unique isoforms; and (E) FSM and ISM isoforms. (F) Identified novel isoforms, NIC, NNC, antisense, fusion, genic genomic, intergenic, and genic intron isoforms. (G) Coefficient of variation (CV) calculated for each type of isoform (A–F) for determining the distribution variability of detected isoforms among individuals, which is a value indicative of physiological diversity among individuals. The coefficient of variation was defined as the ratio of the standard deviation to the average of distribution [27]. (H) Absolute numbers of “Known canonical SJs”, “Known non-canonical SJs”, “Novel canonical SJs”, and “Novel non-canonical SJs” quantification by GRCh38 and T2T-CHM13. (I) Detail of splice junctions’ percentage by type of SJs for known and novel subcategories of isoforms. (B–F,H) N = 4; white boxes show the interquartile range (25–75% percentile range), the median is designated by the horizontal line, and the mean is indicated by the white square. Boundaries of the whiskers are based on the 1.5 interquartile range value, and all observed data points outside the limit of the whiskers are considered outliers.
We found that the Iso-Seq whole blood data analyzed with GRCh38 could identify about 46,000 genes, 1.3-fold more genes than T2T-CHM13 (Figure 1B). This difference is not due to the identified known genes, but to the difference in identified novel genes (Figure 1C). Moreover, a similar increase was confirmed for unique isoforms (Figure 1D)—about 185,000 isoforms were identified with GRCh38 versus 140,000 isoforms with T2T-CHM13. This difference was observed for FSM and ISM (Figure 1E) isoforms, respectively, with a ratio of about 1.3- and 1.5-fold in favor of GRCH38. The total number of novel isoforms was about 90,000 for GRCh38 and 70,000 for T2T-CHM13 (Figure 1F). Of these, 47% were the novel isoforms identified with GRCh38, and 50% were the novel isoforms identified with T2T-CHM13. When we analyzed the average numbers of novel isoforms, similar amounts were identified for “NIC”, “NNC”, “Antisense”, and “Fusion”, while the largest difference in isoform numbers between GRCh38 and T2T-CHM13 were identified for the subcategories “Genic Genomic”, “Intergenic”, and “Genic Intron”. When we focused on the variability of the identified numbers of isoforms among individuals, a value indicative of physiological differences, we observed a high degree of variation in the number of “Antisense” isoforms, independent of the reference genome used (Figure 1G), followed also by a moderate level of variation in “NIC”, “Genic Genomic”, and “Intergenic” isoforms.
3.2. Comparison of Splice Junction Profiles and Isoform Characteristics Using GRCh38 and T2T-CHM13 References in SQANTI Analysis
SQANTI identifies isoforms based on their splice junctions—the base pair GT at the beginning and AG at the end of the intron is located in 98.9% of all introns in the human genome [28]. Specifically, GT–AG, GC–AG, and AT–AC are canonical splicing acceptor/donor pairs found in most human introns [28,29], while the remaining combinations are defined as non-canonical splice junctions [25]. In our data, no difference was observed between GRCh38 and T2T-CHM13 for the absolute numbers of “Known canonical SJs”, “Known non-canonical SJs”, “Novel canonical SJs”, and “Novel non-canonical SJs” (Figure 1H). When we verified in detail the splice junctions percentage by type of SJs, GRCh38 and T2T-CHM13 performed similarly (Figure 1I); among patients, FSM, ISM, and NIC isoforms were characterized mainly by “Known canonical SJs”; “NNC”, and “Fusion” isoforms by about 70–80% of “Known canonical SJs”; and the remainder by “Novel canonical SJs” and “Novel non-canonical SJs”. “Genic Genomic”, “Intergenic”, and “Antisense” isoforms all had a high percentage of “Novel canonical SJs” and “Novel non-canonical SJs”. In the case of “Antisense” variants, we also detected about 20% of “Known non-canonical SJs” (Figure 1I).
No difference was observed between individuals regarding the read counts for all isoform length distributions (Figure 2A). However, in general, the GRCh38 number of counts per isoform was higher than the one obtained with T2T-CHM13. Moreover, we confirmed a difference in the read counts of the “ISM” and “Intergenic” variant subcategories among individuals based on the GRCh38 reference, whereas we did not see the same difference when we used T2T-CHM13 (Figure 2B). Finally, in the analysis of read counts of the categories “Multi-Exon” and “Mono-Exon”, we confirmed a higher number of counts for “Mono-Exon” variants when the GRCh38 reference was used (Figure 2C). A similar overall pattern was confirmed for the distribution density of isoforms when GRCh38 or T2T-CHM13 were used (Figure 2D).
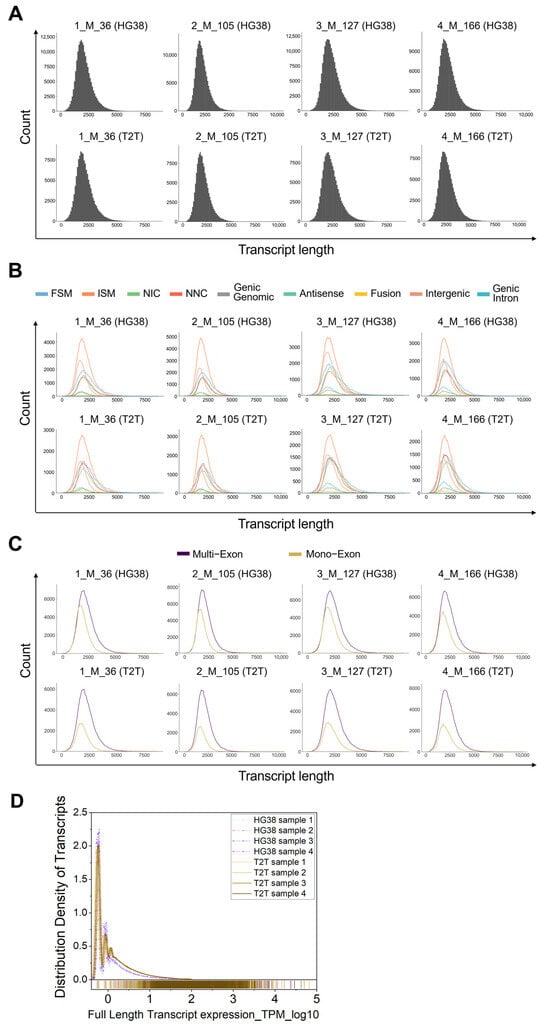
Figure 2.
Count quantification per isoform length in peripheral blood using GRCh38 (HG38) and T2T-CHM13 (T2T) genome references. (A) Reading counts for all isoform length distributions for each sample, HG38 versus T2T-CHM13. (B) Reading counts of isoforms for each subcategory among individuals by GRCh38 and T2T-CHM13 references. (C) Analysis of reading counts of “Multi-Exon” and “Mono-Exon”. (A–C) The plots were depicted as obtained from the SQANTI3 output. (D) Distribution density of isoforms when GRCh38 or T2T-CHM13 were used for each sample.
3.3. Features of Coding and Non-Coding Isoforms
When we analyzed the number of isoforms identified per gene, we found the main difference between GRCh38 and T2T-CHM13 to be in the number of genes with 1 isoform, while a similar number of isoforms were obtained for those genes producing 2–3, 4–5, or more than 5 isoforms (Figure 3A). Variability of the number of isoforms per gene among blood samples remained low for both GRCh38 and T2T-CHM13 references (Figure 3B).
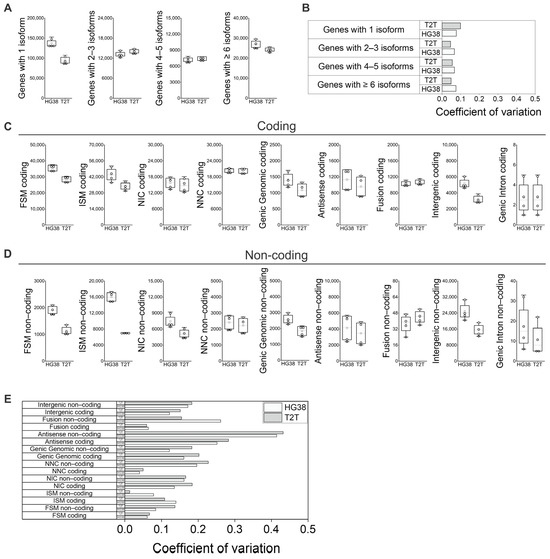
Figure 3.
Features of coding and non-coding isoforms in peripheral blood using GRCh38 (HG38) and T2T-CHM13 (T2T) genome references. (A) Number of isoforms per gene; (B) variability of number of isoforms per gene among samples for both GRCh38 and T2T-CHM13 references. (C) Number of coding isoforms by subcategories, and (D) number of non-coding isoforms by subcategories. (E) Identified isoform number variability among individuals in the context of differences between “Coding” and “Non-coding” subcategories for both GRCh38 and T2T-CHM13 references. (A,C,D) N = 4; white boxes show the interquartile range (25–75% percentile range), the median is designated by the horizontal line, and the mean is indicated by the white square. Boundaries of the whiskers are based on the 1.5 interquartile range value; all observed data points outside the limit of the whiskers are considered outliers. (B,E) The coefficient of variation was defined as the ratio of standard deviation to the average of distribution [27].
When the main variant categories are further separated into “Coding” and “Non-coding” we observe further differences between the references and among individuals (Figure 3C,D): the differences between GRCh38 and T2T-CHM13 in the number of variants identified for all categories in the “Coding” context (Figure 3C) show no differences from the overall counts (“Coding” plus “Non-coding”; Figure 1E,F) However, when the “Non-coding” subcategory was analyzed separately for each variant category (Figure 3D), GRCh38 generally identified a higher number of isoforms. Specifically, we see a higher difference in “FSM Non-coding”, “ISM Non-coding”, “NIC Non-coding”, and “Genic Intron Non-coding” when compared to the “Coding” subcategories (Figure 3C,D). When we analyzed the identified isoform number variability among individuals in the context of differences between the “Coding” and “Non-coding” subcategories, we observed an increase in the coefficient of variation in the “Non-coding” subcategories of “FSM”, “NNC”, “Antisense”, and “Fusion” when compared to the “Coding” subcategories (Figure 3C–E).
3.4. Features of Isoform Subcategories in Whole Blood
Following the subcategorization analysis of FSM isoforms (Figure 4A) [25,26], we observed that the “Alternative 5′ end”, the “Alternative 5′ and 3′ end, and the “Mono-exon” are identified 1.7 to 2 times more often when we used GRCh38 than when we used T2T-CHM13; this points to limitations in the identification of alternative transcription start site (TSS) sequences of alternative 5′ ends when the T2T-CHM13 reference is used (Figure 4B). Regarding the ISM subcategories (Figure 4C), we observed a tendency to identify a higher number of isoforms when the GRCh38 reference was used. More strikingly, the number of recorded “Mono-exons” in the ISM subcategory was on average 2.8 higher when GRCh38 was compared to T2T-CHM13 (Figure 4D). Then, we analyzed the subcategories applied to novel isoforms (Figure 5A). No difference was observed between GRCh38 and T2T-CHM13 regarding the NIC subcategories “Combination of known SJs”, “Combination of known splice sites”, and “Intron retention”; while for the NIC subcategories “Mono-exon by intron retention” and “Mono-exon”, GRCh38 identified 1.4 and 1.6 times more isoforms than the T2T-CHM13 reference (Figure 5B). No difference was found in the number of identified NNC isoforms with “At least one novel splice site” or “Intron retention” (Figure 5C). Finally, regarding the categories “Genic Genomic”, “Antisense”, “Fusion”, and “Intergenic”, the main differences were generally related to the higher number of identified “mono-exons” by using GRCH38 (Figure 5D,E,G), while the “multi-exon” and “Fusion: intron retention” were characterized by similar numbers for both GRCh38 and T2T-CHM13 references (Figure 5D–G). In addition, we identified genes showing the highest diversity of full-length transcript isoforms in each sample and compared the results between the GRCh38 and T2T-CHM13 reference genomes. We found that the genes with the greatest isoform diversity were largely consistent across healthy individuals and between the two references (Supplementary Table S2). Notably, genes such as AKAP13, PTPRC, LRRK2, and IQGAP1 are associated with the MAPK cascade, FYB1, FCGR3B, ADGRE5, LILRB2, CTSS, and HLA-E are involved in immune responses, and PTPRC, LRRK2, and HLA-E participate in the positive regulation of tumor necrosis factor production (Supplementary Table S3).
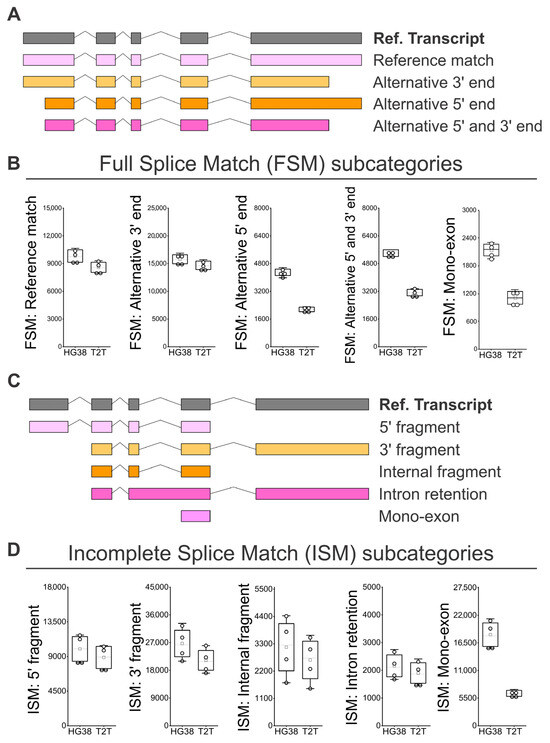
Figure 4.
Identification of FSM and ISM isoform subcategories in peripheral blood using GRCh38 (HG38) and T2T-CHM13 (T2T) genome references. (A) SQANTI classification for subcategories of FSM isoforms [25,26]: “Alternative 5′ end”, “Alternative 5′ and 3′ end, and “Mono-exon”. (B) Number of isoforms by subcategories of FSM. (C) SQANTI classification for subcategories of ISM isoforms [25,26]: “5′ fragment”, “3′ fragment”, “Internal fragment”, “Intron retention”, and “Mono-exon”. (D) Number of isoforms by subcategories of ISM. (B,D) N = 4; white boxes show the interquartile range (25–75% percentile range), the median is designated by the horizontal line, and the mean is indicated by the white square. Boundaries of the whiskers are based on the 1.5 interquartile range value, and all observed data points outside the limit of the whiskers are considered outliers.
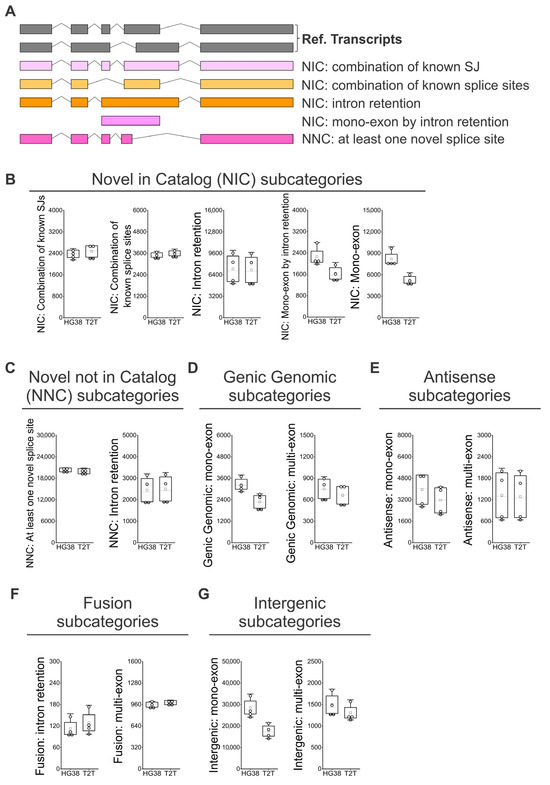
Figure 5.
Identification of NIC and NNC isoform subcategories in peripheral blood using GRCh38 (HG38) and T2T-CHM13 (T2T) genome references. (A) SQANTI classification for subcategories of NIC and NNC isoforms [25,26]: “Combination of known SJs”, “Combination of known splice sites”, “Intron retention”, “Mono-exon by intron retention”, and “At least one novel splice site”. (B) Number of isoforms by subcategories of NIC. (C) Number of isoforms by subcategories of NNC. (D) Number of isoforms by subcategories of Genic Genomic. (E) Number of isoforms by subcategories of Antisense. (F) Number of isoforms by subcategories of Fusion. (G) Number of isoforms by subcategories of Intergenic. (B–G) N = 4; white boxes show the interquartile range (25–75% percentile range), the median is designated by the horizontal line, and the mean is indicated by the white square. Boundaries of the whiskers are based on the 1.5 interquartile range value, and all observed data points outside the limit of the whiskers are considered outliers.
3.5. Expression Levels of lrRNA-Seq Identified Isoforms
We selected eight genes of interest that are related to mRNA splicing and modulation to verify the expression of each known and novel isoform when GRCh38 (Figure 6A) or T2T-CHM13 is used as the reference genome (Figure 6B). The expression in transcripts per million (TPM) of each single isoform for each gene and for each sample was obtained by lrRNA-seq, and the summation of all isoforms’ expressions corresponded to the overall expression of the considered gene. We can observe that the counts and expressions of known and novel isoforms can be very variable among genes. Moreover, within the analysis of the same gene, counts and expressions of isoforms are very different among individuals, even those with no apparent disease. The trend of overall expressions is similar whether the GRCh38 or T2T-CHM13 reference is used (Figure 6A,B). In some cases, for example in the analysis of the FMR1 gene, in two samples (3_M127 and 4_M166), the number of isoforms identified by using GRCh38 (Figure 6A) is higher than when using T2T-CHM13 reference (Figure 6B), but on the other hand, the expression of one of the isoforms for the same gene for both 3_M127 and 4_M166 samples is about 10 times higher when T2T-CHM13 is used. In addition, since fusion genes are critical for the development of new diagnoses and new drugs, we selected four fusion genes for testing the isoform expression (Figure 6C,D): CTBS::GNG5, involved in nonneoplastic hematologic disorders in the lymph node and spleen, as well as in epithelial lesions in bladder, skin, and lung [30]; PRIM1::NACA, related to chronic myeloid leukemia [31]; IFNAR2::IL10RB, involved in lung nonneoplastic epithelial disorder [32]; and DNAJC4::NUDT22, related to lung adenocarcinoma [33]. Since the blood samples were from healthy individuals, we did not expect high levels of isoform expression related to these fusion genes. Nonetheless, we could confirm that the trend of detected expressions also approximately matched whether GRCh38 or T2T-CHM13 was used (Figure 6C,D). For CTBS::GNG5 and PRIM1::NACA, the results were very similar, while for IFNAR2::IL10RB and DNAJC4::NUDT22, a certain degree of difference was noticed. We could also confirm the detection in blood of other isoforms derived from fusion genes specific to different organs, such as: FOXP1::EIF4E3, ABI2::CYP20A1, and CTSD::IFITM10, related to breast adenocarcinoma [33,34,35,36]; HAVCR1::TIMD4 and JAK3::INSL3, related to kidney adenocarcinoma [34,36,37]; and HACL1::COLQ and SIDT2::TAGLN, related to lymph node nonneoplastic disorders [30,36].
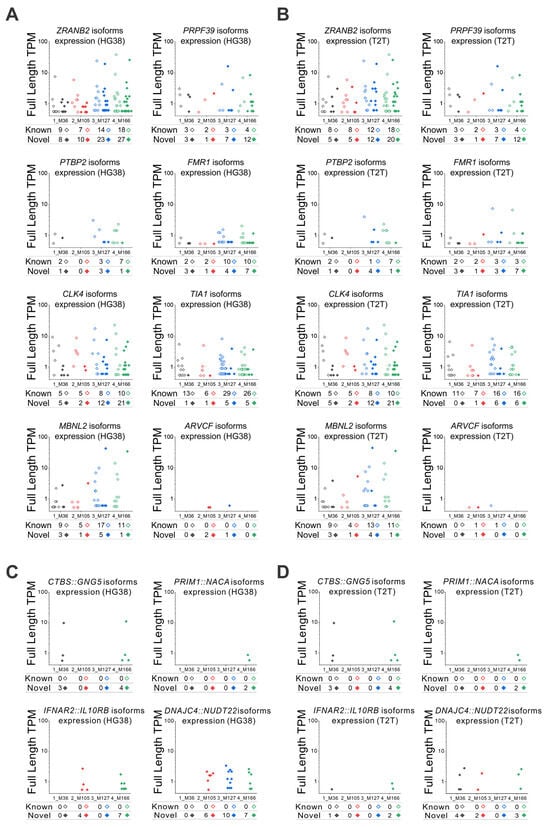
Figure 6.
Long-read RNA-seq identified isoforms and expression levels. (A,B) Expression levels obtained by lrRNA-seq for each known and novel isoform of the selected eight genes, using both (A) GRCh38 (HG38) and (B) T2T-CHM13 (T2T) methods. Isoform expression of four fusion genes by (C) HG38 and (D) T2T methods: CTBS::GNG5, PRIM1::NACA, IFNAR2::IL10RB, and DNAJC4::NUDT22. The total number of isoforms and the TPM expression of each single isoform are shown in the graphs. The total numbers of known and novel isoforms are annotated for each blood sample at the base of the graphs.
These preliminary analyses need to be validated, since some of the fusion genes detected in healthy individuals might be artifacts. The expression identified by lrRNA-seq methods should be checked in terms of expression variations with other methods, such as DNA microarray or short-read RNA-seq. In this context, for the gene expression validation of the eight genes depicted in Figure 6A,B, we performed short-read RNA sequencing for all four samples, annotated with both GRCh38 and T2T-CHM13 references, and calculated the TPM values (Supplementary Figure S2).
The correlation between the genes’ expressions identified by short-read RNA-seq and the extent of expression identified for each gene/sample by lrRNA-seq (TPM expression of each single isoform together with the number of isoforms in their totality) was confirmed. These results confirm that the summation of isoforms’ expressions (overall gene expression) identified by lrRNA-seq methods can be cross-validated in terms of expression variations with short-read RNA-seq methods. In addition, for the eight novel isoforms shown in Figure 6A,B, we always used the sequence with the highest TPM value detected in each sample as the reference for mapping short-read RNA-seq data against both GRCh38 and T2T-CHM13 references. In cases where the sequence information was incomplete, we used regions shared across samples as the reference. The results of this mapping are summarized in Supplementary Figure S3. It should be noted that the novel isoforms identified by long-read sequencing largely overlap with the genomic regions of known genes. Therefore, most of these isoforms can also be detected by short-read sequencing, although detection may be limited for certain low-abundance isoforms, such as MBNL2 (Supplementary Figure S3). To validate the fusion genes depicted in Figure 6C,D, we used the longest isoform detected for each of the four fusion genes, annotated against both GRCh38 and T2T-CHM13, as the reference sequence for short-read RNA-seq analysis. We confirmed that the long fusion isoforms identified by long-read sequencing were also detectable with short reads, as the reads mapped to multiple regions within the fusion (Supplementary Figure S4).
4. Discussion
Blood flows all through the body and is in constant contact with every organ via the cardiovascular system. It consists of many different cell types, which differ in the amount of RNA they contain; this influences the phenotype and function of these cells [38], and these changes reflect the physiological or pathological condition of the host [39,40]. A large percentage of potential novel RNA isoforms are still undiscovered and need to be characterized [3]. Isoform variations and differences in splicing are key factors to be investigated. The lrRNA-seq analysis allows us to identify new isoforms as well as their expression, which could be associated with disease risk [41,42], clarifying how the transcriptome performs in response to pathological stimuli [43]. In this context, the identification of new RNA isoforms and their relative expression analysis in whole blood is an important means to evaluate an individual’s biological condition and assess unmanifested diseases. Therefore, we used full blood from four healthy individuals with no apparent disease and examined their RNA variant composition and expression using long-read RNA-seq. Two different human reference genome mapping techniques, GRCh38 and T2T-CHM13, were used to assess the lrRNA-seq analysis performance and downstream results. We found that the Iso-Seq data analyzed with GRCh38 could identify about 46,000 genes, 1.3-fold more genes than T2T-CHM13. The difference was mainly due to the higher number of identified novel genes; a similar increase was confirmed for identified unique isoforms. Among all the isoforms identified, 47% were the novel isoforms when GRCh38 was used, and 50% were the novel isoforms when T2T-CHM13 was used. Independent of the reference gene used, a large number of novel isoforms can still be discovered and characterized in full blood. Regarding novel isoforms, similar numbers were identified for “NIC”, “NNC”, “Antisense”, and “Fusion”, while the largest difference in isoform numbers between GRCh38 and T2T-CHM13 were for the subcategories “Genic Genomic”, “Intergenic”, “Genic Intron”, and “Mono-Exon”. For all of these, GRCh38 allowed the identification of a higher number when compared to T2T-CHM13. When T2T-CHM13 was used as the reference, we noticed a reduced ability to identify isoforms aligning in the intron and intergenic regions. This was unexpected, since we thought T2T-CHM13 would perform better than GRCh38 due to being a more gapless sequence reference of the human genome [8]. Interestingly, we observed no discrepancies in the recognition of splice junctions, where both GRCh38 and T2T-CHM13 performed similarly, so we deduced that the above differences were a matter of sequence alignment in the non-exonic areas. Another difference between GRCh38 and T2T-CHM13 was observed when following the subcategorization analysis of FSM and ISM isoforms. In particular, T2T-CHM13 was outperformed by GRCh38 in the identification of isoforms with alternative 5′ ends or with a missing exon at the 5′ end. The subcategories “Antisense”, “NIC”, “Genic Genomic”, and “Intergenic” had the highest variability in the number of isoforms detected among individuals; it follows that the isoforms within these subcategories were the most related to physiological differences among individuals. In this context, “Antisense” isoforms could be used as drug targets since they regulate gene expression and are involved in cancer promotion or suppression [44,45], while novel “Genic Genomic”, and “Intergenic” isoforms may also regulate gene expression as well as chromatin remodeling [46,47]. In addition, similar statistics for both GRCh38 and T2T-CHM13 were obtained, indicating that the interindividual differences were due to the blood features and not to the selected reference genome. Lastly, we studied the expression of eight selected genes of interest related to mRNA splicing, maturation, and process stability as well as four fusion genes related to diseases. Among genes, the counts and expressions of known and novel isoforms were very variable, but unexpectedly, also within the same gene, counts and expressions of isoforms were very different among individuals even in the absence of apparent disease. In terms of gene expression, similar levels could be observed independent of the reference genome used. For some genes, the known/novel ratio of the number of isoforms is very high; one such example is TIA1, which is an extensively studied gene (1210 hits on PubMed [27 August 2024]) [48]. Meanwhile, for the gene ZRANB2, a much less investigated gene (39 hits on PubMed [27 August 2024]) [48], many novel isoforms were identified, in contrast to the known isoforms. We confirmed that blood examination would allow the identification of isoforms derived from fusion genes for both hematological diseases and organ-specific diseases. For long-read RNA-seq, it is also necessary to assess and confirm that the RNA information is correct by using other techniques, and to identify which reference assembly performs better and in which conditions. GRCh38 generally identifies a higher number of isoforms when compared to T2T-CHM13, but regions of the GRCh38 reference are sometimes artificial, with incomplete assembly or incorrect sequences [49,50,51], and this may have led to the identification of a higher number of false positives. In this regard, we confirmed that the validation of long-read RNA-seq information can be performed by short-read RNA-seq with both GRCh38 and T2T-CHM13 references for both the detection and the quantification of genes and the detection of fusion genes. The correlation between the genes’ expressions identified by short-read RNA-seq and the expressions identified for each gene/sample by lrRNA-seq was confirmed. When the sequence information was incomplete, we used regions shared across samples as the reference. Short-read RNA-seq may fail to fully detect the isoform due to insufficient read length or fragmentation; nonetheless, even low-abundance isoforms are still detectable, and the detection sensitivity may differ between GRCh38 and T2T-CHM13 references. Issues related to isoform identification can occur due to blood cell heterogeneity—cell type composition varies between individuals, especially when patients are in disease states, so the proportions of immune cell types might affect the transcriptome and potentially induce mistakes in biomarker identification. This complexity means that analysis in the future needs to take account of cellular heterogeneity for proper interpretation. Computational deconvolution methods, such as BayesPrism, can estimate the proportions of different cell types and help to interpret bulk RNA-seq data more accurately [1,52]. Gender differences might also impact the analysis of long-read RNA sequencing. However, with only four samples in this pilot study (two males and two females), it is premature to perform any stratified analysis. Previous transcriptome studies with short-read RNA-seq of whole blood [53] and in immune cells [54] have reported variation in expression of immune response genes, cell cycle regulation, and oxidative stress. Future studies with larger sample sizes will be needed to assess potential gender effects reliably. Another limitation is the matter of selection bias—since blood samples are derived from participants of Japanese ethnicity, the data obtained should be carefully examined before conclusions are generalized for an international context. From a statistical perspective, four samples represent a small cohort, and these findings should be considered an initial pilot study. Broader validation across larger and more diverse populations is needed to definitively determine which reference genome, GRCh38 or T2T-CHM13, is preferable.
5. Conclusions
In this preliminary assessment of the whole blood full transcriptome, GRCh38 annotation appears to identify a higher number of isoforms when compared to T2T-CHM13. These differences are more evident for regions related to intron and intergenic sequencies, as well as for the identification of mono-exons. Moreover, knowing how these gene products are generated is crucial, for example in cases of isoforms derived from fusion genes, which are important when investigating diseases. We performed this first analysis to create a database that will allow users to view different isoforms, associated exons, and the endpoints of transcription on a map with two different genome references.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16091075/s1, Table S1: Clinical parameters of the 4 individuals whose blood was examined for gene expression and isoform identification; Table S2: Identified genes with the highest number of diverse full-length transcript isoforms for each sample: comparison between the two reference genomes GRCh38 and T2T-CHM13; Table S3: Enrichment Analysis by Gene Ontology Biological Process of identified genes with the highest number of diverse full-length transcript isoforms in blood of healthy individuals (Supplementary Table S2); Figure S1: Transcriptomic profiles depicted by PCA; Figure S2: Gene expression validation by short-read RNA sequencing of the 8 genes depicted in Figure 6A,B; Figure S3: Short-read RNA-seq isoform validation of the 8 genes depicted in Figure 6A,B; Figure S4: Short-read RNA-seq validation of the 4 fusion genes depicted in Figure 6C,D.
Author Contributions
Conceptualization, H.O., A.N., Y.S., Y.T., S.I., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; methodology, H.O., A.N., Y.S., Y.T., S.I., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; software, S.I. and T.T. (Takuro Tamura); validation, H.O., A.N., Y.S., Y.T., S.I., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; formal analysis, H.O., A.N., Y.T., S.I., T.T. (Takuro Tamura) and S.K.; investigation, H.O., A.N., Y.T., T.T. (Takuro Tamura) and S.K.; resources, H.O., A.N., Y.S., T.H., N.T., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; data curation, H.O., A.N., Y.T., S.I., H.Y., T.H. and N.T.; writing—original draft preparation, H.O., A.N., Y.S., Y.T., H.Y., T.H., N.T., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; writing—review and editing, H.O., A.N., Y.S., Y.T., S.I., H.Y., T.H., N.T., T.-A.S., T.U., S.N., T.T. (Toshinari Takamura), T.Y., T.T. (Takuro Tamura), K.M. and S.K.; visualization, A.N., S.I. and H.Y.; supervision, Y.S. and S.K.; project administration, S.K.; funding acquisition, S.K. All authors contributed to the drafting of the work and reviewed the draft critically for the intellectual content, have read and agreed to the published version of the manuscript, and agreed to be accountable for all aspects of the work to ensure accuracy and integrity. H.O. and A.N. contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the department of Information-Based Medicine Development, Graduate School of Medical Sciences, Kanazawa University.
Institutional Review Board Statement
The study protocol was approved by the review committees of Kanazawa University (Human Genome and Genetic Analysis Committee; protocol number: 2015-007; approval date: 25 November 2015, https://www.med.kanazawa-u.ac.jp/staff/ethics/genome/index.html) and by Matto Ishikawa Public Central Hospital. The principles of the Declaration of Helsinki were followed during the study, and the clinical trial was registered in the University Hospital Medical Information Network (UMIN; Clinical Trials Registry, no.: UMIN000051647).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Original dataset available upon reasonable request from the authors.
Acknowledgments
We sincerely thank Nami Nishiyama, Mai Nakayama, Yui Kawamura, Riei Tsurumi, and Miu Awaki for their excellent technical assistance.
Conflicts of Interest
Taka-Aki Sato and Kenichi Matsubara are affiliated with the company iLAC Co., Ltd. (Tsukuba 305–0821, Japan). Seiji Nakamura is affiliated with the company DNA Chip Research Inc. (Tokyo 105–0022, Japan).
Abbreviations
The following abbreviations are used in this manuscript:
| CV | Coefficient of Variation |
| FSM | Full Splice Match |
| GRCh38 | Genome Reference Consortium Human Build 38 |
| GTF | Gene Transfer Format |
| ISM | Incomplete Splice Match |
| lrRNA-seq | long-read RNA sequencing |
| NIC | Novel in Catalog |
| NNC | Novel Not in Catalog |
| SJs | Splice Junctions |
| T2T | Telomere-to-Telomere |
| T2T-CHM13 | Telomere-to-Telomere assembly of the CHM13 cell line |
| TPM | Transcripts per Million |
| TSS | Transcription Start Site |
References
- Sakai, Y.; Nasti, A.; Takeshita, Y.; Okumura, M.; Kitajima, S.; Honda, M.; Wada, T.; Nakamura, S.; Takamura, T.; Tamura, T.; et al. Eight-Year Longitudinal Study of Whole Blood Gene Expression Profiles in Individuals Undergoing Long-Term Medical Follow-Up. Sci. Rep. 2021, 11, 16564. [Google Scholar] [CrossRef]
- Pardo-Palacios, F.J.; Wang, D.; Reese, F.; Diekhans, M.; Carbonell-Sala, S.; Williams, B.; Loveland, J.E.; De María, M.; Adams, M.S.; Balderrama-Gutierrez, G.; et al. Systematic Assessment of Long-Read RNA-Seq Methods for Transcript Identification and Quantification. Nat. Methods 2024, 21, 1349–1363. [Google Scholar] [CrossRef]
- De Paoli-Iseppi, R.; Gleeson, J.; Clark, M.B. Isoform Age-Splice Isoform Profiling Using Long-Read Technologies. Front. Mol. Biosci. 2021, 8, 711733. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Liu, Q.; Perdomo, J.E.; Fang, L.; Wang, K. A Survey of Algorithms for the Detection of Genomic Structural Variants from Long-Read Sequencing Data. Nat. Methods 2023, 20, 1143–1158. [Google Scholar] [CrossRef]
- Christofi, T.; Zaravinos, A. RNA Editing in the Forefront of Epitranscriptomics and Human Health. J. Transl. Med. 2019, 17, 319. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.D.; Nam, S.W. Pathogenic Diversity of RNA Variants and RNA Variation-Associated Factors in Cancer Development. Exp. Mol. Med. 2020, 52, 582–593. [Google Scholar] [CrossRef]
- National Library of Medicine Homo Sapiens Genome Assembly GRCh38. Available online: https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000001405.26/ (accessed on 4 September 2024).
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- National Human Genome Research Institute Telomere-to-Telomere. Available online: https://www.genome.gov/about-genomics/telomere-to-telomere (accessed on 4 September 2024).
- Perez, G.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser Database: 2025 Update. Nucleic Acids Res. 2025, 53, D1243–D1249. [Google Scholar] [CrossRef] [PubMed]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.-C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de Novo Haploid Genome Assemblies Demonstrates the Enduring Quality of the Reference Assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Behera, S.; LeFaive, J.; Orchard, P.; Mahmoud, M.; Paulin, L.F.; Farek, J.; Soto, D.C.; Parker, S.C.J.; Smith, A.V.; Dennis, M.Y.; et al. FixItFelix: Improving Genomic Analysis by Fixing Reference Errors. Genome Biol. 2023, 24, 31. [Google Scholar] [CrossRef] [PubMed]
- Altemose, N.; Logsdon, G.A.; Bzikadze, A.V.; Sidhwani, P.; Langley, S.A.; Caldas, G.V.; Hoyt, S.J.; Uralsky, L.; Ryabov, F.D.; Shew, C.J.; et al. Complete Genomic and Epigenetic Maps of Human Centromeres. Science 2022, 376, eabl4178. [Google Scholar] [CrossRef]
- Aganezov, S.; Yan, S.M.; Soto, D.C.; Kirsche, M.; Zarate, S.; Avdeyev, P.; Taylor, D.J.; Shafin, K.; Shumate, A.; Xiao, C.; et al. A Complete Reference Genome Improves Analysis of Human Genetic Variation. Science 2022, 376, eabl3533. [Google Scholar] [CrossRef] [PubMed]
- Gershman, A.; Sauria, M.E.G.; Guitart, X.; Vollger, M.R.; Hook, P.W.; Hoyt, S.J.; Jain, M.; Shumate, A.; Razaghi, R.; Koren, S.; et al. Epigenetic Patterns in a Complete Human Genome. Science 2022, 376, eabj5089. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Ding, T.; Chang, T.; Ruan, J.; Yang, J.; Ma, M.; Liu, J.; Liu, Z.; Jiao, S.; Wu, J.; et al. Nanopore Sequencing with T2T-CHM13 for Accurate Detection and Preventing the Transmission of Structural Rearrangements in Highly Repetitive Heterochromatin Regions in Human Embryos. Clin. Transl. Med. 2024, 14, e1612. [Google Scholar] [CrossRef]
- Alkan, C.; Carbone, L.; Dennis, M.; Ernst, J.; Evrony, G.; Girirajan, S.; Leung, D.C.Y.; Cheng, C.C.; MacAlpine, D.; Ni, T. Implications of the First Complete Human Genome Assembly. Genome Res. 2022, 32, 595–598. [Google Scholar] [CrossRef]
- Hu, Y.; Fang, L.; Chen, X.; Zhong, J.F.; Li, M.; Wang, K. LIQA: Long-Read Isoform Quantification and Analysis. Genome Biol. 2021, 22, 182. [Google Scholar] [CrossRef] [PubMed]
- Grigorev, K.; Nelson, T.M.; Overbey, E.G.; Houerbi, N.; Kim, J.; Najjar, D.; Damle, N.; Afshin, E.E.; Ryon, K.A.; Thierry-Mieg, J.; et al. Direct RNA Sequencing of Astronaut Blood Reveals Spaceflight-Associated m6A Increases and Hematopoietic Transcriptional Responses. Nat. Commun. 2024, 15, 4950. [Google Scholar] [CrossRef]
- Cornaby, C.; Montgomery, M.C.; Liu, C.; Weimer, E.T. Unique Molecular Identifier-Based High-Resolution HLA Typing and Transcript Quantitation Using Long-Read Sequencing. Front. Genet. 2022, 13, 901377. [Google Scholar] [CrossRef]
- Schwenk, V.; Leal Silva, R.M.; Scharf, F.; Knaust, K.; Wendlandt, M.; Häusser, T.; Pickl, J.M.A.; Steinke-Lange, V.; Laner, A.; Morak, M.; et al. Transcript Capture and Ultradeep Long-Read RNA Sequencing (CAPLRseq) to Diagnose HNPCC/Lynch Syndrome. J. Med. Genet. 2023, 60, 747–759. [Google Scholar] [CrossRef]
- Hoyt, S.J.; Storer, J.M.; Hartley, G.A.; Grady, P.G.S.; Gershman, A.; de Lima, L.G.; Limouse, C.; Halabian, R.; Wojenski, L.; Rodriguez, M.; et al. From Telomere to Telomere: The Transcriptional and Epigenetic State of Human Repeat Elements. Science 2022, 376, eabk3112. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Zou, Y.; Zhang, S.; Xia, M.; Fu, L.; Vollger, M.R.; Chen, N.-C.; Taylor, D.J.; Harvey, W.T.; et al. Characterization of Large-Scale Genomic Differences in the First Complete Human Genome. Genome Biol. 2023, 24, 157. [Google Scholar] [CrossRef]
- Nasti, A.; Okumura, M.; Takeshita, Y.; Ho, T.T.B.; Sakai, Y.; Sato, T.-A.; Nomura, C.; Goto, H.; Nakano, Y.; Urabe, T.; et al. The Declining Insulinogenic Index Correlates with Inflammation and Metabolic Dysregulation in Non-Obese Individuals Assessed by Blood Gene Expression. Diabetes Res. Clin. Pract. 2024, 208, 111090. [Google Scholar] [CrossRef]
- Tardaguila, M.; De La Fuente, L.; Marti, C.; Pereira, C.; Pardo-Palacios, F.J.; Del Risco, H.; Ferrell, M.; Mellado, M.; Macchietto, M.; Verheggen, K.; et al. SQANTI: Extensive Characterization of Long-Read Transcript Sequences for Quality Control in Full-Length Transcriptome Identification and Quantification. Genome Res. 2018, 28, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Palacios, F.J.; Arzalluz-Luque, A.; Kondratova, L.; Salguero, P.; Mestre-Tomás, J.; Amorín, R.; Estevan-Morió, E.; Liu, T.; Nanni, A.; McIntyre, L.; et al. SQANTI3: Curation of Long-Read Transcriptomes for Accurate Identification of Known and Novel Isoforms. Nat. Methods 2024, 21, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Everitt, B.S.; Skrondal, A. The Cambridge Dictionary of Statistics; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Parada, G.E.; Munita, R.; Cerda, C.A.; Gysling, K. A Comprehensive Survey of Non-Canonical Splice Sites in the Human Transcriptome. Nucleic Acids Res. 2014, 42, 10564–10578. [Google Scholar] [CrossRef]
- Cocquet, J.; Chong, A.; Zhang, G.; Veitia, R.A. Reverse Transcriptase Template Switching and False Alternative Transcripts. Genomics 2006, 88, 127–131. [Google Scholar] [CrossRef]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent Chimeric Fusion RNAs in Non-Cancer Tissues and Cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef]
- Levin, J.Z.; Berger, M.F.; Adiconis, X.; Rogov, P.; Melnikov, A.; Fennell, T.; Nusbaum, C.; Garraway, L.A.; Gnirke, A. Targeted Next-Generation Sequencing of a Cancer Transcriptome Enhances Detection of Sequence Variants and Novel Fusion Transcripts. Genome Biol. 2009, 10, R115. [Google Scholar] [CrossRef]
- Pintarelli, G.; Dassano, A.; Cotroneo, C.E.; Galvan, A.; Noci, S.; Piazza, R.; Pirola, A.; Spinelli, R.; Incarbone, M.; Palleschi, A.; et al. Read-through Transcripts in Normal Human Lung Parenchyma Are down-Regulated in Lung Adenocarcinoma. Oncotarget 2016, 7, 27889–27898. [Google Scholar] [CrossRef]
- Yoshihara, K.; Wang, Q.; Torres-Garcia, W.; Zheng, S.; Vegesna, R.; Kim, H.; Verhaak, R.G.W. The Landscape and Therapeutic Relevance of Cancer-Associated Transcript Fusions. Oncogene 2015, 34, 4845–4854. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liang, W.-W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.-W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238.e3. [Google Scholar] [CrossRef]
- Varley, K.E.; Gertz, J.; Roberts, B.S.; Davis, N.S.; Bowling, K.M.; Kirby, M.K.; Nesmith, A.S.; Oliver, P.G.; Grizzle, W.E.; Forero, A.; et al. Recurrent Read-through Fusion Transcripts in Breast Cancer. Breast Cancer Res. Treat. 2014, 146, 287–297. [Google Scholar] [CrossRef]
- Mitelman, F.; Johansson, B.; Mertens, F. Mitelman Database Chromosome Aberrations and Gene Fusions in Cancer. Available online: https://mitelmandatabase.isb-cgc.org/ (accessed on 3 January 2025).
- The Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar] [CrossRef]
- Sumitomo, S.; Nagafuchi, Y.; Tsuchida, Y.; Tsuchiya, H.; Ota, M.; Ishigaki, K.; Suzuki, A.; Kochi, Y.; Fujio, K.; Yamamoto, K. Transcriptome Analysis of Peripheral Blood from Patients with Rheumatoid Arthritis: A Systematic Review. Inflamm. Regen. 2018, 38, 21. [Google Scholar] [CrossRef]
- De Sota, R.E.; Quake, S.R.; Sninsky, J.J.; Toden, S. Decoding Bioactive Signals of the RNA Secretome: The Cell-Free Messenger RNA Catalogue. Expert Rev. Mol. Med. 2024, 26, e12. [Google Scholar] [CrossRef]
- Cao, X.; Zhou, X.; Chen, S.; Xu, C. Integration of Transcriptomics and Metabolomics Reveals the Responses of the Maternal Circulation and Maternal-Fetal Interface to LPS-Induced Preterm Birth in Mice. Front. Immunol. 2023, 14, 1213902. [Google Scholar] [CrossRef]
- Humphrey, J.; Brophy, E.; Kosoy, R.; Zeng, B.; Coccia, E.; Mattei, D.; Ravi, A.; Efthymiou, A.G.; Navarro, E.; Muller, B.Z.; et al. Long-Read RNA-Seq Atlas of Novel Microglia Isoforms Elucidates Disease-Associated Genetic Regulation of Splicing. medRxiv 2023. [Google Scholar] [CrossRef]
- Aguzzoli Heberle, B.; Brandon, J.A.; Page, M.L.; Nations, K.A.; Dikobe, K.I.; White, B.J.; Gordon, L.A.; Fox, G.A.; Wadsworth, M.E.; Doyle, P.H.; et al. Mapping Medically Relevant RNA Isoform Diversity in the Aged Human Frontal Cortex with Deep Long-Read RNA-Seq. Nat. Biotechnol. 2025, 43, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Ament, I.H.; DeBruyne, N.; Wang, F.; Lin, L. Long-Read RNA Sequencing: A Transformative Technology for Exploring Transcriptome Complexity in Human Diseases. Mol. Ther. 2025, 33, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J.; Zhang, W. Antisense RNA: The New Favorite in Genetic Research. J. Zhejiang Univ. Sci. B 2018, 19, 739–749. [Google Scholar] [CrossRef]
- Liu, B.; Xiang, W.; Liu, J.; Tang, J.; Wang, J.; Liu, B.; Long, Z.; Wang, L.; Yin, G.; Liu, J. The Regulatory Role of Antisense lncRNAs in Cancer. Cancer Cell Int. 2021, 21, 459. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- National Library of Medicine Pubmed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 3 January 2025).
- Miga, K.H.; Newton, Y.; Jain, M.; Altemose, N.; Willard, H.F.; Kent, W.J. Centromere Reference Models for Human Chromosomes X and Y Satellite Arrays. Genome Res. 2014, 24, 697–707. [Google Scholar] [CrossRef]
- Chaisson, M.J.P.; Huddleston, J.; Dennis, M.Y.; Sudmant, P.H.; Malig, M.; Hormozdiari, F.; Antonacci, F.; Surti, U.; Sandstrom, R.; Boitano, M.; et al. Resolving the Complexity of the Human Genome Using Single-Molecule Sequencing. Nature 2015, 517, 608–611. [Google Scholar] [CrossRef]
- Wagner, J.; Olson, N.D.; Harris, L.; McDaniel, J.; Cheng, H.; Fungtammasan, A.; Hwang, Y.-C.; Gupta, R.; Wenger, A.M.; Rowell, W.J.; et al. Curated Variation Benchmarks for Challenging Medically Relevant Autosomal Genes. Nat. Biotechnol. 2022, 40, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.; Wang, Z.; Pe’er, D.; Danko, C.G. Cell Type and Gene Expression Deconvolution with BayesPrism Enables Bayesian Integrative Analysis across Bulk and Single-Cell RNA Sequencing in Oncology. Nat. Cancer 2022, 3, 505–517. [Google Scholar] [CrossRef]
- Aoki, Y.; Taguchi, K.; Anzawa, H.; Kawashima, J.; Ishida, N.; Otsuki, A.; Hasegawa, A.; Baird, L.; Suzuki, T.; Motoike, I.N.; et al. Whole Blood Transcriptome Analysis for Age- and Gender-Specific Gene Expression Profiling in Japanese Individuals. J. Biochem. 2024, 175, 611–627. [Google Scholar] [CrossRef]
- Gal-Oz, S.T.; Maier, B.; Yoshida, H.; Seddu, K.; Elbaz, N.; Czysz, C.; Zuk, O.; Stranger, B.E.; Ner-Gaon, H.; Shay, T. ImmGen Report: Sexual Dimorphism in the Immune System Transcriptome. Nat. Commun. 2019, 10, 4295. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).