Bulk and Single-Cell Transcriptomes Reveal Exhausted Signature in Prognosis of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Classification Based on Exhausted T Cell Signatures
2.3. Identification of Differentially Expressed Genes
2.4. Construction of the TEX Risk Score Model
2.5. Establishment and Validation of the TEX Diagnostic Model
2.6. Functional Enrichment Analysis
2.7. Construction of PPI Network
3. Results
3.1. Grouping of Cells and Samples Based on TEX Genes
3.2. Prognosis of T Cell Exhaustion Gene Signature
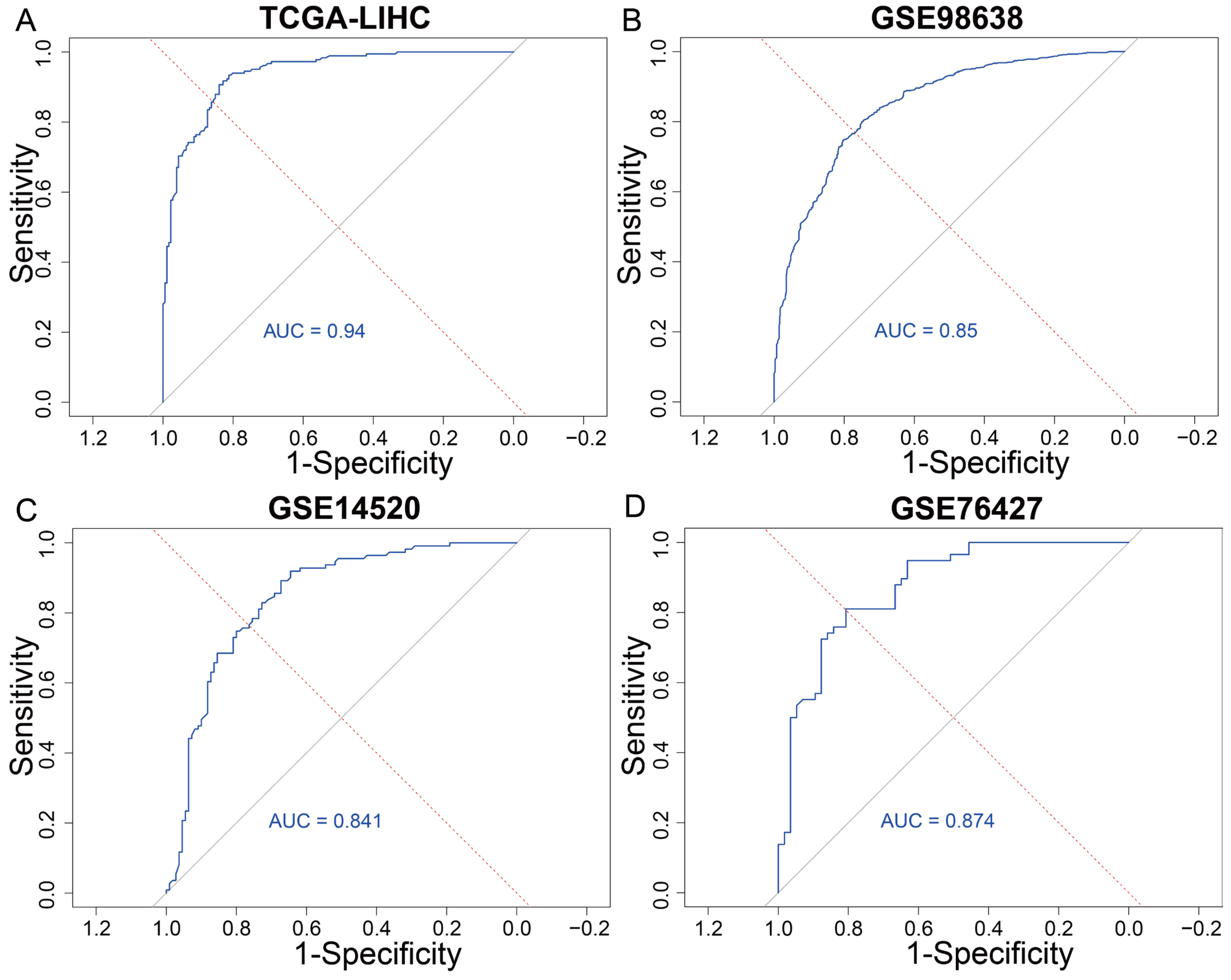
3.3. The Predictive Modeling of the T Cell Exhaustion Gene Signature
3.4. Functional Enrichment Analysis of the T Cell Exhaustion Gene Signature
3.5. Analysis of Immune Infiltration for the T Cell Exhaustion Gene Signature
3.6. The Construction of PPI Network
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.Y.; Danpanichkul, P.; Yong, J.N.; Yu, Z.; Tan, D.J.H.; Lim, W.H.; Koh, B.; Lim, R.Y.Z.; Tham, E.K.J.; Mitra, K.; et al. Liver cancer in 2021: Global Burden of Disease study. J. Hepatol. 2025, 82, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Llovet, J.; Kelley, R.; Villanueva, A.; Singal, A.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Llovet, J.M.; De Baere, T.; Kulik, L.; Haber, P.K.; Greten, T.F.; Meyer, T.; Lencioni, R. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 293–313. [Google Scholar] [CrossRef] [PubMed]
- Zahn, L.M. Effects of the tumor microenvironment. Science 2017, 355, 1386–1388. [Google Scholar] [CrossRef]
- Bhat, A.A.; Yousuf, P.; Wani, N.A.; Rizwan, A.; Chauhan, S.S.; Siddiqi, M.A.; Bedognetti, D.; El-Rifai, W.; Frenneaux, M.P.; Batra, S.K.; et al. Tumor microenvironment: An evil nexus promoting aggressive head and neck squamous cell carcinoma and avenue for targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 12. [Google Scholar] [CrossRef]
- Glaviano, A.; Lau, H.S.-H.; Carter, L.M.; Lee, E.H.C.; Lam, H.Y.; Okina, E.; Tan, D.J.J.; Tan, W.; Ang, H.L.; Carbone, D.; et al. Harnessing the tumor microenvironment: Targeted cancer therapies through modulation of epithelial-mesenchymal transition. J. Hematol. Oncol. 2025, 18, 6. [Google Scholar] [CrossRef]
- Gewandter, J.S.; McDermott, M.P.; He, H.; Gao, S.; Cai, X.; Farrar, J.T.; Katz, N.P.; Markman, J.D.; Senn, S.; Turk, D.C.; et al. Demonstrating Heterogeneity of Treatment Effects Among Patients: An Overlooked but Important Step Toward Precision Medicine. Clin. Pharmacol. Ther. 2019, 106, 204–210. [Google Scholar] [CrossRef]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef]
- Lucca, L.E.; Dominguez-Villar, M. Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat. Rev. Immunol. 2020, 20, 680–693. [Google Scholar] [CrossRef]
- Chihara, N.; Madi, A.; Kondo, T.; Zhang, H.; Acharya, N.; Singer, M.; Nyman, J.; Marjanovic, N.D.; Kowalczyk, M.S.; Wang, C.; et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 2018, 558, 454–459. [Google Scholar] [CrossRef]
- Barsch, M.; Salié, H.; Schlaak, A.E.; Zhang, Z.; Hess, M.; Mayer, L.S.; Tauber, C.; Otto-Mora, P.; Ohtani, T.; Nilsson, T.; et al. T-cell exhaustion and residency dynamics inform clinical outcomes in hepatocellular carcinoma. J. Hepatol. 2022, 77, 397–409. [Google Scholar] [CrossRef]
- Ma, J.; Zheng, B.; Goswami, S.; Meng, L.; Zhang, D.; Cao, C.; Li, T.; Zhu, F.; Ma, L.; Zhang, Z.; et al. PD1(Hi) CD8(+) T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.P.; Mullin, N.K.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell RNA sequencing in vision research: Insights into human retinal health and disease. Prog. Retin. Eye Res. 2021, 83, 100934. [Google Scholar] [CrossRef] [PubMed]
- Wahlsten, M.; Shaffer, S.M. Unveiling heterogeneity in rare cells by combining RNA-based sorting and scRNA-seq. Nat. Genet. 2025, 57, 283–284. [Google Scholar] [CrossRef] [PubMed]
- Mohr, R.; Jost-Brinkmann, F.; Özdirik, B.; Lambrecht, J.; Hammerich, L.; Loosen, S.H.; Luedde, T.; Demir, M.; Tacke, F.; Roderburg, C. Lessons From Immune Checkpoint Inhibitor Trials in Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 652172. [Google Scholar] [CrossRef]
- Wang, W.-J.; Wang, H.; Hua, T.-Y.; Song, W.; Zhu, J.; Wang, J.-J.; Huang, Y.-Q.; Ding, Z.-L. Establishment of a Prognostic Model Using Immune-Related Genes in Patients With Hepatocellular Carcinoma. Front. Genet. 2020, 11, 15. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, L.; Liu, C.; Wang, Z.; Xu, W.; Lu, J.; Wang, C.; Xu, X. Identification and validation of immune-related gene signature models for predicting prognosis and immunotherapy response in hepatocellular carcinoma. Front. Immunol. 2024, 15, 1371829. [Google Scholar] [CrossRef]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023, 51, D870–D876. [Google Scholar] [CrossRef]
- González-Blas, C.B.; De Winter, S.; Hulselmans, G.; Hecker, N.; Matetovici, I.; Christiaens, V.; Poovathingal, S.; Wouters, J.; Aibar, S.; Aerts, S. SCENIC+: Single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 2023, 20, 1355–1367. [Google Scholar] [CrossRef]
- Steen, C.B.; Liu, C.L.; Alizadeh, A.A.; Newman, A.M. Profiling Cell Type Abundance and Expression in Bulk Tissues with CIBERSORTx. Methods Mol. Biol. 2020, 2117, 135–157. [Google Scholar]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Sheshachalam, A.; Srivastava, N.; Mitchell, T.; Lacy, P.; Eitzen, G. Granule protein processing and regulated secretion in neutrophils. Front. Immunol. 2014, 5, 448. [Google Scholar] [CrossRef]
- Roberts, T.L.; Idris, A.; Dunn, J.A.; Kelly, G.M.; Burnton, C.M.; Hodgson, S.; Hardy, L.L.; Garceau, V.; Sweet, M.J.; Ross, I.L.; et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009, 323, 1057–1060. [Google Scholar] [CrossRef]
- Massa, D.; Baran, M.; Bengoechea, J.A.; Bowie, A.G. PYHIN1 regulates pro-inflammatory cytokine induction rather than innate immune DNA sensing in airway epithelial cells. J. Biol. Chem. 2020, 295, 4438–4450. [Google Scholar] [CrossRef]
- Wang, L.; Wei, Y.; Jin, Z.; Liu, F.; Li, X.; Zhang, X.; Bai, X.; Jia, Q.; Zhu, B.; Chu, Q. IFN-alpha/beta/IFN-gamma/IL-15 pathways identify GBP1-expressing tumors with an immune-responsive phenotype. Clin. Exp. Med. 2024, 24, 102. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, W.; Chen, M.; Chen, K.; Xia, X.; Zhou, S.; Yang, H. GBP1 Facilitates Indoleamine 2,3-Dioxygenase Extracellular Secretion to Promote the Malignant Progression of Lung Cancer. Front. Immunol. 2020, 11, 622467. [Google Scholar] [CrossRef] [PubMed]
- Solitano, V.; Facciorusso, A.; McGovern, D.P.; Nguyen, T.; Colman, R.J.; Zou, L.; Boland, B.S.; Syversen, S.W.; Jørgensen, K.K.; Ma, C.; et al. HLA-DQA1 *05 Genotype and Immunogenicity to Tumor Necrosis Factor-alpha Antagonists: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2023, 21, 3019–3029.e5. [Google Scholar] [CrossRef] [PubMed]
- Agresta, L.; Hoebe, K.H.; Janssen, E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018, 9, 2809. [Google Scholar] [CrossRef]
- Heinonen, M.T.; Kanduri, K.; Lähdesmäki, H.J.; Lahesmaa, R.; Henttinen, T.A. Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol. Cell Biol. 2015, 93, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yuan, H.; Sun, L.; Zhao, L.; Wang, Y.; Hou, C.; Zhang, H.; Lv, P.; Yang, G.; Zhang, N.; et al. Tumor-intrinsic PRMT5 upregulates FGL1 via methylating TCF12 to inhibit CD8(+) T-cell-mediated antitumor immunity in liver cancer. Acta Pharm. Sin. B 2025, 15, 188–204. [Google Scholar] [CrossRef] [PubMed]
- O’cOnnell, P.; Hyslop, S.; Blake, M.K.; Godbehere, S.; Amalfitano, A.; A Aldhamen, Y. SLAMF7 Signaling Reprograms T Cells toward Exhaustion in the Tumor Microenvironment. J. Immunol. 2021, 206, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Suryandari, D.A.; Miftahuzzakiyah, M.; Yunaini, L.; Kodariah, R.; Sukmawati, D.; Rustamadji, P.; Sari, P.; Ningsih, S.S. Identification of AKNA Gene and Its Role for Genetic Susceptibility in Epithelial Ovarian Cancer. Curr. Issues Mol. Biol. 2025, 47, 78. [Google Scholar] [CrossRef]
- Hernandez-Chacon, J.A.; Li, Y.; Wu, R.C.; Bernatchez, C.; Wang, Y.; Weber, J.S.; Hwu, P.; Radvanyi, L.G. Costimulation through the CD137/4-1BB pathway protects human melanoma tumor-infiltrating lymphocytes from activation-induced cell death and enhances antitumor effector function. J. Immunol. 2011, 34, 236–250. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Veena Shankaran, V. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Chi, H.; Zhao, S.; Yang, J.; Gao, X.; Peng, G.; Zhang, J.; Xie, X.; Song, G.; Xu, K.; Xia, Z.; et al. T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing. Front. Immunol. 2023, 14, 1137025. [Google Scholar] [CrossRef]
- E Ramirez, D.; Dragnev, C.P.C.; Searles, T.G.; Spicer, N.; Chen, T.; Lines, J.L.; Hawkes, A.R.; Davis, W.L.; Mohamed, A.; Shirai, K.; et al. Depletion of conventional CD4(+) T cells is required for robust priming and dissemination of tumor antigen-specific CD8(+) T cells in the setting of anti-CD4 therapy. J. Immunother. Cancer 2024, 12, e010170. [Google Scholar] [CrossRef]
- de Mare-Bredemeijer, E.L.; Shi, X.L.; Mancham, S.; van Gent, R.; van der Heide-Mulder, M.; De Boer, R.; Heemskerk, M.H.M.; de Jonge, J.; van der Laan, L.I.W.; Metselaar, H.J. Cytomegalovirus-Induced Expression of CD244 after Liver Transplantation Is Associated with CD8+ T Cell Hyporesponsiveness to Alloantigen. J. Immunol. 2015, 195, 1838–1848. [Google Scholar] [CrossRef]
- Hsu, C.L.; Ou, D.L.; Bai, L.Y.; Chen, C.W.; Lin, L.; Huang, S.F.; Cheng, A.-L.; Jeng, Y.-M.; Hsu, C. Exploring Markers of Exhausted CD8 T Cells to Predict Response to Immune Checkpoint Inhibitor Therapy for Hepatocellular Carcinoma. Liver Cancer 2021, 10, 346–359. [Google Scholar] [CrossRef]
- Li, W.; Mei, M.; Liu, T.; Zhang, S.; Wang, Z.; Suo, Y.; Wang, S.; Liu, Y.; Zhang, N.; Lu, W. Identification of PDCD1 and PDCD1LG2 as Prognostic Biomarkers and Associated with Immune Infiltration in Hepatocellular Carcinoma. Int. J. Gen. Med. 2022, 15, 437–449. [Google Scholar] [CrossRef]
- Canale, F.P.; Ramello, M.C.; Núñez, N.; Furlan, C.L.A.; Bossio, S.N.; Serrán, M.G.; Boari, J.T.; del Castillo, A.; Ledesma, M.; Sedlik, C.; et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8(+) T Cells. Cancer Res. 2018, 78, 115–128. [Google Scholar] [CrossRef]
- Reolo, M.J.Y.; Otsuka, M.; Seow, J.J.W.; Lee, J.; Lee, Y.H.; Nguyen, P.H.D.; Lim, C.J.; Wasser, M.; Chua, C.; Lim, T.K.H.; et al. CD38 marks the exhausted CD8+ tissue-resident memory T cells in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1182016. [Google Scholar] [CrossRef] [PubMed]
- Bahri, R.; Bollinger, A.; Bollinger, T.; Orinska, Z.; Bulfone-Paus, S. Ectonucleotidase CD38 demarcates regulatory, memory-like CD8+ T cells with IFN-gamma-mediated suppressor activities. PLoS ONE 2012, 7, e45234. [Google Scholar] [CrossRef] [PubMed]
- Raziorrouh, B.; Heeg, M.; Kurktschiev, P.; Schraut, W.; Zachoval, R.; Wendtner, C.; Wächtler, M.; Spannagl, M.; Denk, G.; Ulsenheimer, A.; et al. Inhibitory phenotype of HBV-specific CD4+ T-cells is characterized by high PD-1 expression but absent coregulation of multiple inhibitory molecules. PLoS ONE 2014, 9, e105703. [Google Scholar] [CrossRef]
- Huang, S.; Liang, C.; Zhao, Y.; Deng, T.; Tan, J.; Lu, Y.; Liu, S.; Li, Y.; Chen, S. Increased TOX expression concurrent with PD-1, Tim-3, and CD244 in T cells from patients with non-Hodgkin lymphoma. Asia Pac. J. Clin. Oncol. 2022, 18, 143–149. [Google Scholar] [CrossRef]
- Han, H.S.; Jeong, S.; Kim, H.; Kim, H.-D.; Kim, A.; Kwon, M.; Park, S.-H.; Woo, C.G.; Kim, H.K.; Lee, K.H.; et al. TOX-expressing terminally exhausted tumor-infiltrating CD8(+) T cells are reinvigorated by co-blockade of PD-1 and TIGIT in bladder cancer. Cancer Lett. 2021, 499, 137–147. [Google Scholar] [CrossRef]
- Wang, M.; Bu, J.; Zhou, M.; Sido, J.; Lin, Y.; Liu, G.; Lin, Q.; Xu, X.; Leavenworth, J.W.; Shen, E. CD8(+)T cells expressing both PD-1 and TIGIT but not CD226 are dysfunctional in acute myeloid leukemia (AML) patients. Clin. Immunol. 2018, 190, 64–73. [Google Scholar] [CrossRef]
- Beck, R.J.; Sloot, S.; Matsushita, H.; Kakimi, K.; Beltman, J.B. Mathematical modeling identifies LAG3 and HAVCR2 as biomarkers of T cell exhaustion in melanoma. iScience 2023, 26, 106666. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, X.; Qiu, J.; Cai, Y.; Ma, L.; Zhao, P.; Jiang, Y. Increased numbers of circulating ICOS(+) follicular helper T and CD38(+) plasma cells in patients with newly diagnosed primary biliary cirrhosis. Dig. Dis. Sci. 2015, 60, 405–413. [Google Scholar] [CrossRef]
- Ehrlich, A.K.; Pennington, J.M.; Tilton, S.; Wang, X.; Marshall, N.B.; Rohlman, D.; Funatake, C.; Punj, S.; O’Donnell, E.; Zhen Yu, Z.; et al. AhR activation increases IL-2 production by alloreactive CD4(+) T cells initiating the differentiation of mucosal-homing Tim3(+) Lag3(+) Tr1 cells. Eur. J. Immunol. 2017, 47, 1989–2001. [Google Scholar] [CrossRef] [PubMed]
- Lozano, E.; Dominguez-Villar, M.; Kuchroo, V.; A Hafler, D. The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 2012, 188, 3869–3875. [Google Scholar] [CrossRef]
- Tauriainen, J.; Scharf, L.; Frederiksen, J.; Naji, A.; Ljunggren, H.-G.; Sönnerborg, A.; Lund, O.; Reyes-Terán, G.; Hecht, F.M.; Deeks, S.G.; et al. Perturbed CD8(+) T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals. Sci. Rep. 2017, 7, 40354. [Google Scholar] [CrossRef]
- Liu, K.; Liu, J.; Zhang, X.; Liu, D.; Yao, W.; Bu, Y.; Chen, B. Identification of a Novel CD8+ T cell exhaustion-related gene signature for predicting survival in hepatocellular carcinoma. BMC Cancer 2023, 23, 1185. [Google Scholar] [CrossRef]
- Shi, J.; Li, G.; Liu, L.; Yuan, X.; Wang, Y.; Gong, M.; Li, C.; Ge, X.; Lu, S. Establishment and validation of exhausted CD8+ T cell feature as a prognostic model of HCC. Front. Immunol. 2023, 14, 1166052. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, A.; Quan, C.; Pan, Y.; Zhang, H.; Li, Y.; Gao, C.; Lu, H.; Wang, X.; Cao, P.; et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. NAT Commun. 2022, 13, 4594. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Perica, K.; Klebanoff, C.A.; Wolchok, J.D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2022, 19, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.Y.; Lee, S.H.; Baek, S.-W.; Sohn, B.; Jeong, Y.S.; Kang, S.-H.; Park, K.; Park, H.; Lee, S.S.; Kaseb, A.O.; et al. Genomic biomarkers to predict response to atezolizumab plus bevacizumab immunotherapy in hepatocellular carcinoma: Insights from the IMbrave150 trial. Clin. Mol. Hepatol. 2024, 30, 807–823. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, L.; Khatib, S.A.; Chang, C.-W.; Heinrich, S.; Dominguez, D.A.; Forgues, M.; Candia, J.; Hernandez, M.O.; Kelly, M.; et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 2021, 75, 1397–1408. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chun, R.; Ni, H.; Zhao, Z.; Zhang, C. Bulk and Single-Cell Transcriptomes Reveal Exhausted Signature in Prognosis of Hepatocellular Carcinoma. Genes 2025, 16, 1034. https://doi.org/10.3390/genes16091034
Chun R, Ni H, Zhao Z, Zhang C. Bulk and Single-Cell Transcriptomes Reveal Exhausted Signature in Prognosis of Hepatocellular Carcinoma. Genes. 2025; 16(9):1034. https://doi.org/10.3390/genes16091034
Chicago/Turabian StyleChun, Ruixin, Haisen Ni, Ziyi Zhao, and Chunlong Zhang. 2025. "Bulk and Single-Cell Transcriptomes Reveal Exhausted Signature in Prognosis of Hepatocellular Carcinoma" Genes 16, no. 9: 1034. https://doi.org/10.3390/genes16091034
APA StyleChun, R., Ni, H., Zhao, Z., & Zhang, C. (2025). Bulk and Single-Cell Transcriptomes Reveal Exhausted Signature in Prognosis of Hepatocellular Carcinoma. Genes, 16(9), 1034. https://doi.org/10.3390/genes16091034





