Genotype–Phenotype Correlations in PRPH2 Retinopathies: A Comprehensive Analysis of 36 Patients from the Oxford Eye Hospital, UK
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment
2.2. Clinical Phenotype
2.3. Genetic Analysis
2.4. Statistical Analysis
3. Results
3.1. Clinical Findings
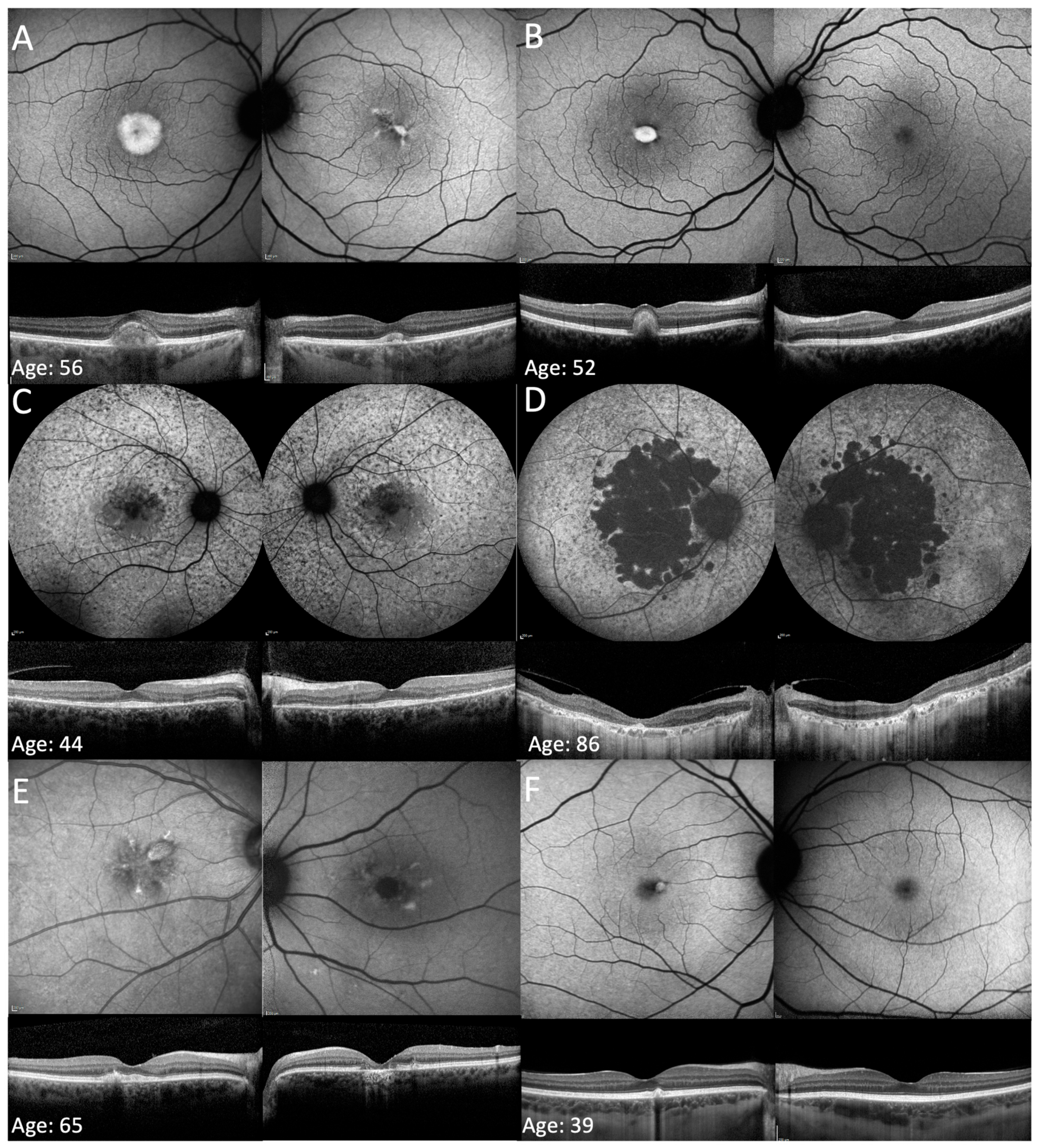
3.2. Phenotype on FAF and OCT Imaging
3.3. Genetic Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | Autofluorescence |
| adRP | Autosomal dominant retinitis pigmentosa |
| AVMD | Adult-onset vitelliform macular dystrophy |
| BCVA | Best corrected visual acuities |
| EZ | Ellipsoid zone |
| FAF | Fundus autofluorescence |
| IRD | Inherited retinal disease |
| LoF | Loss of function |
| OCT | Optical coherence tomography |
| NGS | Next-generation sequencing |
| NMD | Nonsense mediated decay |
| OD | Right eye |
| OS | Left eye |
| PD | Pattern dystrophy |
| PRPH2 | Peripherin 2 |
| PSPD | Pseudo-Stargardt pattern dystrophy |
| RDS | Retinal degeneration slow |
| RP | Retinitis pigmentosa |
| RPE | Retinal pigment epithelium |
| SNV | Single-nucleotide variant |
| VUS | Variant of unknown significance |
References
- Broadgate, S.; Yu, J.; Downes, S.M.; Halford, S. Unravelling the genetics of inherited retinal dystrophies: Past, present and future. Prog. Retin. Eye Res. 2017, 59, 53–96. [Google Scholar] [CrossRef] [PubMed]
- Liew, G.; Michaelides, M.; Bunce, C. A comparison of the causes of blindness certifications in England and Wales in working age adults (16–64 years), 1999–2000 with 2009–2010. BMJ Open 2014, 4, e004015. [Google Scholar] [CrossRef] [PubMed]
- Boon, C.J.; den Hollander, A.I.; Hoyng, C.B.; Cremers, F.P.; Klevering, B.J.; Keunen, J.E. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog. Retin. Eye Res. 2008, 27, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Connell, G.J.; Molday, R.S. Molecular cloning, primary structure, and orientation of the vertebrate photoreceptor cell protein peripherin in the rod outer segment disk membrane. Biochemistry 1990, 29, 4691–4698. [Google Scholar] [CrossRef]
- Pontikos, N.; Arno, G.; Jurkute, N.; Schiff, E.; Ba-Abbad, R.; Malka, S.; Gimenez, A.; Georgiou, M.; Wright, G.; Armengol, M.; et al. Genetic Basis of Inherited Retinal Disease in a Molecularly Characterized Cohort of More Than 3000 Families from the United Kingdom. Ophthalmology 2020, 127, 1384–1394. [Google Scholar] [CrossRef]
- Koyanagi, Y.; Akiyama, M.; Nishiguchi, K.M.; Momozawa, Y.; Kamatani, Y.; Takata, S.; Inai, C.; Iwasaki, Y.; Kumano, M.; Murakami, Y.; et al. Genetic characteristics of retinitis pigmentosa in 1204 Japanese patients. J. Med. Genet. 2019, 56, 662–670. [Google Scholar] [CrossRef]
- Falsini, B.; Placidi, G.; De Siena, E.; Chiurazzi, P.; Minnella, A.M.; Savastano, M.C.; Ziccardi, L.; Parisi, V.; Iarossi, G.; Percio, M.; et al. Genetic characteristics of 234 Italian patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2022, 12, 3774. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Shaw, K.; Phillips, A.; Cooper, D.N. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014, 133, 1–9. [Google Scholar] [CrossRef]
- Shankar, S.P.; Hughbanks-Wheaton, D.K.; Birch, D.G.; Sullivan, L.S.; Conneely, K.N.; Bowne, S.J.; Stone, E.M.; Daiger, S.P. Autosomal Dominant Retinal Dystrophies Caused by a Founder Splice Site Mutation, c.828+3A>T, in PRPH2 and Protein Haplotypes in trans as Modifiers. Investig. Ophthalmol. Vis. Sci. 2016, 57, 349–359. [Google Scholar] [CrossRef]
- Boon, C.J.; Klevering, B.J.; Cremers, F.P.; Zonneveld-Vrieling, M.N.; Theelen, T.; Den Hollander, A.I.; Hoyng, C.B. Central areolar choroidal dystrophy. Ophthalmology 2009, 116, 771–782.e1. [Google Scholar] [CrossRef]
- Alapati, A.; Goetz, K.; Suk, J.; Navani, M.; Al-Tarouti, A.; Jayasundera, T.; Tumminia, S.J.; Lee, P.; Ayyagari, R. Molecular diagnostic testing by eyeGENE: Analysis of patients with hereditary retinal dystrophy phenotypes involving central vision loss. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5510–5521. [Google Scholar] [CrossRef]
- Birtel, J.; Eisenberger, T.; Gliem, M.; Müller, P.L.; Herrmann, P.; Betz, C.; Zahnleiter, D.; Neuhaus, C.; Lenzner, S.; Holz, F.G.; et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018, 8, 4824. [Google Scholar] [CrossRef]
- Coco-Martin, R.M.; Sanchez-Tocino, H.T.; Desco, C.; Usategui-Martín, R.; Tellería, J.J. PRPH2-Related Retinal Diseases: Broadening the Clinical Spectrum and Describing a New Mutation. Genes 2020, 11, 773. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Holder, G.E.; Bradshaw, K.; Hunt, D.M.; Moore, A.T. Cone-rod dystrophy, intrafamilial variability, and incomplete penetrance associated with the R172W mutation in the peripherin/RDS gene. Ophthalmology 2005, 112, 1592–1598. [Google Scholar] [CrossRef] [PubMed]
- Weleber, R.G.; Carr, R.E.; Murphey, W.H.; Sheffield, V.C.; Stone, E.M. Phenotypic Variation Including Retinitis Pigmentosa, Pattern Dystrophy, and Fundus Flavimaculatus in a Single Family with a Deletion of Codon 153 or 154 of the Peripherin/RDS Gene. Arch. Ophthalmol. 1993, 111, 1531–1542. [Google Scholar] [CrossRef]
- Khan, A.O.; Al Rashaed, S.; Neuhaus, C.; Bergmann, C.; Bolz, H.J. Peripherin mutations cause a distinct form of recessive Leber congenital amaurosis and dominant phenotypes in asymptomatic parents heterozygous for the mutation. Br. J. Ophthalmol. 2016, 100, 209–215. [Google Scholar] [CrossRef]
- Dryja, T.P.; Hahn, L.B.; Kajiwara, K.; Berson, E.L. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 1997, 38, 1972–1982. [Google Scholar]
- Kajiwara, K.; Berson, E.L.; Dryja, T.P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 1994, 264, 1604–1608. [Google Scholar] [CrossRef]
- Molday, R.S.; Hicks, D.; Molday, L. Peripherin. A rim-specific membrane protein of rod outer segment discs. Investig. Ophthalmol. Vis. Sci. 1987, 28, 50–61. [Google Scholar]
- Travis, G.H.; Brennan, M.B.; Danielson, P.E.; Kozak, C.A.; Sutcliffe, J.G. Identification of a photoreceptor-specific mRNA encoded by the gene responsible for retinal degeneration slow (rds). Nature 1989, 338, 70–73. [Google Scholar] [CrossRef]
- Travis, G.H.; Christerson, L.; Danielson, P.E.; Klisak, I.; Sparkes, R.S.; Hahn, L.B.; Dryja, T.P.; Sutcliffe, J.G. The human retinal degeneration slow (RDS) gene: Chromosome assignment and structure of the mRNA. Genomics 1991, 10, 733–739. [Google Scholar] [CrossRef]
- Travis, G.H.; Groshan, K.R.; Lloyd, M.; Bok, D. Complete rescue of photoreceptor dysplasia and degeneration in transgenic retinal degeneration slow (rds) mice. Neuron 1992, 9, 113–119. [Google Scholar] [CrossRef]
- Sanyal, S.; Jansen, H.G. Absence of receptor outer segments in the retina of rds mutant mice. Neurosci. Lett. 1981, 21, 23–26. [Google Scholar] [CrossRef]
- Arikawa, K.; Molday, L.L.; Molday, R.S.; Williams, D.S. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: Relationship to disk membrane morphogenesis and retinal degeneration. J. Cell Biol. 1992, 116, 659–667. [Google Scholar] [CrossRef]
- Cheng, T.; Peachey, N.S.; Li, S.; Goto, Y.; Cao, Y.; Naash, M.I. The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J. Neurosci. 1997, 17, 8118–8128. [Google Scholar] [CrossRef]
- Chakraborty, D.; Conley, S.M.; Fliesler, S.J.; Naash, M.I. The function of oligomerization-incompetent RDS in rods. Adv. Exp. Med. Biol. 2010, 664, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.F.; Loewen, C.J.; Molday, R.S. Cysteine residues of photoreceptor peripherin/rds: Role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry 1998, 37, 680–685. [Google Scholar] [CrossRef] [PubMed]
- El Mazouni, D.; Gros, P. Cryo-EM structures of peripherin-2 and ROM1 suggest multiple roles in photoreceptor membrane morphogenesis. Sci. Adv. 2022, 8, eadd3677. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.Q.; Stricker, H.M.; Naash, M.I. Role of the second intradiscal loop of peripherin/rds in homo and hetero associations. Biochemistry 2005, 44, 4897–4904. [Google Scholar] [CrossRef]
- Conley, S.M.; Naash, M.I. Gene therapy for PRPH2-associated ocular disease: Challenges and prospects. Cold Spring Harb. Perspect. Med. 2014, 4, a017376. [Google Scholar] [CrossRef]
- Sanyal, S.; De Ruiter, A.; Hawkins, R.K. Development and degeneration of retina in rds mutant mice: Light microscopy. J. Comp. Neurol. 1980, 194, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Jansen, H.G.; Sanyal, S. Development and degeneration of retina in rds mutant mice: Electron microscopy. J. Comp. Neurol. 1984, 224, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Zulliger, R.; Conley, S.M.; Mwoyosvi, M.L.; Al-Ubaidi, M.R.; Naash, M.I. Oligomerization of Prph2 and Rom1 is essential for photoreceptor outer segment formation. Hum. Mol. Genet. 2018, 27, 3507–3518. [Google Scholar] [CrossRef]
- Reeves, M.J.; Goetz, K.E.; Guan, B.; Ullah, E.; Blain, D.; Zein, W.M.; Tumminia, S.J.; Hufnagel, R.B. Genotype-phenotype associations in a large PRPH2-related retinopathy cohort. Hum. Mutat. 2020, 41, 1528–1539. [Google Scholar] [CrossRef]
- Oishi, A.; Fujinami, K.; Mawatari, G.; Naoi, N.; Ikeda, Y.; Ueno, S.; Kuniyoshi, K.; Hayashi, T.; Kondo, H.; Mizota, A.; et al. Genetic and Phenotypic Landscape of PRPH2-Associated Retinal Dystrophy in Japan. Genes 2021, 12, 1817. [Google Scholar] [CrossRef]
- Peeters, M.; Khan, M.; Rooijakkers, A.; Mulders, T.; Haer-Wigman, L.; Boon, C.J.F.; Klaver, C.C.W.; van den Born, L.I.; Hoyng, C.B.; Cremers, F.P.M.; et al. PRPH2 mutation update: In silico assessment of 245 reported and 7 novel variants in patients with retinal disease. Hum. Mutat. 2021, 42, 1521–1547. [Google Scholar] [CrossRef]
- Fernández-Caballero, L.; Martín-Merida, I.; Blanco-Kelly, F.; Avila-Fernandez, A.; Carreño, E.; Jose, P.F.-S.; Irigoyen, C.; Jimenez-Rolando, B.; Lopez-Grondona, F.; Mahillo, I.; et al. PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort. Int. J. Mol. Sci. 2024, 25, 2913. [Google Scholar] [CrossRef]
- van Lith-Verhoeven, J.J.; van den Helm, B.; Deutman, A.F.; Bergen, A.A.; Cremers, F.P.; Hoyng, C.B.; de Jong, P.T. A peculiar autosomal dominant macular dystrophy caused by an asparagine deletion at codon 169 in the peripherin/RDS gene. Arch. Ophthalmol. 2003, 121, 1452–1457. [Google Scholar] [CrossRef]
- Manes, G.; Guillaumie, T.; Vos, W.L.; Devos, A.; Audo, I.; Zeitz, C.; Marquette, V.; Zanlonghi, X.; Defoort-Dhellemmes, S.; Puech, B.; et al. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in france and characterization of biochemical and clinical features. Am. J. Ophthalmol. 2015, 159, 302–314. [Google Scholar] [CrossRef]
- Heath Jeffery, R.C.; Thompson, J.A.; Lo, J.; Chelva, E.S.; Armstrong, S.; Pulido, J.S.; Procopio, R.; Vincent, A.L.; Bianco, L.; Battaglia Parodi, M.; et al. Retinal Dystrophies Associated With Peripherin-2: Genetic Spectrum and Novel Clinical Observations in 241 Patients. Investig. Ophthalmol. Vis. Sci. 2024, 65, 22. [Google Scholar] [CrossRef]
- Cheng, J.; Fu, J.; Zhou, Q.; Xiang, X.; Wei, C.; Yang, L.; Fu, S.; Khan, M.A.; Lv, H.; Fu, J. A novel splicing mutation in the PRPH2 gene causes autosomal dominant retinitis pigmentosa in a Chinese pedigree. J. Cell. Mol. Med. 2019, 23, 3776–3780. [Google Scholar] [CrossRef] [PubMed]
- Daftarian, N.; Mirrahimi, M.; Sabbaghi, H.; Moghadasi, A.; Zal, N.; Dehghan Banadaki, H.; Ahmadieh, H.; Suri, F. PRPH2 mutation as the cause of various clinical manifestations in a family affected with inherited retinal dystrophy. Ophthalmic Genet. 2019, 40, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Sun, V.; Tuan, H.F.; Keser, V.; Wang, K.; Ren, H.; Lopez, I.; Zaneveld, J.E.; Siddiqui, S.; et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J. Med. Genet. 2013, 50, 674–688. [Google Scholar] [CrossRef]
- Marmor, M.F.; McNamara, J.A. Pattern Dystrophy of the Retinal Pigment Epithelium and Geographic Atrophy of the Macula. Am. J. Ophthalmol. 1996, 122, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Garibaldi, D.C.; Li, Y.; Green, W.R.; Zack, D.J. Butterfly-Shaped Pattern Dystrophy: A Genetic, Clinical, and Histopathological Report. Arch. Ophthalmol. 2002, 120, 485–490. [Google Scholar] [CrossRef]
- Marano, F.; Deutman, A.F.; Leys, A.; Aandekerk, A.L. Hereditary retinal dystrophies and choroidal neovascularization. Graefe’s Arch. Clin. Exp. Ophthalmol. 2000, 238, 760–764. [Google Scholar] [CrossRef]
- Lee, C.S.; Leys, M. A Family Affected by Novel C213W Mutation in PRPH2: Long-Term Follow-Up of CNV Secondary to Pattern Dystrophy. Ophthalmic Surg. Lasers Imaging Retin. 2020, 51, 354–362. [Google Scholar] [CrossRef]
- Antonelli, G.; Parravano, M.; Barbano, L.; Costanzo, E.; Bertelli, M.; Medori, M.C.; Parisi, V.; Ziccardi, L. Multimodal Study of PRPH2 Gene-Related Retinal Phenotypes. Diagnostics 2022, 12, 1851. [Google Scholar] [CrossRef]
- McCulloch, D.L.; Marmor, M.F.; Brigell, M.G.; Hamilton, R.; Holder, G.E.; Tzekov, R.; Bach, M. ISCEV Standard for full-field clinical electroretinography (2015 update). Doc. Ophthalmol. 2015, 130, 1–12. [Google Scholar] [CrossRef]
- Heath Jeffery, R.C.; Lo, J.; Thompson, J.A.; Lamey, T.M.; McLaren, T.L.; De Roach, J.N.; Ayton, L.N.; Vincent, A.L.; Sharma, A.; Chen, F.K. Analysis of the Outer Retinal Bands in ABCA4 and PRPH2-Associated Retinopathy using OCT. Ophthalmol. Retin. 2024, 8, 174–183. [Google Scholar] [CrossRef]
- Shah, M.; Shanks, M.; Packham, E.; Williams, J.; Haysmoore, J.; MacLaren, R.E.; Németh, A.H.; Clouston, P.; Downes, S.M. Next generation sequencing using phenotype-based panels for genetic testing in inherited retinal diseases. Ophthalmic Genet. 2020, 41, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Nassisi, M.; Mohand-Saïd, S.; Dhaenens, C.M.; Boyard, F.; Démontant, V.; Andrieu, C.; Antonio, A.; Condroyer, C.; Foussard, M.; Méjécase, C.; et al. Expanding the Mutation Spectrum in ABCA4: Sixty Novel Disease Causing Variants and Their Associated Phenotype in a Large French Stargardt Cohort. Int. J. Mol. Sci. 2018, 19, 2196. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Jane Farrar, G.; Kenna, P.; Jordan, S.A.; Kumar-Singh, R.; Humphries, M.M.; Sharp, E.M.; Sheils, D.; Humphries, P. Autosomal dominant retinitis pigmentosa: A novel mutation at the peripherin/RDS locus in the original 6p-linked pedigree. Genomics 1992, 14, 805–807. [Google Scholar] [CrossRef]
- Bianco, L.; Arrigo, A.; Antropoli, A.; Saladino, A.; Spiga, I.; Patricelli, M.G.; Bandello, F.; Carrera, P.; Battaglia Parodi, M. PRPH2-Associated Retinopathy: Novel Variants and Genotype-Phenotype Correlations. Ophthalmol. Retin. 2023, 7, 450–461. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Jiang, Y.; Zhu, D.; Ouyang, J.; Yi, Z.; Li, S.; Jia, X.; Xiao, X.; Sun, W.; et al. New Insight into the Genotype-Phenotype Correlation of PRPH2-Related Diseases Based on a Large Chinese Cohort and Literature Review. Int. J. Mol. Sci. 2023, 24, 6728. [Google Scholar] [CrossRef]
- Ekström, U.; Ponjavic, V.; Andréasson, S.; Ehinger, B.; Nilsson-Ehle, P.; Abrahamson, M. Detection of alterations in all three exons of the peripherin/RDS gene in Swedish patients with retinitis pigmentosa using an efficient DGGE system. Mol. Pathol. 1998, 51, 287–291. [Google Scholar] [CrossRef]
- Souied, E.H.; Rozet, J.M.; Gerber, S.; Dufier, J.L.; Soubrane, G.; Coscas, G.; Munnich, A.; Kaplan, J. Two novel missense mutations in the peripherin/RDS gene in two unrelated French patients with autosomal dominant retinitis pigmentosa. Eur. J. Ophthalmol. 1998, 8, 98–101. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Parmann, R.; Tsang, S.H.; Allikmets, R.; Chang, S.; Jauregui, R. Shared Features in Retinal Disorders With Involvement of Retinal Pigment Epithelium. Investig. Ophthalmol. Vis. Sci. 2021, 62, 15. [Google Scholar] [CrossRef]
- Boon, C.J.; van Schooneveld, M.J.; den Hollander, A.I.; van Lith-Verhoeven, J.J.; Zonneveld-Vrieling, M.N.; Theelen, T.; Cremers, F.P.; Hoyng, C.B.; Klevering, B.J. Mutations in the peripherin/RDS gene are an important cause of multifocal pattern dystrophy simulating STGD1/fundus flavimaculatus. Br. J. Ophthalmol. 2007, 91, 1504–1511. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Curcio, C.A. Anatomical correlates to the bands seen in the outer retina by optical coherence tomography: Literature review and model. Retina 2011, 31, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Staurenghi, G.; Sadda, S.; Chakravarthy, U.; Spaide, R.F. Proposed Lexicon for Anatomic Landmarks in Normal Posterior Segment Spectral-Domain Optical Coherence Tomography: The IN•OCT Consensus. Ophthalmology 2014, 121, 1572–1578. [Google Scholar] [CrossRef]
- Lewis, T.R.; Makia, M.S.; Kakakhel, M.; Al-Ubaidi, M.R.; Arshavsky, V.Y.; Naash, M.I. Photoreceptor Disc Enclosure Occurs in the Absence of Normal Peripherin-2/rds Oligomerization. Front. Cell. Neurosci. 2020, 14, 92. [Google Scholar] [CrossRef]
- Stuck, M.W.; Conley, S.M.; Naash, M.I. PRPH2/RDS and ROM-1: Historical context, current views and future considerations. Prog. Retin. Eye Res. 2016, 52, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Downes, S.M.; Fitzke, F.W.; Holder, G.E.; Payne, A.M.; Bessant, D.A.; Bhattacharya, S.S.; Bird, A.C. Clinical features of codon 172 RDS macular dystrophy: Similar phenotype in 12 families. Arch. Ophthalmol. 1999, 117, 1373–1383. [Google Scholar] [CrossRef]
- Payne, A.M.; Downes, S.M.; Bessant, D.A.; Bird, A.C.; Bhattacharya, S.S. Founder effect, seen in the British population, of the 172 peripherin/RDS mutation—And further refinement of genetic positioning of the peripherin/RDS gene. Am. J. Hum. Genet. 1998, 62, 192–195. [Google Scholar] [CrossRef]
- Stricker, H.M.; Ding, X.Q.; Quiambao, A.; Fliesler, S.J.; Naash, M.I. The Cys214⟶Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice. Biochem. J. 2005, 388, 605–613. [Google Scholar] [CrossRef]
- Salinas, R.Y.; Baker, S.A.; Gospe, S.M., 3rd; Arshavsky, V.Y.; Barnes, S. A single valine residue plays an essential role in peripherin/rds targeting to photoreceptor outer segments. PLoS ONE 2013, 8, e54292. [Google Scholar] [CrossRef] [PubMed]
- Loewen, C.J.; Moritz, O.L.; Tam, B.M.; Papermaster, D.S.; Molday, R.S. The role of subunit assembly in peripherin-2 targeting to rod photoreceptor disk membranes and retinitis pigmentosa. Mol. Biol. Cell 2003, 14, 3400–3413. [Google Scholar] [CrossRef] [PubMed]
- Conley, S.M.; Naash, M.I. Focus on molecules: RDS. Exp. Eye Res. 2009, 89, 278–279. [Google Scholar] [CrossRef]
- Ding, X.Q.; Nour, M.; Ritter, L.M.; Goldberg, A.F.; Fliesler, S.J.; Naash, M.I. The R172W mutation in peripherin/rds causes a cone-rod dystrophy in transgenic mice. Hum. Mol. Genet. 2004, 13, 2075–2087. [Google Scholar] [CrossRef]
- Chakraborty, D.; Strayve, D.G.; Makia, M.S.; Conley, S.M.; Kakahel, M.; Al-Ubaidi, M.R.; Naash, M.I. Novel molecular mechanisms for Prph2—Associated pattern dystrophy. FASEB J. 2020, 34, 1211–1230. [Google Scholar] [CrossRef]
- Nakazawa, M.; Wada, Y.; Tamai, M. Macular dystrophy associated with monogenic Arg172Trp mutation of the peripherin/RDS gene in a Japanese family. Retina 1995, 15, 518–523. [Google Scholar] [CrossRef]
- Sodi, A.; Mucciolo, D.P.; Giorgio, D.; Passerini, I.; Pacini, B.; Bruschi, M.; Verdina, T.; Virgili, G.; Giansanti, F.; Murro, V. Clinical and molecular findings in patients with pattern dystrophy. Ophthalmic Genet. 2021, 42, 577–587. [Google Scholar] [CrossRef]
- Khan, A.O. Recessive pediatric-onset cone-rod dysfunction or dominant maculopathy in a consanguineous family harboring the peripherin mutation p.Arg220Gln. Ophthalmic Genet. 2019, 40, 60–63. [Google Scholar] [CrossRef]
- Albertos-Arranz, H.; Sánchez-Sáez, X.; Martínez-Gil, N.; Pinilla, I.; Coco-Martin, R.M.; Delgado, J.; Cuenca, N. Phenotypic Differences in a PRPH2 Mutation in Members of the Same Family Assessed with OCT and OCTA. Diagnostics 2021, 11, 777. [Google Scholar] [CrossRef]
- Simonelli, F.; Testa, F.; Marini, V.; Interlandi, E.; Rossi, S.; Pognuz, D.R.; Virgili, G.; Garrè, C.; Bandello, F. Intrafamilial clinical heterogeneity associated with a novel mutation of the retinal degeneration slow/peripherin gene. Ophthalmic Res. 2007, 39, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Sanlialp, A.; Escher, P.; Schaller, A.; Todorova, M. Clinical Heterogeneity in Two Siblings Harbouring a Heterozygous PRPH2 Pathogenic Variant. Klin. Monatsblatter Augenheilkd. 2023, 240, 536–543. [Google Scholar] [CrossRef]
- Piguet, B.; Héon, E.; Munier, F.L.; Grounauer, P.A.; Niemeyer, G.; Butler, N.; Schorderet, D.F.; Sheffield, V.C.; Stone, E.M. Full characterization of the maculopathy associated with an Arg-172-Trp mutation in the RDS/peripherin gene. Ophthalmic Genet. 1996, 17, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Soucy, M.; Kolesnikova, M.; Kim, A.H.; Tsang, S.H. Phenotypic variability in PRPH2 as demonstrated by a family with incomplete penetrance of autosomal dominant cone-rod dystrophy. Doc. Ophthalmol. 2023, 146, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Poloschek, C.M.; Bach, M.; Lagrèze, W.A.; Glaus, E.; Lemke, J.R.; Berger, W.; Neidhardt, J. ABCA4 and ROM1: Implications for modification of the PRPH2-associated macular dystrophy phenotype. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4253–4265. [Google Scholar] [CrossRef]
- Samardzija, M.; Wenzel, A.; Naash, M.; Remé, C.E.; Grimm, C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur. J. Neurosci. 2006, 23, 1028–1034. [Google Scholar] [CrossRef]
- Hanany, M.; Rivolta, C.; Sharon, D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. USA 2020, 117, 2710–2716. [Google Scholar] [CrossRef]
- Paskowitz, D.M.; LaVail, M.M.; Duncan, J.L. Light and inherited retinal degeneration. Br. J. Ophthalmol. 2006, 90, 1060–1066. [Google Scholar] [CrossRef]
- Ayyagari, R.; Borooah, S.; Durham, T.; Gelfman, C.; Bowman, A. Current and Future Directions in Developing Effective Treatments for PRPH2-Associated Retinal Diseases: A Workshop Report. Transl. Vis. Sci. Technol. 2024, 13, 16. [Google Scholar] [CrossRef]
- Ali, R.R.; Sarra, G.M.; Stephens, C.; Alwis, M.D.; Bainbridge, J.W.; Munro, P.M.; Fauser, S.; Reichel, M.B.; Kinnon, C.; Hunt, D.M.; et al. Restoration of photoreceptor ultrastructure and function in retinal degeneration slow mice by gene therapy. Nat. Genet. 2000, 25, 306–310. [Google Scholar] [CrossRef]
- Daich Varela, M.; Georgiadis, A.; Michaelides, M. Genetic treatment for autosomal dominant inherited retinal dystrophies: Approaches, challenges and targeted genotypes. Br. J. Ophthalmol. 2023, 107, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
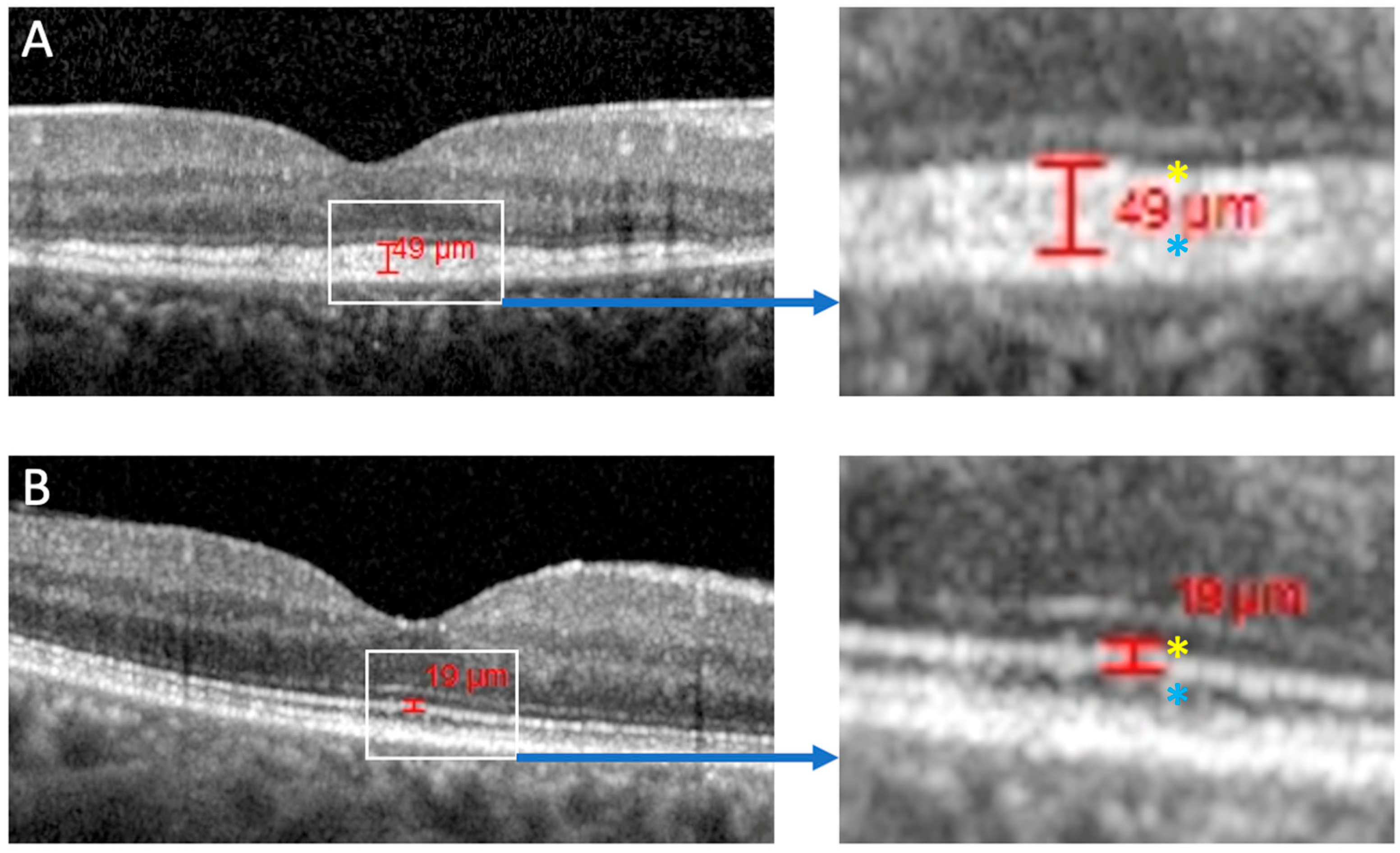
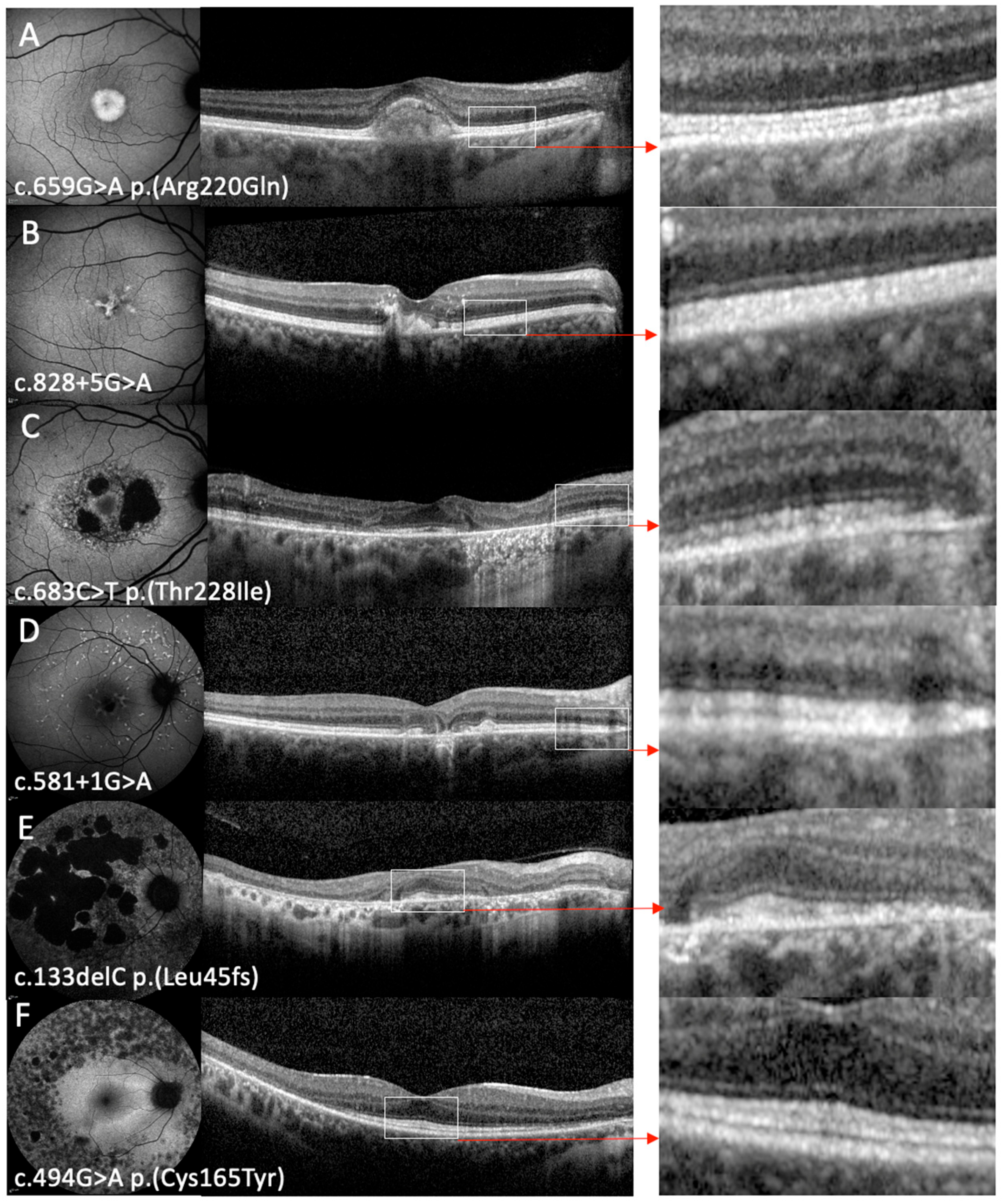

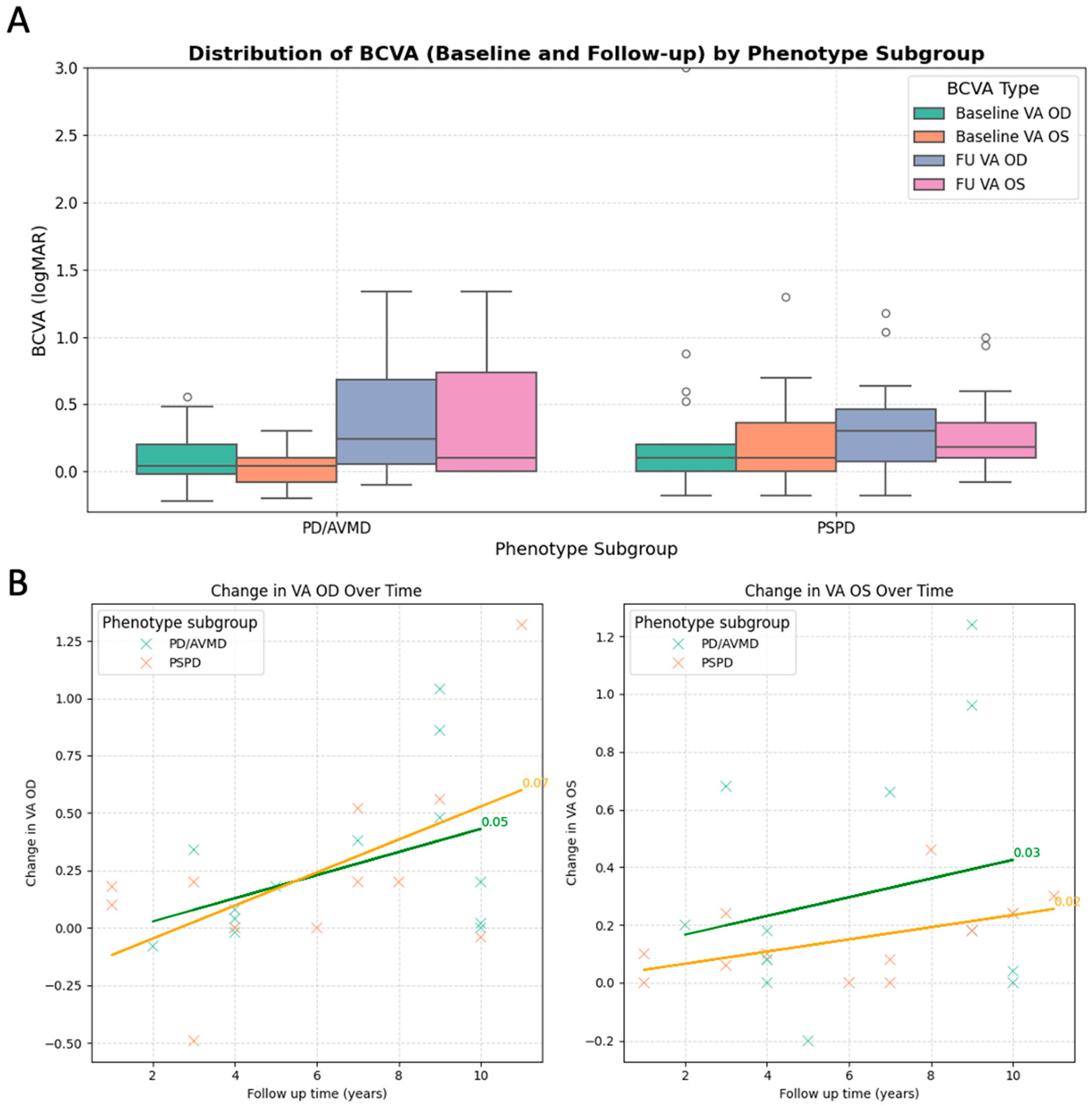
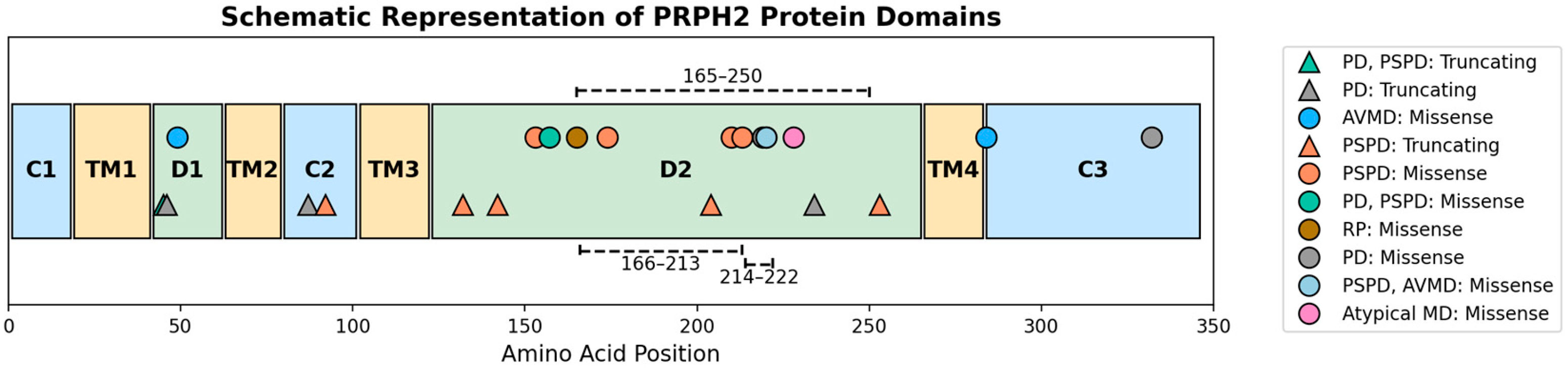

| Case ID | Variant | Protein | Sex | Baseline Age | Onset | Symptoms | Inheritance | Baseline VA OD | Baseline VA OS | FU VA OD | FU VA OS | FU Time | FAF Phenotype | OCT Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.656C>G | p.(Pro219Arg) | F | 26 | 24 | Visual distortion | AD | −0.02 | −0.2 | −0.10 | 0.00 | 2 | PD | PD |
| 2 | c.828+5G>A | F | 41 | 41 | Slightly blurred vision at night | Sporadic | 0.04 | 0.02 | 0.24 | 0.02 | 10 | PD | PD | |
| 3 | c.995T>A | p.(Val332Glu) | F | 49 | NR | NR | Sporadic | 0.08 | −0.14 | 0.10 | 0.10 | 10 | PD | PD |
| 4 | c.259_266del | p.(Asp87Glnfs*87) | F | 52 | asym | asymptomatic | Sporadic | −0.10 | 0.14 | 0.24 | 0.82 | 3 | PD | PD |
| 5 | c.629C>G | p.(Pro210Arg) | F | 36 | 26 | Photosensitivity and reduced night vision | AD | −0.22 | 0.00 | NA | NA | 0 | PD | PD |
| 6 | c.700dup | p.(Tyr234fs) | F | 43 | 43 | Blurred vision | Sporadic | 0.20 | 0.10 | NA | NA | 0 | PD | PD |
| 7 | c.136C>T | p.(Arg46*) | F | 41 | 41 | Photosensitivity and delayed dark adaptation | AD | −0.04 | 0.04 | 1 | 1 | 9 | PD | PD |
| 8 | c.133delC | p.(Leu45fs) | F | 47 | NR | NR | AD | 0.10 | 0.06 | 0.1 | 0.1 | 10 | PD | PD |
| 9A | c.469G>A | p.(Asp157Asn) | F | 39 | asym | Asymptomatic | AD (mother 9B) | −0.08 | −0.08 | 0.00 | 0.00 | 4 | PD | PD |
| 9B | c.469G>A | p.(Asp157Asn) | F | 65 | NR | NR | AD (daughter 9A) | 0.20 | 0.54 | 0.40 | 1.00 | 8 | PD->PSPD | Macular atrophy + flecks |
| 10 | exon 3 deletion | M | 47 | 38 | NR | Sporadic (AMD) | 0.02 | −0.08 | 0 | 0 | 4 | PD | AVMD | |
| 11 | c.828G>A | No AA change | M | 45 | 45 | Difficulty reading in low light | Sporadic | 0.12 | 0.3 | 0.3 | 0.1 | 5 | AVMD | AVMD |
| 12 | exon 3 deletion | M | 46 | NR | NR | Sporadic (AMD-mother) | 0.02 | −0.10 | NA | NA | 0 | Atypical PD | Foveal EZ disruption OD and minimal AVMD OS | |
| 13A | c.659G>A | p.(Arg220Gln) | F | 56 | 50 | Visual distortion | AD (sister 13B) | 0.56 | 0.30 | 0.94 | 0.96 | 7 | AVMD OD PD → AVMD OS | AVMD |
| 13B | c.659G>A | p.(Arg220Gln) | F | 52 | 52 | Blurred vision | AD (sister 13A) | 0.30 | 0.10 | 0.78 | 0.28 | 9 | AVMD | AVMD |
| 14 | c.850C>T | p.(Arg284Cys) | M | 85 | NR | NR | NR | 0.40 | 0.30 | 0.40 | 0.48 | 4 | PD | AVMD |
| 15 | c.147C>G | p.(Ser49Arg) | F | 85 | NR | Blurred vision | Sporadic | 0.48 | 0.1 | 1.34 | 1.34 | 9 | PD | AVMD + bilateral CNV |
| 16 | c.658C>T | p.(Arg220Trp) | M | 47 | 47 | NR | Sporadic | 0 | 0 | 0.04 | 0 | 4 | AVMD OD PD OS | AVMD OD PD OS |
| 17 | c.457A>G | p.(Lys153Glu) | F | 52 | asym | Asymptomatic | AD | 0.1 | 0 | 0.1 | 0.1 | 4 | PSPD | Flecks |
| 18 | c.581+1G>A | Splicing | M | 52 | 52 | Central visual distortion and delayed dark adaptation | Sporadic | −0.08 | −0.06 | NA | NA | 0 | PSPD | Macular atrophy + flecks |
| 19 | c.520T>C | p.(Trp174Arg) | M | 50 | 58 | Photosensitivity and blurred vision | AD | −0.14 | 0.36 | −0.18 | 0.60 | 10 | PSPD | Macular atrophy + spared fovea + flecks |
| 20 | c.394del | p.(Gln132fs) | F | 50 | 50 | Blurred vision, central scotomas, delayed dark adaptation, photosensitivity, and reduced night vision | AD | 0.1 | 0.7 | 0.30 | 0.94 | 3 | PSPD | Macular atrophy + foveal sparing |
| 21 | c.394del | p.(Gln132fs) | F | 50 | 40 | Blurred vision | AD | 0.1 | 0.3 | NR | NR | 0 | PSPD | Foveal atrophy + hyper-reflective material extending into ONL |
| 22 | c.756dup | p.(Ala253fs) | F | 62 | 22 | Blurred vision and metamorphopsia | Sporadic | 0.88 | 0.04 | 0.39 | 0.1 | 3 | PSPD | Macular atrophy OD + flecks OU |
| 23 | c.659G>A | p.(Arg220Gln) | M | 78 | NR | NR | AD | 0.1 | 0.6 | NA | NA | 0 | PSPD | Flecks |
| 24 | c.272_273insAT | p.(Ala92fs) | F | 69 | 55 | Photophobia and delayed dark adaptation | AD | 0.52 | 0.22 | 1.04 | 0.22 | 7 | PSPD + heterogenous background AF | Macular atrophy |
| 25 | c.638G>A | p.(Cys213Tyr) | M | 52 | 52 | Visual distortion | Sporadic | −0.18 | −0.18 | −0.08 | −0.08 | 1 | PSPD | Flecks |
| 26 | c.424delC | p.(Arg142fs) | M | 78 | 30 | Blurred vision | AD | 0.60 | 0.30 | NA | NA | 0 | PSPD | Macular atrophy |
| 27A | c.612C>G | p.(Tyr204*) | F | 44 | 44 | Delayed dark adaptation, constricted visual fields and photosensitivity | AD (mother 27B) | 0 | 0.1 | 0 | 0.1 | 6 | PSPD + heterogenous background AF | Macular atrophy + foveal sparing |
| 27B | c.612C>G | p.(Tyr204*) | F | 86 | 50 | Blurred vision | AD (daughter 27A) | HM | 1.3 | NA | NA | 0 | PSPD + heterogenous background AF | Macular atrophy |
| 28 | c.629C>G | p.(Pro210Arg) | M | 56 | 54 | Blurred vision and delayed dark adaptation | Sporadic | −0.14 | −0.12 | 1.18 | 0.18 | 11 | PSPD | Flecks + vitelliform lesion |
| 29 | c.659G>A | p.(Arg220Gln) | M | 66 | asym | asymptomatic | AD | 0 | 0 | 0.18 | 0 | 1 | PSPD | Macular atrophy + foveal sparing + flecks |
| 30 | c.394delC | p.(Gln132fs) | F | 63 | 54 | Distortion, delayed dark adaptation, photosensitivity, and central scotoma | AD | 0.08 | 0.1 | 0.64 | 0.28 | 9 | PSPD | Macular atrophy + foveal sparing OS |
| 31 | c.133del | p.(Leu45fs) | F | 61 | 57 | Delayed dark adaptation, photosensitivity, constriction of peripheral fields, and Charles Bonnet | Sporadic | 0.1 | 0.1 | 0.3 | 0.18 | 7 | PSPD + heterogenous background AF | Macular atrophy + foveal sparing |
| 32 | c.494G>A | p.(Cys165Tyr) | M | 41 | 22 | Nyctalopia and constricted visual fields | AD | 0.00 | 0.00 | 0 | 0.00 | 6 | RP | RP |
| 33 | c.683C>T | p.(Thr228Ile) | M | 53 | 53 | NR | Sporadic | 0.08 | 0.00 | −0.20 | 0.18 | 1 | Atypical mottled perifoveal AF with patches of retinal atrophy | Macular atrophy + foveal sparing |
| PD/AVMD | PSPD | p= | |||
|---|---|---|---|---|---|
| N= | 17 | 17 | - | ||
| Sex | 5 M | 12 F | 7 M | 10 F | 0.721 |
| Age of onset (mean) | 41.5 ± 9.1 | 49.1 ± 11.2 | 0.069 | ||
| Age at baseline examination (mean) | 49.2 ± 15.1 | 60.8 ± 11.8 | 0.018 * | ||
| Vision at baseline examination logMAR BCVA OD (mean) | 0.11 ± 0.21 | 0.31 ± 0.75 | 0.300 | ||
| Vision at baseline examination logMAR BCVA OS (mean) | 0.05 ± 0.15 | 0.25 ± 0.37 | 0.045 * | ||
| Variant | Amino Acid | Mutation Type | Exon/Intron | Protein Domain | GnomAD | Mutation Taster | Polyphen-2 | SIFT | CADD Score | SpliceAI Score | Novel | ACMG Class | Clinical Phenotype | Allele Count |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.133delC | p.(Leu45fs) | Frameshift | Ex 1 | D1 | 1.24 × 10−6 | Disease causing | - | - | - | 0 | Y | 5 | PD, PSPD | 2 |
| c.136C>T | p.(Arg46*) | Nonsense | Ex 1 | D1 | 6.20 × 10−6 | Disease causing | - | - | 37 | 0 | N | 5 | PD | 1 |
| c.147C>G | p.(Ser49Arg) | Missense | Ex 1 | D1 | 1.24 × 10−6 | Disease causing | benign | Tolerated | 4.18 | 0 | Y | 3 | AVMD | 1 |
| c.259_266del | p.(Asp87Glnfs*87) | Frameshift | Ex 1 | C2 | 4.96 × 10−6 | Disease causing | - | - | - | 0 | N | 5 | PD | 1 |
| c.272_273insAT | p.(Ala92fs) | Frameshift | Ex 1 | C2 | NR | Disease causing | - | - | - | 0 | Y | 5 | PSPD | 1 |
| c.394delc | p.(Gln132fs) | Nonsense | Ex 1 | D2 | 8.05 × 10−6 | - | - | - | - | 0 | N | 5 | PSPD | 2 |
| c.424delC | p.(Arg142fs) | Frameshift | Ex 1 | D2 | NR | Disease causing | - | - | - | 0 | Y | 5 | PSPD | 1 |
| c.457A>G | p.(Lys153Glu) | Missense | Ex 1 | D2 | NR | Disease causing | Probably damaging | Not tolerated | 27.3 | 0 | N | 3 | PSPD | 1 |
| c.469G>A | p.(Asp157Asn) | Missense | Ex 1 | D2 | NR | Disease causing | Probably damaging | Not tolerated | 32 | 0 | N | 4 | PD, PSPD | 2 |
| c.494G>A | p.(Cys165Tyr) | Missense | Ex 1 | D2 | NR | Disease causing | Probably damaging | Not tolerated | 30 | 0 | N | 5 | RP | 1 |
| c.520T>C | p.(Trp174Arg) | Missense | Ex 1 | D2 | 6.19 × 10−7 | Disease causing | Probably damaging | Not tolerated | 29.2 | 0 | Y | 3 | PSPD | 1 |
| c.581+1G>A | Splicing | - | 6.20 × 10−7 | - | - | - | 29.3 | 0.99 | N | 5 | PSPD | 1 | ||
| c.612C>G | p.(Tyr204*) | Nonsense | Ex 2 | D2 | 6.20 × 10−7 | Disease causing | - | - | 40 | 0 | N | 5 | PSPD | 2 |
| c.629C>G | p.(Pro210Arg) | Missense | Ex 2 | D2 | 1.24 × 10−6 | Disease causing | Probably damaging | Not tolerated | 33 | 0 | N | 4 | PD, PSPD | 1 |
| c.638G>A | p.(Cys213Tyr) | Missense | Ex 2 | D2 | 4.96 × 10−6 | Disease causing | Probably damaging | Not tolerated | 33 | 0 | N | 4 | PSPD | 1 |
| c.656C>G | p.(Pro219Arg) | Missense | Ex 2 | D2 | 9.29 × 10−6 | Disease causing | Probably damaging | Not tolerated | 29.1 | 0 | N | 3 | PD | 1 |
| c.658C>T | p.(Arg220Trp) | Missense | Ex 2 | D2 | 4.96 × 10−6 | Disease causing | Probably damaging | Not tolerated | 25.2 | 0 | N | 4 | PD, AVMD | 1 |
| c.659G>A | p.(Arg220Gln) | Missense | Ex 2 | D2 | 5.58 × 10−6 | Disease causing | Probably damaging | Not tolerated | 32 | 0 | N | 3 | PSPD, AVMD | 4 |
| c.683C>T | p.(Thr228Ile) | Missense | Ex 2 | D2 | 5.33 × 10−5 | Disease causing | Possibly damaging | Not tolerated | 26.4 | 0 | N | 3 | Atypical macular dystrophy | 1 |
| c.700dup | p.(Tyr234fs) | Frameshift | Ex 2 | D2 | 1.24 × 10−6 | - | - | - | - | 0 | N | 5 | PD | 1 |
| c.756dup | p.(Ala253fs) | Frameshift | Ex 2 | D2 | NR | Disease causing | - | - | - | 0 | Y | 5 | PSPD | 1 |
| c.828G>A | No AA change | Seemingly synonymous | Ex 2 | - | Disease causing | - | - | 25.3 | 0.91 | Y | 3 | AVMD | 1 | |
| c.850C>T | p.(Arg284Cys) | Missense | Ex 2 | C3 | 2.48 × 10−6 | Disease causing | Probably damaging | Not tolerated | 34 | 0 | N | 3 | AVMD | 1 |
| c.828+5G>A | Splicing | NR | - | - | - | 33 | 0.79 | Y | 3 | PD | 1 | |||
| c.995T>A | p.(Val332Glu) | Missense | Ex 3 | C3 | 4.34 × 10−6 | Disease causing | Probably damaging | Tolerated | 25.6 | 0 | N | 3 | PD | 1 |
| exon 3 deletion | Exon deletion | Ex 3 | - | - | - | - | - | - | - | N | 5 | AVMD | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Khuzaei, S.; Shah, M.; Reginald, A.; Baba, E.; Shanks, M.; Clouston, P.; MacLaren, R.E.; Halford, S.; De Silva, S.R.; Downes, S.M. Genotype–Phenotype Correlations in PRPH2 Retinopathies: A Comprehensive Analysis of 36 Patients from the Oxford Eye Hospital, UK. Genes 2025, 16, 1016. https://doi.org/10.3390/genes16091016
Al-Khuzaei S, Shah M, Reginald A, Baba E, Shanks M, Clouston P, MacLaren RE, Halford S, De Silva SR, Downes SM. Genotype–Phenotype Correlations in PRPH2 Retinopathies: A Comprehensive Analysis of 36 Patients from the Oxford Eye Hospital, UK. Genes. 2025; 16(9):1016. https://doi.org/10.3390/genes16091016
Chicago/Turabian StyleAl-Khuzaei, Saoud, Mital Shah, Arun Reginald, Edna Baba, Morag Shanks, Penny Clouston, Robert E. MacLaren, Stephanie Halford, Samantha R. De Silva, and Susan M. Downes. 2025. "Genotype–Phenotype Correlations in PRPH2 Retinopathies: A Comprehensive Analysis of 36 Patients from the Oxford Eye Hospital, UK" Genes 16, no. 9: 1016. https://doi.org/10.3390/genes16091016
APA StyleAl-Khuzaei, S., Shah, M., Reginald, A., Baba, E., Shanks, M., Clouston, P., MacLaren, R. E., Halford, S., De Silva, S. R., & Downes, S. M. (2025). Genotype–Phenotype Correlations in PRPH2 Retinopathies: A Comprehensive Analysis of 36 Patients from the Oxford Eye Hospital, UK. Genes, 16(9), 1016. https://doi.org/10.3390/genes16091016






