Complete Chloroplast Genome Features and Phylogenetic Analysis of Linum usitatissimum L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sequencing
2.2. Chloroplast Genome Assembly and Functional Annotation
2.3. Analysis of Dispersed and Simple Sequence Repeats
2.4. Analysis of Chloroplast Genome Nucleotide Diversity and Boundary Regions
2.5. Methods for Phylogenetic Analysis
3. Results
3.1. General Characteristics of the Flax Chloroplast Genome
3.2. Functional Annotation of Flax Chloroplast Genes
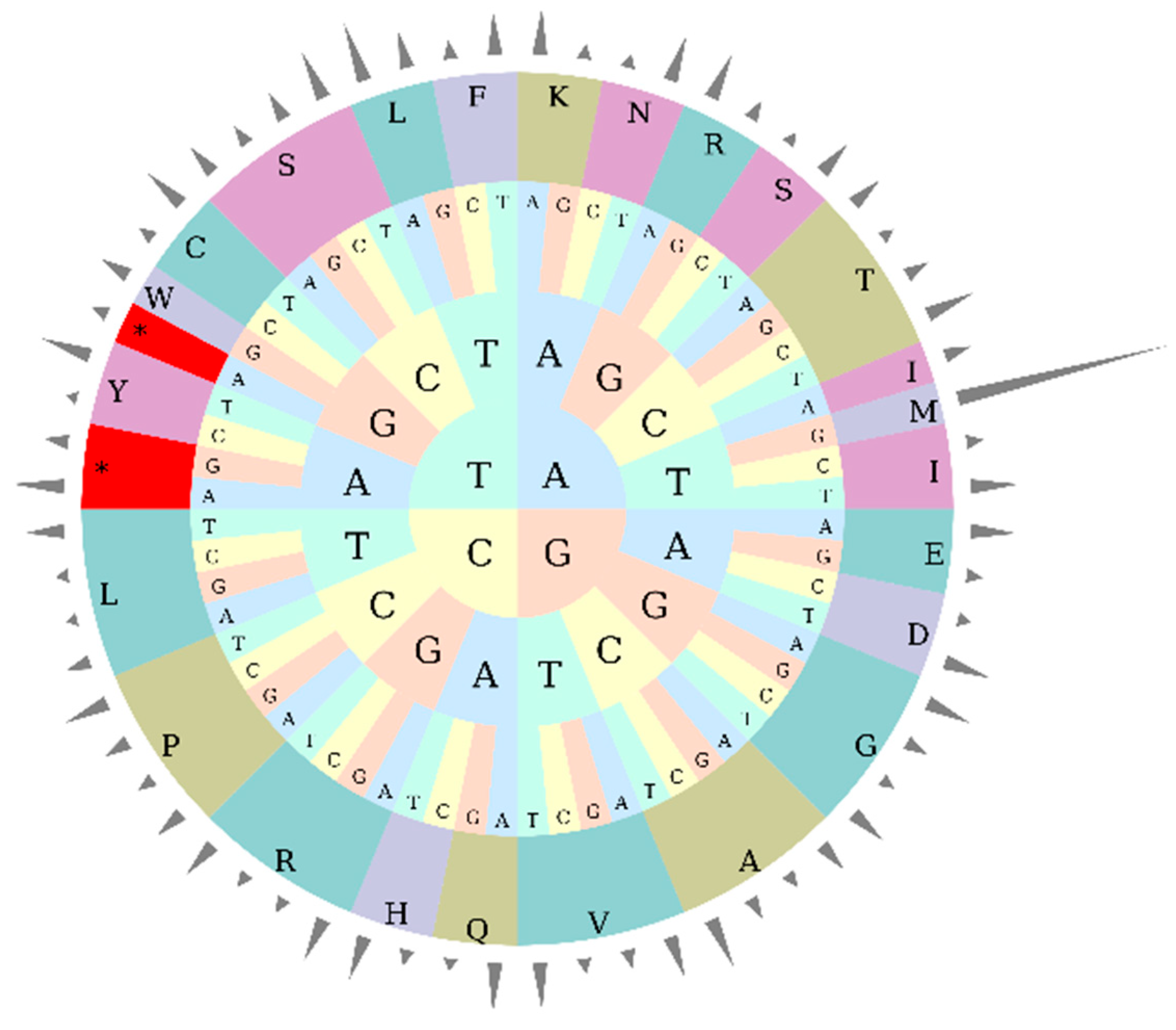
3.3. Codon Usage Bias Analysis
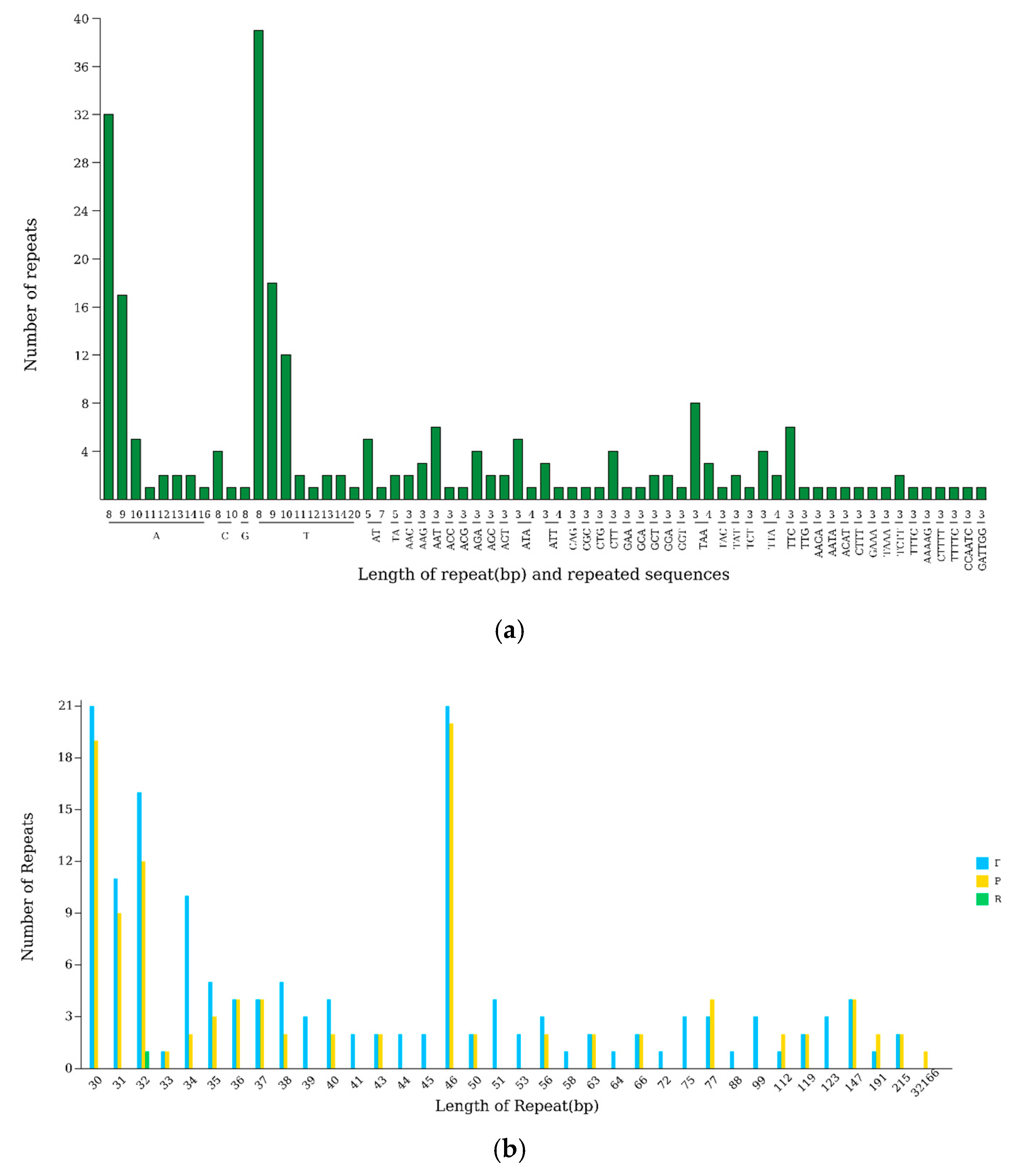
3.4. Repeat Sequence Analysis
3.5. Nucleotide Diversity and Boundary Analysis
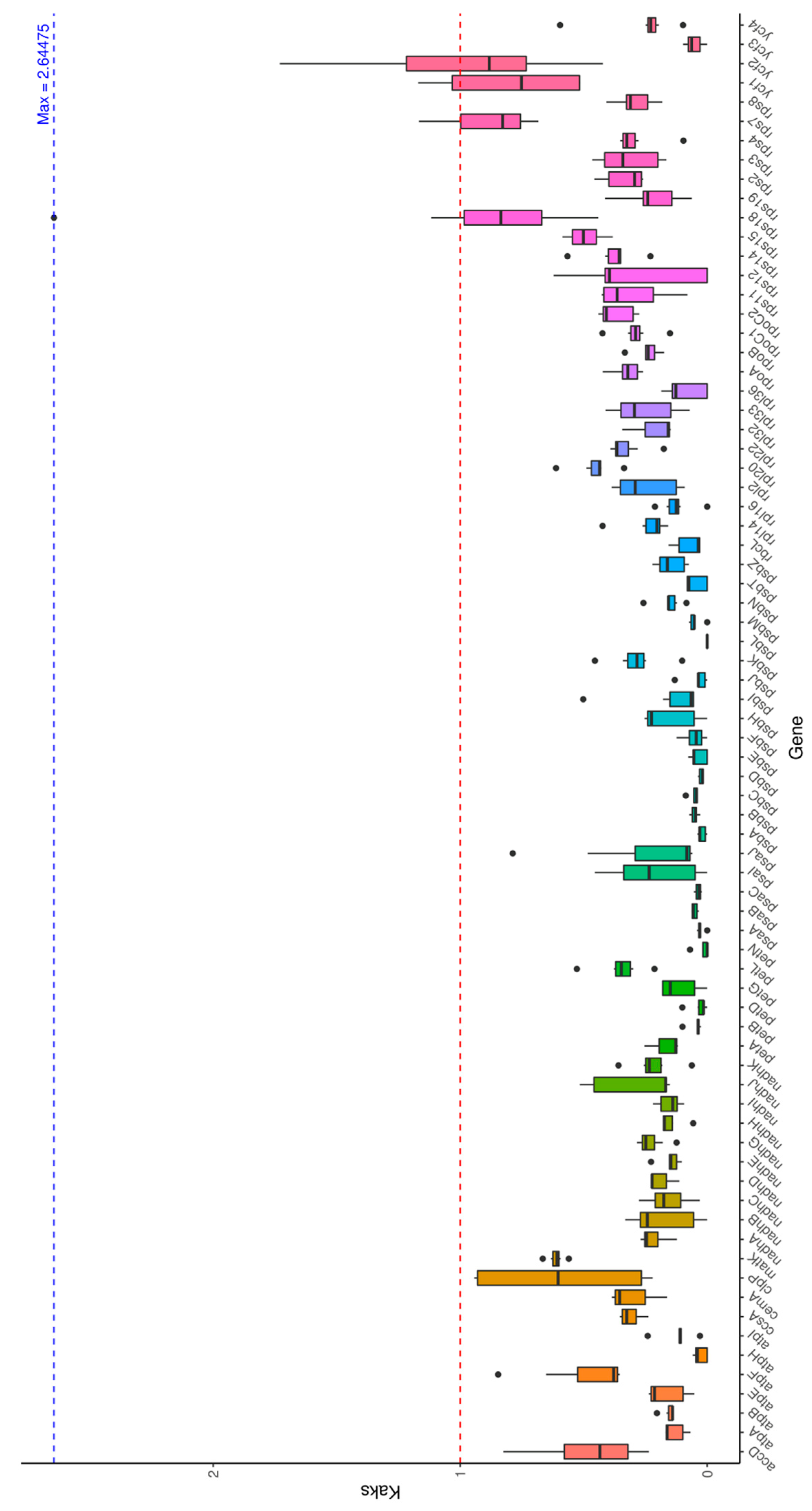
3.6. Ka/Ks Analysis
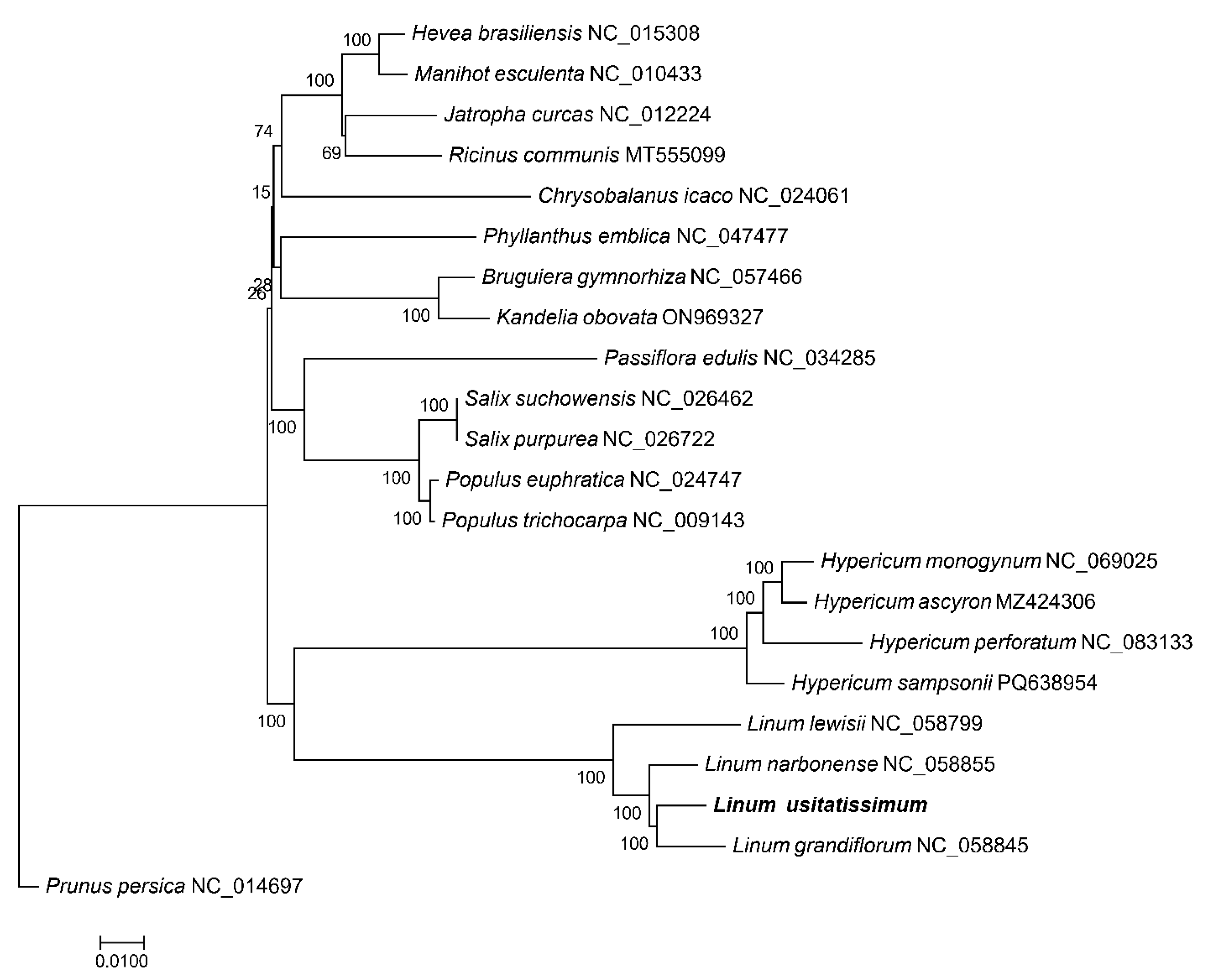
3.7. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Xu, L.Q.; Wang, Q.F. Research Progress on Flax for Different Uses. J. Northeast Agric. Sci. 2018, 43, 16–20. [Google Scholar]
- An, X.; Jin, G.R.; Li, L.F.; Luo, X.H.; Chen, C.L.; Li, W.L.; Zhu, G.L.; Zhang, J.Y. Research Progress on Flax Genetic Engineering. Mol. Plant Breed. Print 2019, 17, 6705–6710. [Google Scholar]
- Dang, Z.; Zhang, J.P.; Wang, L.M. Analysis of Combining Ability of Main Traits in 17 Flax Parents and Their Combinations. Mol. Plant Breed. 2024, 22, 3312–3323. [Google Scholar]
- Guo, C.J. Knowledge Treasury of Flaxseed; Tsinghua University Press: Beijing, China, 2020. [Google Scholar]
- Ji, Q.; Du, G.; Chen, C.; Li, S.; Luo, X.; Liu, T.; Zou, L.; Chen, J.; An, X. Research Progress on the Tolerance Mechanism and Technology of Bast Fiber Crops in Saline Soil. J. Nat. Fibers 2025, 22, 2502654. [Google Scholar] [CrossRef]
- Liu, T.T.; Luo, X.H.; Li, W.L.; Zhu, G.L.; An, X. Differences in Absorption and Accumulation of Heavy Metal Cd Among Different Flax Varieties. J. Zhejiang Agric. Sci. 2022, 63, 2030–2032+2045. [Google Scholar]
- Qiu, C.S.; Kang, Q.H.; Zhu, X.; Wang, Y.F.; Song, X.X.; Chen, X.Y.; Zhao, X.L.; Yao, D.D.; Liu, C.C.; Wu, Z.M.; et al. Research Progress on New Technologies for Flax Breeding. Agric. Technol. 2025, 45, 1–5. [Google Scholar]
- Deng, X. Development of Molecular Markers and Association Analysis of Yield-Related Traits in Flax. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2013. [Google Scholar]
- Hao, D.M.; Qiu, C.S.; Long, S.H. Overview of Molecular Biology Research on Flax in China. Agric. Sci. Technol. 2016, 17, 1767–1772. [Google Scholar]
- Lopes, A.d.S.; Pacheco, T.G.; dos Santos, K.G.; do Nascimento Vieira, L.; Guerra, M.P.; Nodari, R.O.; de Souza, E.M.; de Oliveira Pedrosa, F.; Rogalski, M. The Linum usitatissimum L. Plastome Reveals Atypical Structural Evolution, New Editing Sites, and the Phylogenetic Position of Linaceae within Malpighiales. Plant Cell Rep. 2017, 37, 307–328. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Dong, Y.; Yang, M.-H. Multiplexed Shotgun Sequencing Reveals Congruent Three-Genome Phylogenetic Signals for Four Botanical Sections of the Flax Genus Linum. Mol. Phylogenetics Evol. 2016, 101, 122–132. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.K.; Fang, Y.Z.; Zhou, J.H.; Chen, J.Y. Analysis of Chloroplast Genomes and Divergence Time Estimation of 11 Dendrobium officinale Varieties. J. Zhejiang Univ. Agric. Life Sci. Ed. 2025, 51, 291–302. [Google Scholar]
- Pottosin, I.; Shabala, S. Transport Across Chloroplast Membranes: Optimizing Photosynthesis for Adverse Environmental Conditions. Mol. Plant 2016, 9, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.J.; Guo, X.X. Advances in Research and Application of Plant Chloroplast Genomes. J. Shandong Norm. Univ. Nat. Sci. Ed. 2022, 37, 22–31. [Google Scholar]
- McFadden, G.I. Chloroplast Origin and Integration. Plant Physiol. 2001, 125, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Liu, L.K.; Wang, Z.L.; Yu, L.M.; Li, J.P.; Zeng, Y. Research Progress on Chloroplast Genomes of Orchidaceae Plants. Chin. Wild Plant Resour. 2023, 42, 7. [Google Scholar]
- Jin, D.M. Study on Structural Diversification of Plastid Genomes in Malpighiales. Bachelor’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2020. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a Program to Detect tRNA Genes and tmRNA Genes in Nucleotide Sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S. The Vmatch Large Scale Sequence Analysis Software. 2003. Available online: http://www.vmatch.de/ (accessed on 1 July 2025).
- White, G.L.; Fishbein, S.; Rutsein, J. Passionate Love and the Misattribution of Arousal. J. Pers. Soc. Psychol. 1981, 41, 56–62. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. raxmlGUI: A Graphical Front-End for RAxML. Org. Divers. Evol. 2011, 12, 335–337. [Google Scholar] [CrossRef]
- Bolsheva, N.L.; Melnikova, N.V.; Kirov, I.V.; Dmitriev, A.A.; Krasnov, G.S.; Amosova, A.V.; Samatadze, T.E.; Yurkevich, O.Y.; Zoshchuk, S.A.; Kudryavtseva, A.V.; et al. Characterization of Repeated DNA Sequences in Genomes of Blue-Flowered Flax. BMC Evol. Biol. 2019, 19, 49. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-B.; Allaby, R.G. Phylogenetic Network of Linum Species as Revealed by Non-Coding Chloroplast DNA Sequences. Genet. Resour. Crop Evol. 2009, 57, 667–677. [Google Scholar] [CrossRef]
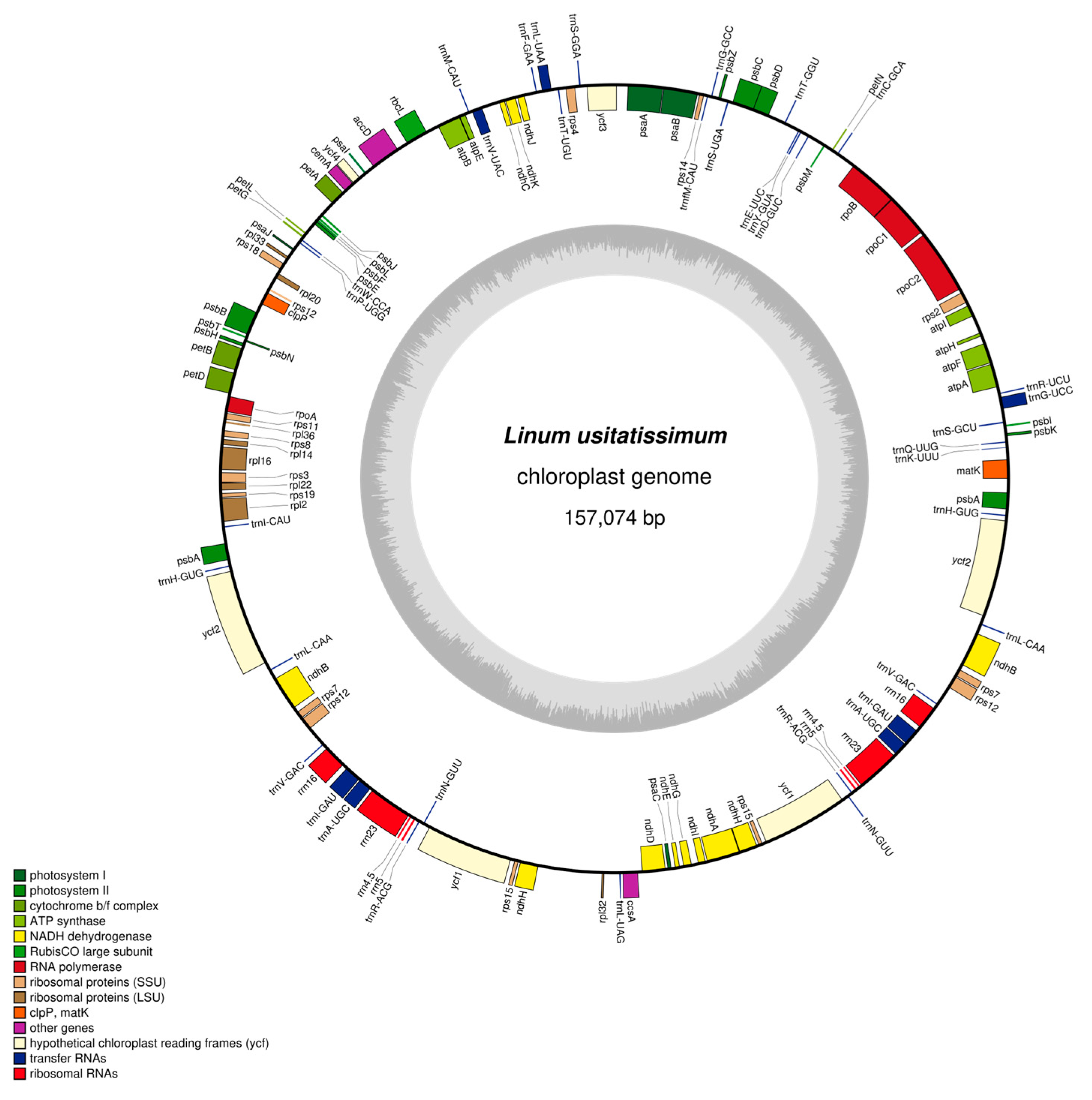


| Region | A Content/% | C Content/% | G Content/% | T Content/% | GC Content/% | Base Length/bp |
|---|---|---|---|---|---|---|
| LSC | 31.03 | 18.45 | 17.43 | 33.08 | 35.89 | 81,769 |
| SSC | 36.71 | 15.97 | 15.94 | 31.39 | 31.91 | 10,973 |
| IRa | 28.89 | 18.58 | 21.72 | 30.80 | 40.31 | 32,166 |
| IRb | 30.80 | 18.58 | 21.72 | 28.89 | 40.31 | 32,166 |
| Total | 30.94 | 18.97 | 18.44 | 31.64 | 37.42 | 157,074 |
| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA(2), psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB *(2), ndhC, ndhD, ndhE, ndhG, ndhH(2), ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Subunits photochlorophyllide reductase | - | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16 *, rpl2 *, rpl20, rpl22, rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12 **(2), rps14, rps15(2), rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RNAs | trnA-UGC *(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG(2), trnI-CAU, trnI-GAU *(2), trnK-UUU *, trnL-CAA(2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC *, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) |
| Transfer RNAs | trnA-UGC *(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG(2), trnI-CAU, trnI-GAU *(2), trnK-UUU *, trnL-CAA(2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC *, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Maturase | matK * | |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | - | |
| Other | - | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1(2), ycf2(2), ycf3 **, ycf4 |
| Amino Acid | Codon | Count | RSCU | Amino Acid | Codon | Count | RSCU | Amino Acid | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ter | UAA | 42 | 1.68 | Ile | AUA | 543 | 0.8952 | Arg | AGA | 332 | 1.56 |
| Ter | UAG | 20 | 0.8001 | Ile | AUC | 333 | 0.549 | Arg | AGG | 146 | 0.6858 |
| Ter | UGA | 13 | 0.5199 | Ile | AUU | 944 | 1.5561 | Arg | CGA | 307 | 1.4424 |
| Ala | GCA | 327 | 1.0388 | Lys | AAA | 957 | 1.5048 | Arg | CGC | 113 | 0.531 |
| Ala | GCC | 231 | 0.734 | Lys | AAG | 315 | 0.4952 | Arg | CGG | 111 | 0.5214 |
| Ala | GCG | 169 | 0.5368 | Leu | CUA | 322 | 0.8106 | Arg | CGU | 268 | 1.2594 |
| Ala | GCU | 532 | 1.6904 | Leu | CUC | 165 | 0.4152 | Ser | AGC | 117 | 0.435 |
| Cys | UGC | 68 | 0.5552 | Leu | CUG | 137 | 0.345 | Ser | AGU | 324 | 1.2054 |
| Cys | UGU | 177 | 1.4448 | Leu | CUU | 491 | 1.236 | Ser | UCA | 292 | 1.086 |
| Asp | GAC | 196 | 0.438 | Leu | UUA | 789 | 1.986 | Ser | UCC | 254 | 0.945 |
| Asp | GAU | 699 | 1.562 | Leu | UUG | 480 | 1.2078 | Ser | UCG | 185 | 0.6882 |
| Glu | GAA | 904 | 1.4688 | Met | AUG | 502 | 6.986 | Ser | UCU | 441 | 1.6404 |
| Glu | GAG | 327 | 0.5312 | Met | GUG | 1 | 0.014 | Thr | ACA | 294 | 1.1104 |
| Phe | UUC | 380 | 0.613 | Asn | AAC | 241 | 0.4662 | Thr | ACC | 209 | 0.7896 |
| Phe | UUU | 860 | 1.387 | Asn | AAU | 793 | 1.5338 | Thr | ACG | 122 | 0.4608 |
| Gly | GGA | 577 | 1.5028 | Pro | CCA | 234 | 1.0344 | Thr | ACU | 434 | 1.6392 |
| Gly | GGC | 185 | 0.4816 | Pro | CCC | 202 | 0.8928 | Val | GUA | 414 | 1.38 |
| Gly | GGG | 288 | 0.75 | Pro | CCG | 133 | 0.588 | Val | GUC | 149 | 0.4968 |
| Gly | GGU | 486 | 1.2656 | Pro | CCU | 336 | 1.4852 | Val | GUG | 183 | 0.61 |
| His | CAC | 133 | 0.5362 | Gln | CAA | 597 | 1.5984 | Val | GUU | 454 | 1.5132 |
| His | CAU | 363 | 1.4638 | Gln | CAG | 150 | 0.4016 | Trp | UGG | 381 | 1 |
| Tyr | UAC | 129 | 0.3458 | ||||||||
| Tyr | UAU | 617 | 1.6542 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Q.; Du, G.; An, X.; Dong, J.; Luo, X.; Chen, C.; Liu, T.; Zou, L.; Li, S.; Chen, J.; et al. Complete Chloroplast Genome Features and Phylogenetic Analysis of Linum usitatissimum L. Genes 2025, 16, 1038. https://doi.org/10.3390/genes16091038
Ji Q, Du G, An X, Dong J, Luo X, Chen C, Liu T, Zou L, Li S, Chen J, et al. Complete Chloroplast Genome Features and Phylogenetic Analysis of Linum usitatissimum L. Genes. 2025; 16(9):1038. https://doi.org/10.3390/genes16091038
Chicago/Turabian StyleJi, Qingqing, Guanghui Du, Xingcai An, Junyuan Dong, Xiahong Luo, Changli Chen, Tingting Liu, Lina Zou, Shaocui Li, Jikang Chen, and et al. 2025. "Complete Chloroplast Genome Features and Phylogenetic Analysis of Linum usitatissimum L." Genes 16, no. 9: 1038. https://doi.org/10.3390/genes16091038
APA StyleJi, Q., Du, G., An, X., Dong, J., Luo, X., Chen, C., Liu, T., Zou, L., Li, S., Chen, J., & An, X. (2025). Complete Chloroplast Genome Features and Phylogenetic Analysis of Linum usitatissimum L. Genes, 16(9), 1038. https://doi.org/10.3390/genes16091038






