1. Introduction
Acid phosphatase (AP) is a water-soluble enzyme secreted by the prostate gland and found in seminal fluid at concentrations that are at least 200 times higher than those in other bodily fluids [
1,
2,
3]. Leveraging this property, forensic scientists have utilised the AP test for presumptive detection of semen since the 1950s [
4,
5]. Like other presumptive tests for bodily fluids such as blood and semen, the AP test can yield false-positive results [
4,
6,
7,
8,
9]. This outcome is not unexpected, as AP is also present in certain vegetable juices and other bodily fluids [
2,
3,
4,
6,
7,
8,
9]. To confirm semen detection after a positive AP result, forensic scientists can examine samples microscopically for spermatozoa and/or conduct immunoassays such as the Seratec
® PSA Semiquant test (Seratec, Göttingen, Germany) to detect prostate-specific antigen and/or the Rapid Stain Identification (RSID)
TM-Semen test (Independent Forensics, Lombard, IL, USA) to detect semenogelin [
10,
11,
12].
Despite its lack of specificity, the AP test has been widely adopted in the forensic community due to its rapid, sensitive, and straightforward methodology, making it especially useful for detecting semen stains in forensic casework. Additionally, the AP test is also more practical for screening large crime exhibits such as bedding compared to other presumptive tests for semen [
3,
4,
6,
9,
13].
The AP test can be applied directly to the surfaces of exhibits such as cotton swabs, but more commonly, it is applied indirectly via a process known as the “press test” [
3,
4,
9,
12,
13]. This process involves pressing moistened blotting or filter paper firmly onto the exhibit’s surface to transfer any semen present to the paper. The paper is then removed and sprayed with AP reagent, which results in a purplish colour in the presence of AP. Specifically, α-naphthyl phosphate is hydrolysed to α-naphthol, which then reacts with Brentamine Fast Blue to form a purple azo dye [
3,
4,
5,
6,
7,
8,
9]. Strong purple colour development indicates a high level of AP activity, suggesting the presence of semen at the corresponding location on the exhibit. The dried filter paper can then be placed over the dried exhibit, and the AP stain is documented on the exhibit using a crayon. This process involves flipping the filter paper back and forth to accurately outline the purple-staining pattern, ensuring it aligns with its original location on the exhibit. Documenting both the paper and the exhibit is essential to enable accurate relocation of AP-positive areas for subsequent confirmatory tests, such as microscopy [
3,
4,
9,
13,
14,
15]. However, this crayon-based documentation process is time-consuming and laborious, particularly when there are multiple stains on a large exhibit.
The present study sought to explore a more practical approach towards documenting AP-positive areas. Saral Wax-Free Transfer Paper (TP) (Saral Paper Corp, Jersey City, NJ, USA), also referred to as graphite paper, has long been used in arts and crafts to transfer designs from one surface to another. Here, we evaluated its use as a tool to outline AP stain boundaries on exhibits. We further assessed whether TP impacts downstream DNA laboratory processes and compared its documenting time against the conventional crayon method.
2. Material and Methods
This study first involved assessing the effects of three different colour pigments (white, yellow and red) of Saral Wax-Free Transfer Paper (TP) on protein immunoassays and DNA processing, followed by an evaluation of TP for documenting AP-positive areas on mocked crime exhibits, consisting of a cotton T-shirt stained with semen.
2.1. Assessing the Effect of TP Pigment on Protein Immunoassays
Seven sections (~1 cm × 1 cm) of TP were prepared for each colour and placed into separate Eppendorf tubes for subsequent protein immunoassays, as described in
Section 2.6.
2.2. Assessing the Effect of TP Pigment on DNA Processing
Seven sections (~1 cm × 1 cm) of TP were prepared for each colour and subjected to Maxwell (
n = 2) and differential DNA extractions (
n = 5), as described in
Section 2.7.
2.3. Assessing the Effect of TP Pigment on Semen Detection via Protein Immunoassays
TP was placed with the pigment side facing down onto a cotton cloth. Firm pressure was applied using shading motions with a pen, causing the pigment from the TP to be transferred to the cotton surface, leaving visible colouration. For each coloured TP, seven cloth sections (~1 cm × 1 cm) were excised and spiked with 5.5 µL of diluted semen (Cat No. IRHUSMS5ML, Innovation Research, Novi, MI, USA) and then allowed to air-dry overnight prior to protein immunoassay testing.
2.4. Assessing the Effect of TP Pigment on DNA Processing from Semen Stains
For each coloured TP, seven cloth sections (~1 cm × 1 cm) coloured with TP pigment (similar to
Section 2.3) were spiked with diluted semen, then subjected to Maxwell (
n = 2) and differential DNA extractions (
n = 5).
2.5. Documentation of AP-Positive Areas on Semen-Stained Cotton T-Shirt
Whatman Grade 1 Cellulose Chromatography Paper (Cat No. 3001-917, Cytiva, Marlborough, MA, USA) was moistened with ultrapure water and placed over the semen stains on the cotton T-shirt. A piece of dry chromatography paper was then placed over the moistened paper for support, and even pressure was applied over the area via a handheld roller. The moistened paper was allowed to air-dry in a fume hood. AP SPOT Test reagent (Lot No. 1969, SERI, Richmond, CA, USA) was sprayed onto the paper, and purple colouration indicated AP activity. Any spreading of the semen stain would have occurred during these wet application phases. The dried paper was placed back over the corresponding area on the dried T-shirt, and the AP-positive areas were documented using either a crayon or TP. For AP mapping using both crayon and TP, competent staff applied moderate, even pressure, consistent with the laboratory’s routine practice for AP documentation. Pressure was not quantitatively measured in this exploratory study but was guided by laboratory SOP to ensure visible markings without fabric damage. For TP documentation, tracing was performed using a standard stylus with a 2 mm tip. The SciPy library (version 1.12.0) was used to conduct a Wilcoxon signed-rank test. Effect size (Cohen’s d) was calculated by the mean difference divided by the pooled standard deviation.
2.6. Protein Immunoassays
Samples were incubated in 100 µL of RSIDTM Universal buffer on a thermomixer at 25 °C for 30 min with agitation at 1000 rpm. After centrifugation (to pellet sperm in semen-containing samples), aliquots of the supernatant were used for testing with the Rapid Stain Identification (RSID)TM-Semen and SERATEC® PSA Semiquant kits.
For the RSIDTM-Semen kit (Lot No. 051023S1), 20 µL of supernatant was diluted to 100 µL with RSIDTM universal buffer.
For the SERATEC® PSA Semiquant kit (Lot No. 230004), 5 µL of supernatant was diluted to 120 µL with PSA Buffer Solution.
Samples were loaded onto their respective test cassettes, and the results were interpretated according to the manufacturer’s protocols. The remaining supernatant and cell pellet were subjected to DNA extraction as described below.
2.7. DNA Extraction
DNA extraction using the DNA IQTM Casework kit (Promega Corporation, Madison, WI, USA) on the Maxwell FSC instrument (Promega) was performed as per the manufacturer’s protocol.
For differential DNA extraction, the cell pellet and the protein supernatant were resuspended in 400 µL of lysis buffer (10 mM Tris, 10 mM EDTA, 2% (
w/
v) SDS, and 20 µL of Proteinase K (20 mg/mL) [
16]. The mixture was incubated for 1 h at 56 °C with agitation. Intact sperm was pelleted by centrifugation, and the supernatant was collected as the epithelial fraction. The sperm pellet was washed and resuspended in 50 µL of phosphate-buffered saline to form the spermic fraction. Both epithelial and spermic fractions were subsequently subjected to Maxwell DNA extraction.
2.8. DNA Quantification, STR-PCR Amplification, and Amplicon Detection
Extracted DNA was quantified using the Quantifiler® Trio DNA Quantification kit (Applied Biosystems, Foster City, CA, USA) on the QuanStudioTM 7 Flex Real-Time PCR System (Applied Biosystems). DNA extracts were amplified in a 29-cycle reaction using the GlobalFilerTM Plus kit (Applied Biosystems) on the ProFlexTM PCR System (Applied Biosystems). Fluorescent amplicons were detected using the ABI PRISM® 3500xL Genetic Analyzer (Applied Biosystems), with maximum injection parameters of 3 µL at 1.2 kV for 24 s. Data were analysed using QuanStudio Software version 1.7 and GeneMapper® ID-X Software version 1.6, following laboratory guidelines. Percent profile recovery was calculated as the number of detected alleles relative to the expected full male profile.
4. Discussion
Saral Wax-Free Transfer Paper (TP) is a type of tracing paper that facilitates the transfer of drawings or markings from one surface to another without leaving behind greasy residues [
17]. While it is commercially available in five colours (white, yellow, red, blue, and graphite), we focused on white, yellow, and red due to their suitability for forensic purposes. White and yellow are ideal for tracing on dark fabric surfaces, while red is good for tracing on light fabric surfaces. While the formulation of TP is proprietary, it is understood to involve a pressure-sensitive pigment layer combined with a binder that releases TP pigment when pressure is applied. Its non-waxy, residue-free nature makes TP a suitable candidate as an alternative to crayons for documenting AP-positive areas on crime exhibits.
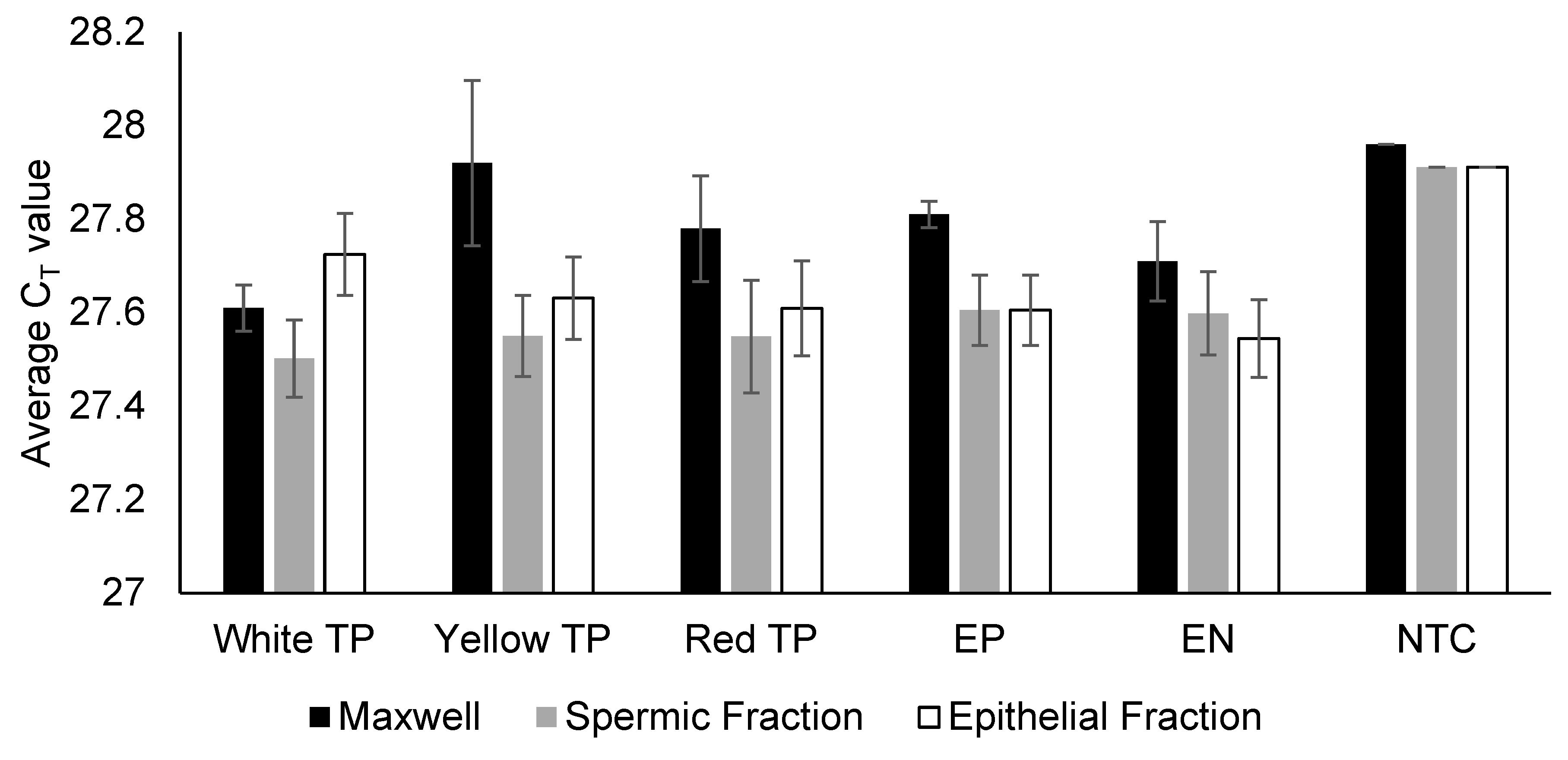
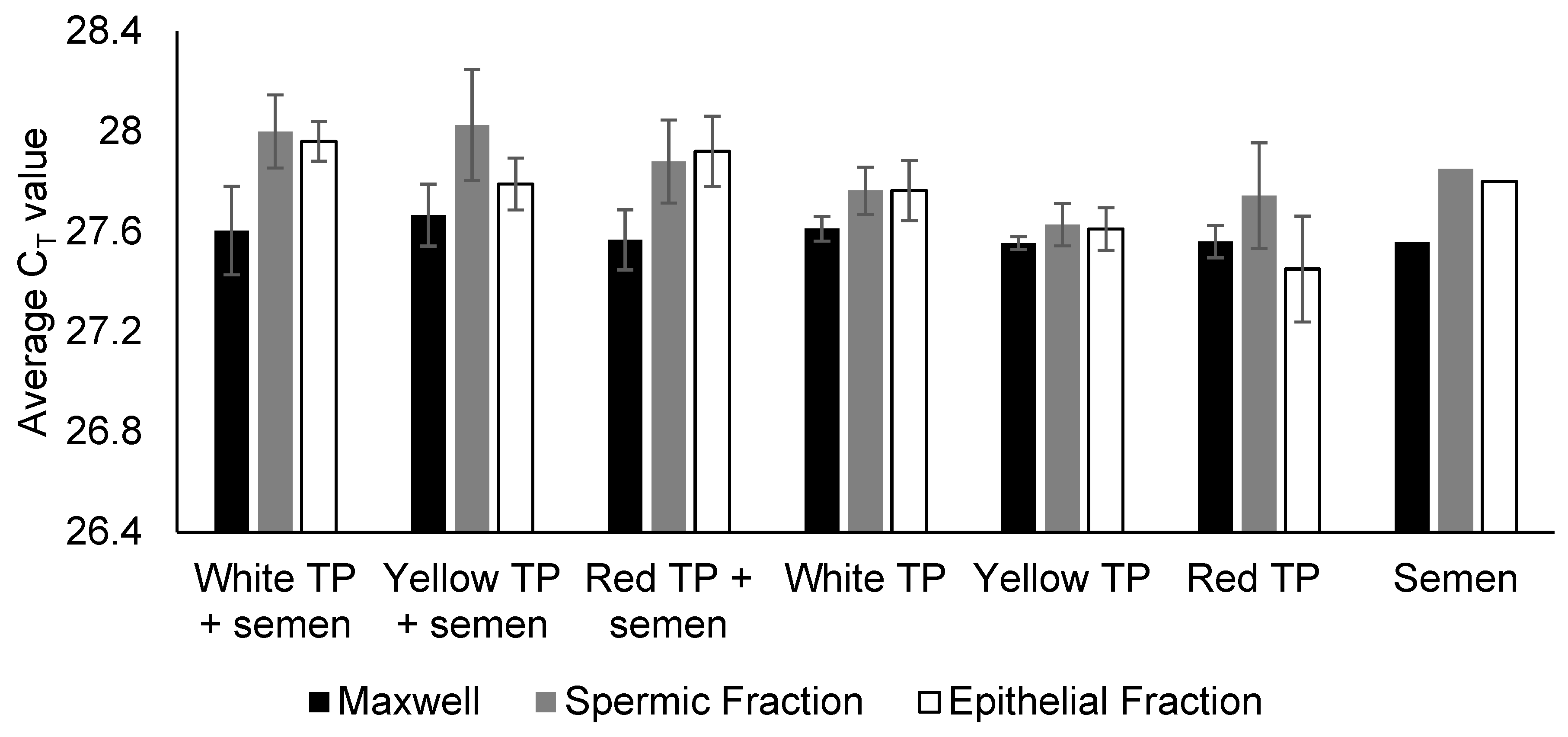
Our results confirm that TP does not interfere with downstream semen immunoassays or DNA processing. Specifically, there were no false-positive or false-negative reactivity RSID-Semen and Seratec PSA assays, no PCR inhibition (based on the IPC CT value during real-time DNA quantification), and no elevated baseline noise during electrophoresis.
When used on semen-spiked fabric, TP pigment reduced DNA yields but did not compromise PCR and allele recovery. It should be noted that the present study intentionally employed a larger amount of TP pigment to create a stress-test condition to rigorously evaluate any potential negative effects on the forensic biology workflow. This finding is highly reassuring as it demonstrates that even direct contact with the pigment does not cause PCR inhibition and that the only risk is a yield reduction (not a failed analysis) if correct protocol is not adopted. In routine practice, only small sections (often as small as ~1 cm × 1 cm) within the AP-marked boundaries (not the boundary line) would typically be excised for DNA extraction, so TP pigment carryover into the DNA extraction process would be minimal or excluded entirely. Thus, the yield reduction observed here reflects a conservative, worst-case scenario rather than the operational reality. Even so, under these exaggerated conditions, the only observed effect was a moderate DNA yield reduction, not allele dropout. This indicates that occasional pigment transfer poses minimal risk to casework outcomes.
Beyond forensic biology workflow, potential interference with other forensic evidence types (e.g., fibres, hairs, latent prints, gunshot residues, or trace DNA) is also considered. In routine casework, AP screening is typically conducted only after these other examinations are completed, thereby minimising or eliminating the risk of interference. Where trace DNA analysis is required, swabbing would precede AP testing, ensuring that the TP-traced boundaries or crayon-marked boundaries do not affect trace recovery. Furthermore, inter-laboratory coordination ensures that distinct areas of an exhibit are assigned to the appropriate discipline. In practice, TP paper can also be cut to approximate sizes to trace only the required boundaries, further reducing any risk of secondary transfer.
In routine laboratory casework, marking AP-positive boundaries is operationally essential to relocate the AP-positive areas, particularly if the purple AP colouration fades over time or if further testing (e.g., RSID-Semen, PSA, or DNA profiling) is needed. For this reason, we did not evaluate cleaning of the TP pigment, as the marked boundary itself was intentionally retained, consistent with existing crayon-based workflows. Mapping was likewise not quantified. In routine practice, AP mapping serves as a visual guide rather than a reportable analytical result. Moreover, excisions for DNA analysis are small and taken from within the visible AP-positive region rather than the traced boundary line, making precise geometric accuracy unnecessary. Compared with crayons, AP mappings with TP were fainter and more susceptible to removal through rubbing or rough handling by staff with less experience in AP documentation. These limitations can be mitigated by re-tracing with TP or by using a thicker stylus.
Finally, the present study provides preliminary evidence that TP can substantially reduce the time required to document AP-positive areas. Mapping time with TP was approximately 5.5 fold faster than with crayons across the three competent staff. Although the small sample size limits formal inference, the consistency of the results and the large effect size (Cohen’s d = 9.13) support the operational significance of TP as a practical alternative.
And while TP markings are less intense and more susceptible to removal due to its wax-free formulation, its ease of use and time-saving benefits outweigh these limitations. While not groundbreaking, this study therefore underscores the potential of TP as a significant time-saving tool. At the time of this manuscript’s writing, we are unaware of any published studies detailing how AP-positive areas are documented on crime scene exhibits.
Overall, TP shows strong potential as a practical alternative to crayons for documenting AP-positive areas, with the important caveats of reduced marked intensity, reduced permanence of markings, and the proof-of-concept nature (limited fabric type, single semen source, small replicate numbers) of this study. Future work may consider expanding the evaluation to different substrates, types of stains, and allele-level DNA quality metrics.