Diagnosis of Periodontitis via Neutrophil Degranulation Signatures Identified by Integrated scRNA-Seq and Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.2. Cell–Cell Communication and Pseudotime Analysis
2.3. hdWGCNA and Pathway Enrichment Analysis
2.4. LASSO Regression and Machine Learning Algorithm
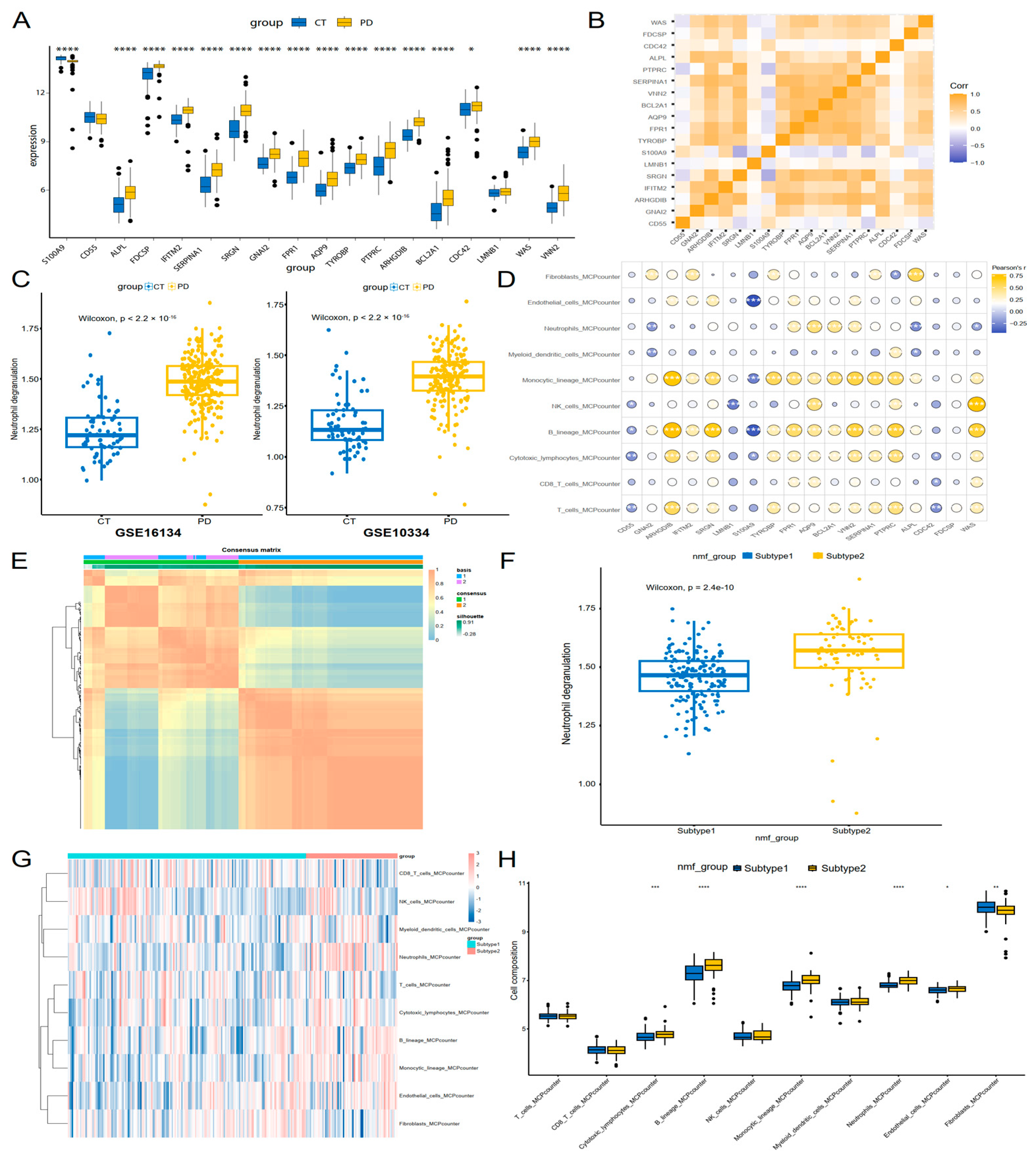
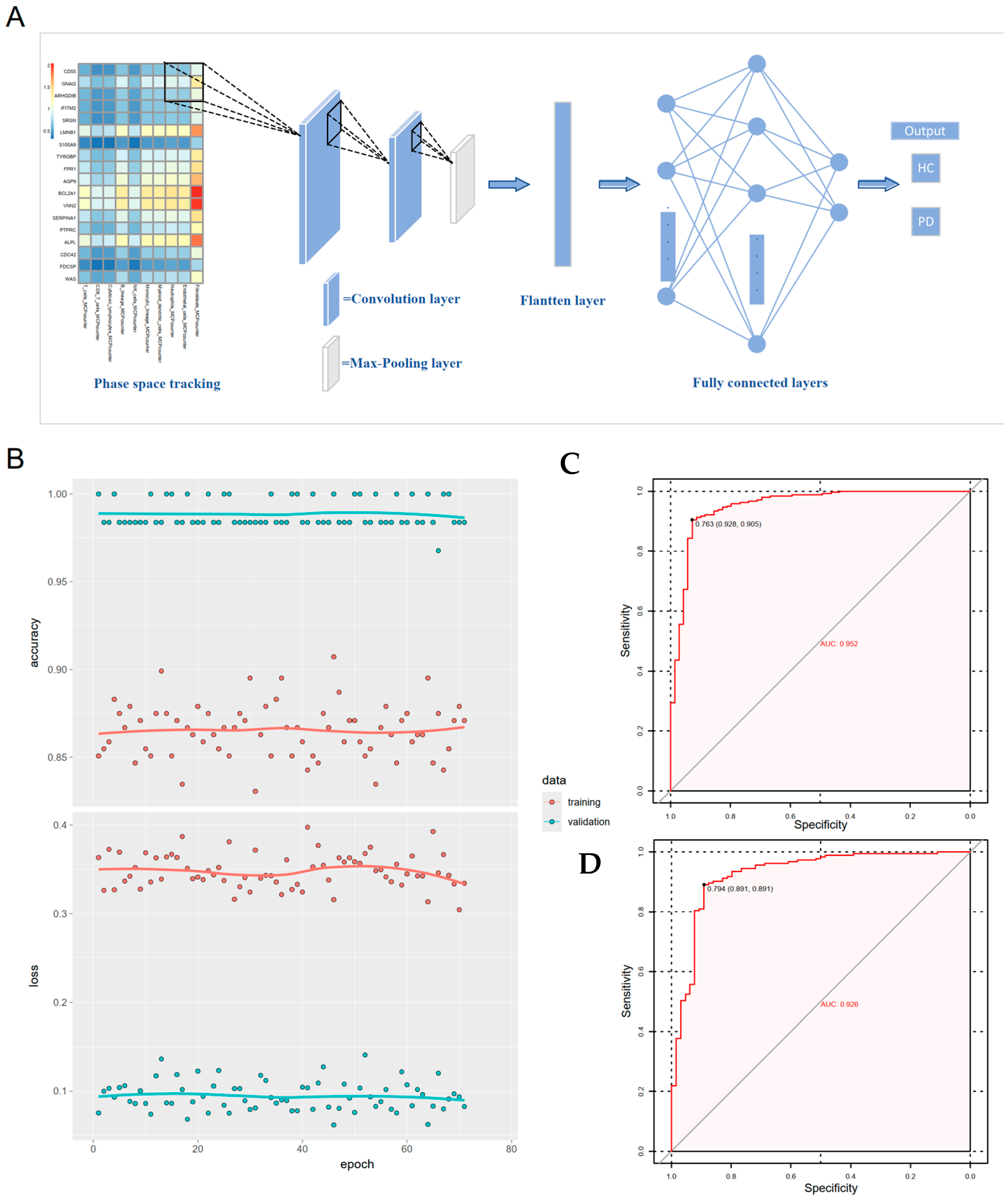
2.5. Immune Infiltration Analysis and Non-Negative Matrix Factorization (NMF)
2.6. Statistical Analysis
3. Results
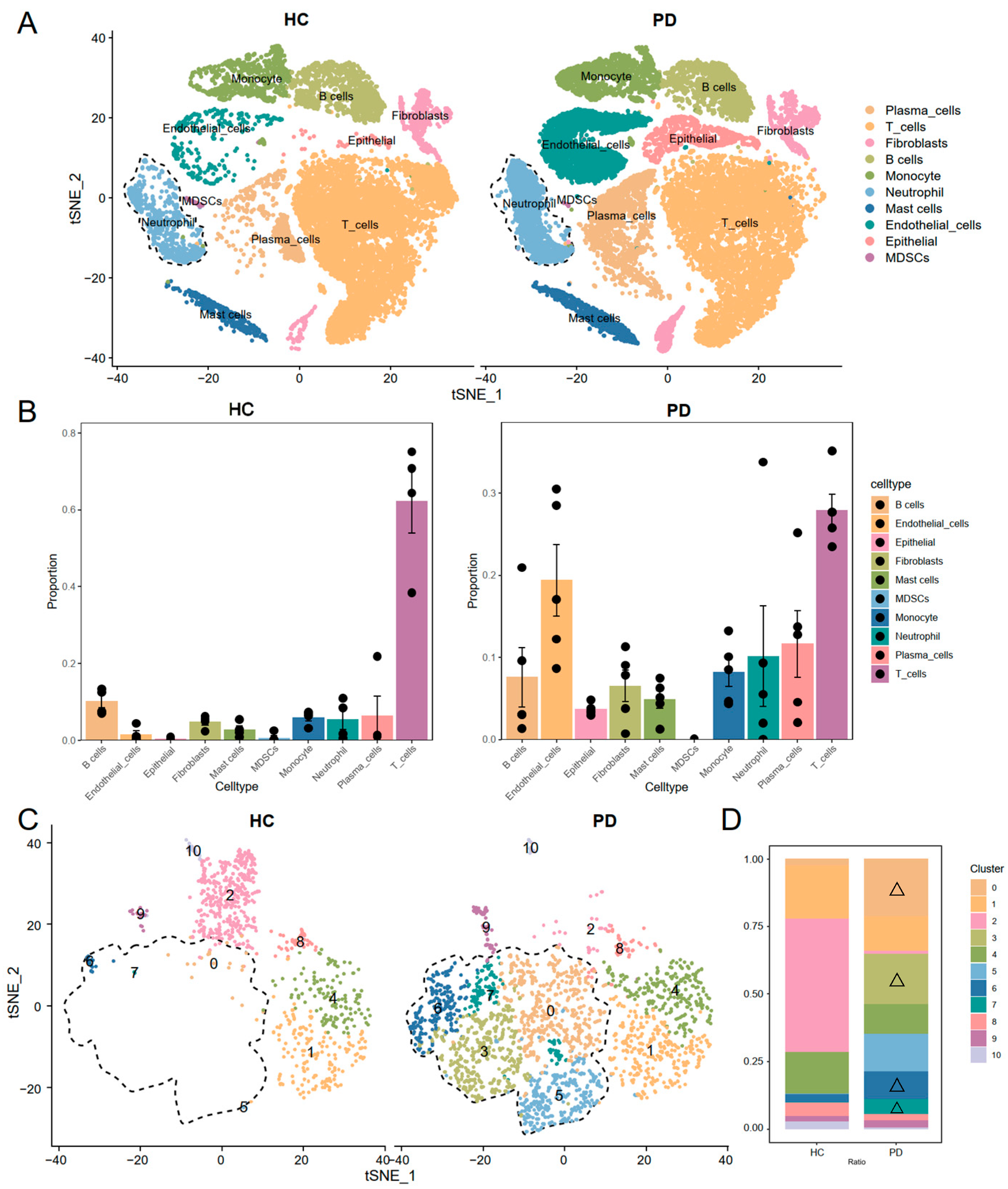
3.1. Single-Cell RNA Sequencing Identified Expanded Neutrophil Subpopulations in Periodontitis
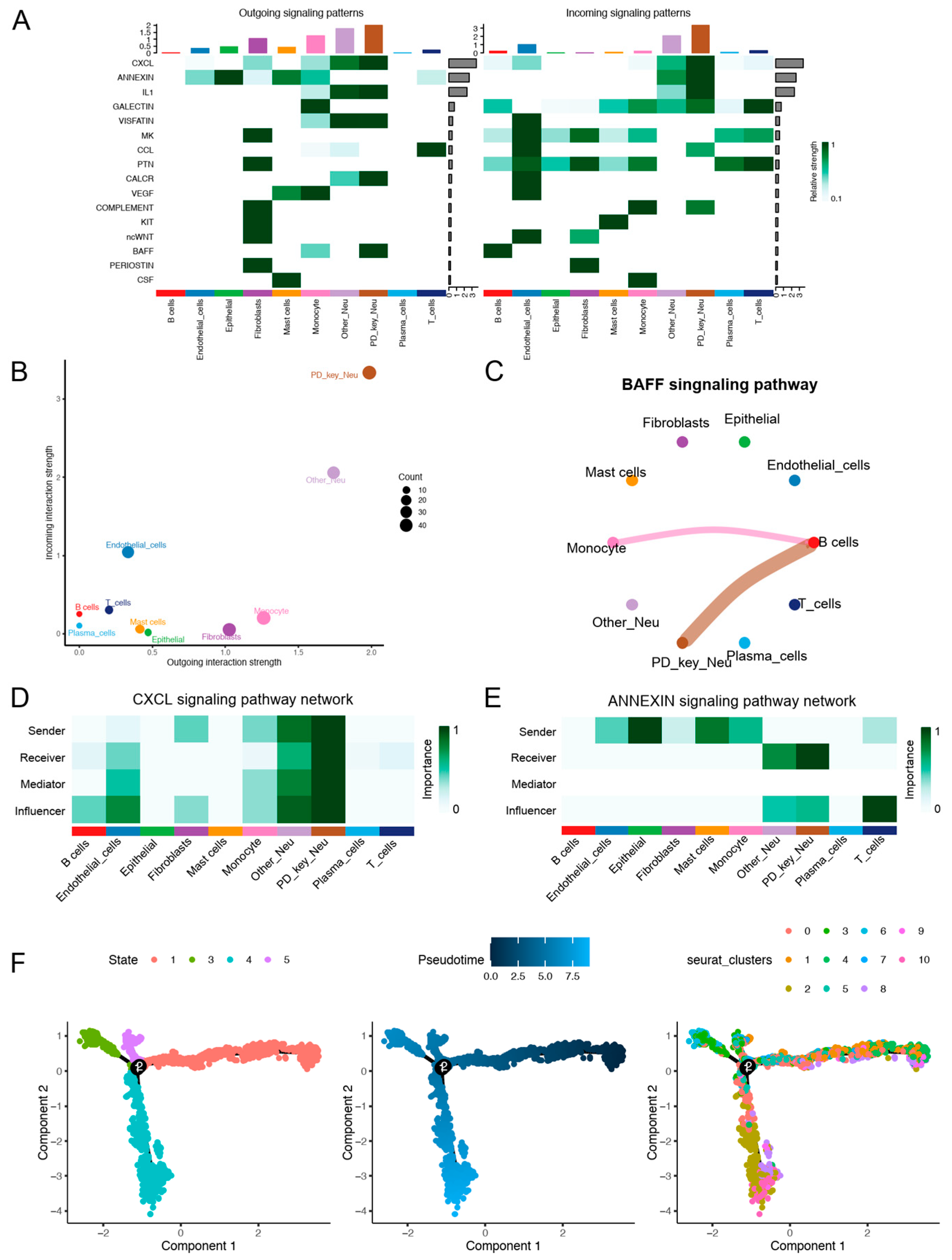
3.2. Cell–Cell Communication and Pseudotime Analysis of PD-Key-Neutrophil
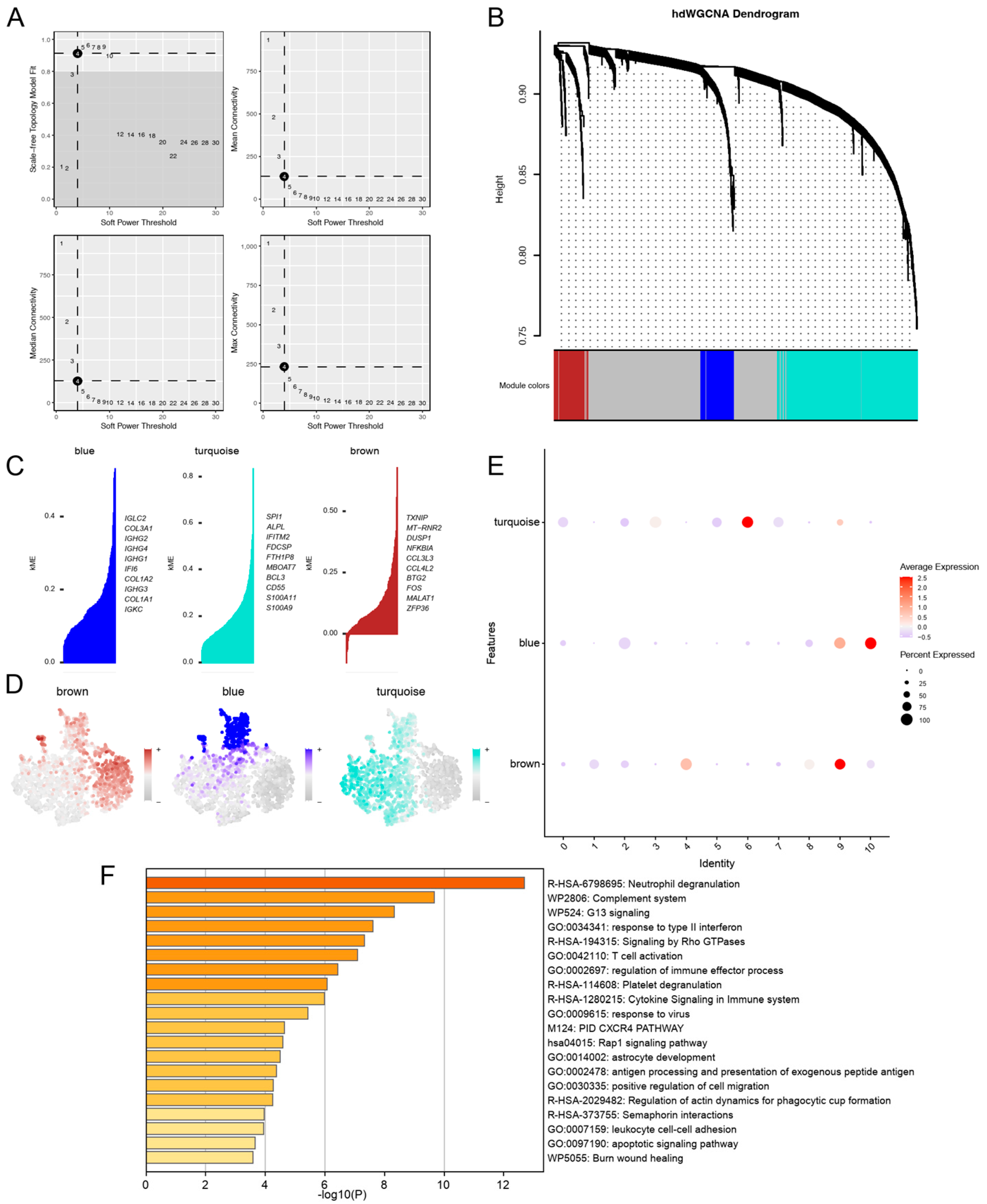
3.3. hdWGCNA Uncovered Turquoise Co-Expression Modules for PD-KEY-Neutrophil
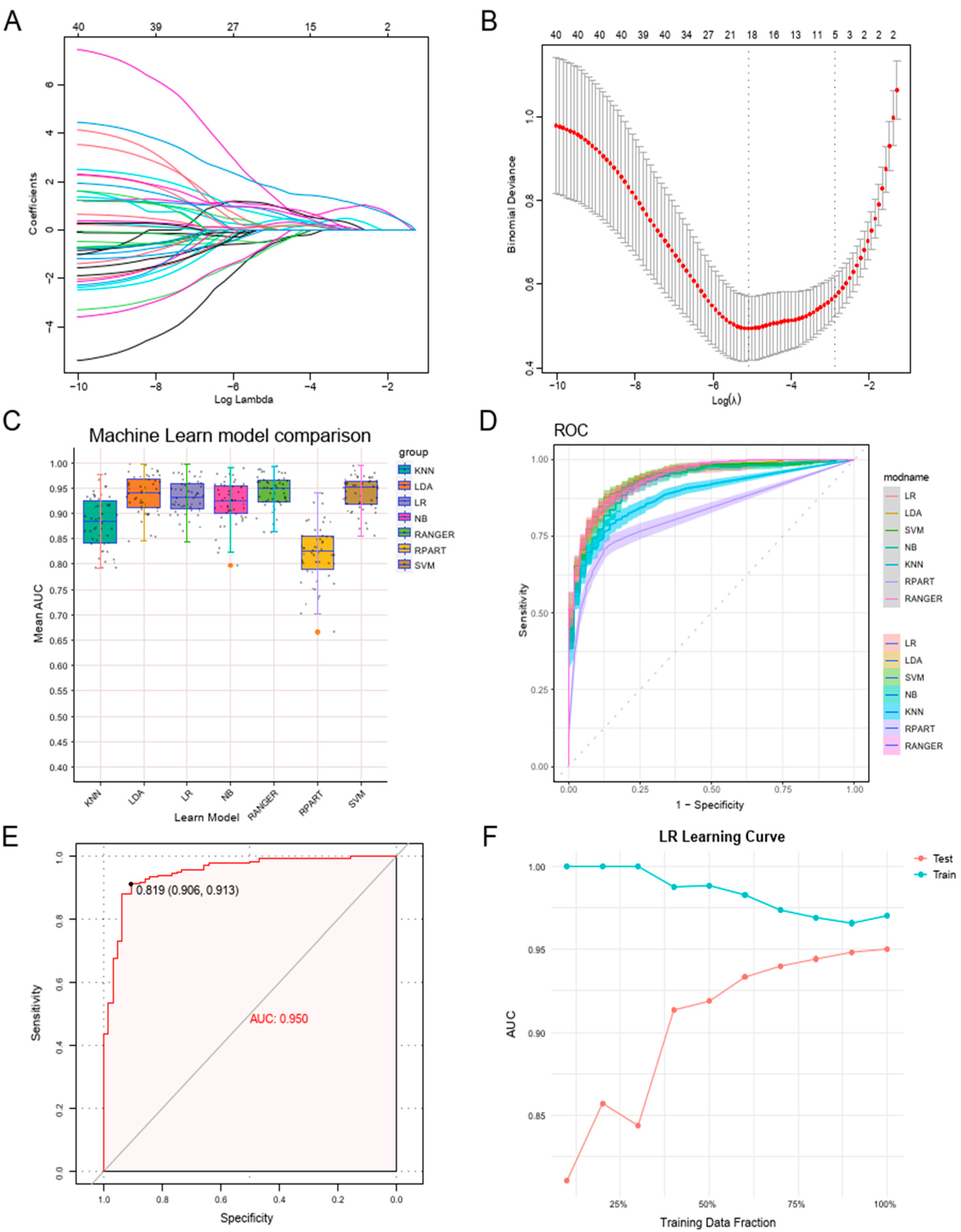
3.4. Constructing a Machine Learning Diagnostic Model Leveraging Neutrophil Degranulation Gene Signatures
3.5. Neutrophil Degranulation Genes Were Linked to Immune Profiles and Molecular Subtyping in Periodontitis
3.6. Gene-Immune Convolutional Neural Network (CNN) for Diagnosis of Periodontitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| hdWGCNA | Hierarchical weighted gene co-expression network analysis |
| CNN | Convolutional neural network |
| scRNA-seq | Single-cell RNA sequencing |
| PD | Periodontitis |
| HC | Healthy controls |
| SVM | Support vector machines |
| KNN | K-nearest neighbor |
| NB | Naïve Bayes |
| RF | Random forest |
| RPART | Recursive partitioning and regression trees |
| LDA | Linear discriminant analysis |
| GEO | Gene Expression Omnibus |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Kassebaum, N.J.; Bernabé, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.; Marcenes, W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef]
- Genco, R.J.; Sanz, M. Clinical and public health implications of periodontal and systemic diseases: An overview. Periodontol. 2000 2020, 83, 7–13. [Google Scholar] [CrossRef]
- Isola, G.; Giudice, A.L.; Polizzi, A.; Alibrandi, A.; Patini, R.; Ferlito, S. Periodontitis and Tooth Loss Have Negative Systemic Impact on Circulating Progenitor Cell Levels: A Clinical Study. Genes 2019, 10, 1022. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Chavakis, T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 2021, 21, 426–440. [Google Scholar] [CrossRef]
- Hajishengallis, G. Interconnection of periodontal disease and comorbidities: Evidence, mechanisms, and implications. Periodontol. 2000 2022, 89, 9–18. [Google Scholar] [CrossRef]
- Zhou, M.; Dong, J.; Zha, L.; Liao, Y. Causal Association between Periodontal Diseases and Cardiovascular Diseases. Genes 2021, 13, 13. [Google Scholar] [CrossRef]
- Butera, A.; Folini, E.; Cosola, S.; Russo, G.; Scribante, A.; Gallo, S.; Stablum, G.; Menchini Fabris, G.B.; Covani, U.; Genovesi, A. Evaluation of the Efficacy of Probiotics Domiciliary Protocols for the Management of Periodontal Disease, in Adjunction of Non-Surgical Periodontal Therapy (NSPT): A Systematic Literature Review. Appl. Sci. 2023, 13, 663. [Google Scholar] [CrossRef]
- Kwon, T.; Lamster, I.B.; Levin, L. Current Concepts in the Management of Periodontitis. Int. Dent. J. 2021, 71, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontol. 2000 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef]
- Cavalla, F.; Biguetti, C.C.; Melchiades, J.L.; Tabanez, A.P.; Azevedo, M.C.S.; Trombone, A.P.F.; Faveri, M.; Feres, M.; Garlet, G.P. Genetic Association with Subgingival Bacterial Colonization in Chronic Periodontitis. Genes 2018, 9, 271. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, W.; Lu, Y.; Li, H.; Yang, Y.; Geng, F.; Liu, J.; Lin, L.; Pan, Y.; Li, C. Association between periodontitis and inflammatory comorbidities: The common role of innate immune cells, underlying mechanisms and therapeutic targets. Int. Immunopharmacol. 2024, 128, 111558. [Google Scholar] [CrossRef]
- Becerra-Ruiz, J.S.; Guerrero-Velázquez, C.; Martínez-Esquivias, F.; Martínez-Pérez, L.A.; Guzmán-Flores, J.M. Innate and adaptive immunity of periodontal disease. From etiology to alveolar bone loss. Oral. Dis. 2022, 28, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Saxena, A.; Yan, W.; Uriarte, S.M.; Siqueira, R.; Li, X. The impact of aging on neutrophil functions and the contribution to periodontitis. Int. J. Oral. Sci. 2025, 17, 10. [Google Scholar] [CrossRef]
- Figueredo, C.M.; Fischer, R.G.; Gustafsson, A. Aberrant neutrophil reactions in periodontitis. J. Periodontol. 2005, 76, 951–955. [Google Scholar] [CrossRef]
- Hajishengallis, G. New developments in neutrophil biology and periodontitis. Periodontol. 2000 2020, 82, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Fan, Q.; Liu, J.; Yin, A.; Wang, P.; Zhang, W. Identification of Key Modules and Genes Associated with Major Depressive Disorder in Adolescents. Genes 2022, 13, 464. [Google Scholar] [CrossRef]
- Fine, N.; Hassanpour, S.; Borenstein, A.; Sima, C.; Oveisi, M.; Scholey, J.; Cherney, D.; Glogauer, M. Distinct Oral Neutrophil Subsets Define Health and Periodontal Disease States. J. Dent. Res. 2016, 95, 931–938. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, T.; Jiang, W.; Ma, Y.; Zhang, Q.; Chen, N.; Chu, M.; Chen, F. Single-Cell Analyses of the Oral Mucosa Reveal Immune Cell Signatures. J. Dent. Res. 2023, 102, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Morabito, S.; Reese, F.; Rahimzadeh, N.; Miyoshi, E.; Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 2023, 3, 100498. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.; Yang, Q.; Zhao, W.; Chen, Y.; Ni, Q.; Li, W.; Shi, J.; Zhang, W.; Li, L.; et al. Single-cell RNA landscape of the osteoimmunology microenvironment in periodontitis. Theranostics 2022, 12, 1074–1096. [Google Scholar] [CrossRef]
- Demmer, R.T.; Behle, J.H.; Wolf, D.L.; Handfield, M.; Kebschull, M.; Celenti, R.; Pavlidis, P.; Papapanou, P.N. Transcriptomes in healthy and diseased gingival tissues. J. Periodontol. 2008, 79, 2112–2124. [Google Scholar] [CrossRef]
- Kebschull, M.; Demmer, R.T.; Grün, B.; Guarnieri, P.; Pavlidis, P.; Papapanou, P.N. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J. Dent. Res. 2014, 93, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Desc ent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Sonabend, R.; Király, F.J.; Bender, A.; Bischl, B.; Lang, M. mlr3proba: An R package for machine learning in survival analysis. Bioinformatics 2021, 37, 2789–2791. [Google Scholar] [CrossRef]
- Ramamurthy, E.; Agarwal, S.; Toong, N.; Sestili, H.; Kaplow, I.M.; Chen, Z.; Phan, B.; Pfenning, A.R. Regression convolutional neural network models implicate peripheral immune regulatory variants in the predisposition to Alzheimer’s disease. PLoS Comput. Biol. 2024, 20, e1012356. [Google Scholar] [CrossRef]
- Hung, J.; Goodman, A.; Ravel, D.; Lopes, S.C.P.; Rangel, G.W.; Nery, O.A.; Malleret, B.; Nosten, F.; Lacerda, M.V.G.; Ferreira, M.U.; et al. Keras R-CNN: Library for cell detection in biological images using deep neural networks. BMC Bioinform. 2020, 21, 300. [Google Scholar] [CrossRef]
- Novac, O.C.; Chirodea, M.C.; Novac, C.M.; Bizon, N.; Oproescu, M.; Stan, O.P.; Gordan, C.E. Analysis of the Application Efficiency of TensorFlow and PyTorch in Convolutional Neural Network. Sensors 2022, 22, 8872. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, T.; Tang, J.; Wang, D.; Zhou, Y.; Gou, H.; Li, L.; Xu, Y. CXCR4-mediated neutrophil dynamics in periodontitis. Cell Signal 2024, 120, 111212. [Google Scholar] [CrossRef]
- Filippi, M.D.; Szczur, K.; Harris, C.E.; Berclaz, P.Y. Rho GTPase Rac1 is critical for neutrophil migration into the lung. Blood 2007, 109, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, J.L.; Dawson, D., 3rd; Emecen-Huja, P.; Nagarajan, R.; Howard, K.; Grady, M.E.; Thompson, K.; Peyyala, R.; Al-Attar, A.; Lethbridge, K.; et al. The periodontal war: Microbes and immunity. Periodontol. 2000 2017, 75, 52–115. [Google Scholar] [CrossRef]
- Uriarte, S.M.; Hajishengallis, G. Neutrophils in the periodontium: Interactions with pathogens and roles in tissue homeostasis and inflammation. Immunol. Rev. 2023, 314, 93–110. [Google Scholar] [CrossRef]
- Vitkov, L.; Muñoz, L.E.; Schoen, J.; Knopf, J.; Schauer, C.; Minnich, B.; Herrmann, M.; Hannig, M. Neutrophils Orchestrate the Periodontal Pocket. Front. Immunol. 2021, 12, 788766. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Ali, Z.S.; Sweezey, N.; Grasemann, H.; Palaniyar, N. Progression of Cystic Fibrosis Lung Disease from Childhood to Adulthood: Neutrophils, Neutrophil Extracellular Trap (NET) Formation, and NET Degradation. Genes 2019, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Machado, V.; Hussain, S.B.; Zehra, S.A.; Proença, L.; Orlandi, M.; Mendes, J.J.; D’Aiuto, F. Periodontitis and circulating blood cell profiles: A systematic review and meta-analysis. Exp. Hematol. 2021, 93, 1–13. [Google Scholar] [CrossRef]
- Vanasi, M.; Shetty, S.; Patel, V.B.; Jagnade Saini, R.; Begum, N.; Singh Baghel, R. Estimation and Co-Relation of the Neutrophil Count and Neutrophil Chemotaxis in Patients With Gingivitis, Chronic Periodontitis, and Localized Aggressive Periodontitis Compared With Healthy Controls. Cureus 2023, 15, e36627. [Google Scholar] [CrossRef]
- Williams, D.W.; Greenwell-Wild, T.; Brenchley, L.; Dutzan, N.; Overmiller, A.; Sawaya, A.P.; Webb, S.; Martin, D.; Hajishengallis, G.; Divaris, K.; et al. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 2021, 184, 4090–4104.e15. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Kuley, R.; Draves, K.E.; Elkon, K.B.; Giltiay, N.V.; Clark, E.A. B cell-activating factor (BAFF) from dendritic cells, monocytes and neutrophils is required for B cell maturation and autoantibody production in SLE-like autoimmune disease. Front. Immunol. 2023, 14, 1050528. [Google Scholar] [CrossRef]
- Yin, X.; Luistro, L.; Zhong, H.; Smith, M.; Nevins, T.; Schostack, K.; Hilton, H.; Lin, T.A.; Truitt, T.; Biondi, D.; et al. RG7212 anti-TWEAK mAb inhibits tumor growth through inhibition of tumor cell proliferation and survival signaling and by enhancing the host antitumor immune response. Clin. Cancer Res. 2013, 19, 5686–5698. [Google Scholar] [CrossRef]
- Hiyari, S.; Green, E.; Pan, C.; Lari, S.; Davar, M.; Davis, R.; Camargo, P.M.; Tetradis, S.; Lusis, A.J.; Pirih, F.Q. Genomewide Association Study Identifies Cxcl Family Members as Partial Mediators of LPS-Induced Periodontitis. J. Bone Miner. Res. 2018, 33, 1450–1463. [Google Scholar] [CrossRef]
- Sochalska, M.; Potempa, J. Manipulation of Neutrophils by Porphyromonas gingivalis in the Development of Periodontitis. Front. Cell Infect. Microbiol. 2017, 7, 197. [Google Scholar] [CrossRef]
- Makkawi, H.; Hoch, S.; Burns, E.; Hosur, K.; Hajishengallis, G.; Kirschning, C.J.; Nussbaum, G. Porphyromonas gingivalis Stimulates TLR2-PI3K Signaling to Escape Immune Clearance and Induce Bone Resorption Independently of MyD88. Front. Cell Infect. Microbiol. 2017, 7, 359. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, J.; Zhang, Y.; Kuang, S.; Shen, Z.; Qin, W. Blocking CXCR1/2 attenuates experimental periodontitis by suppressing neutrophils recruitment. Int. Immunopharmacol. 2024, 128, 111465. [Google Scholar] [CrossRef]
- Ling, M.R.; Chapple, I.L.; Matthews, J.B. Peripheral blood neutrophil cytokine hyper-reactivity in chronic periodontitis. Innate Immun. 2015, 21, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Zimny, A.; Płonczyńska, A.; Jakubowski, W.; Zubrzycka, N.; Potempa, J.; Sochalska, M. Porphyromonas gingivalis affects neutrophil pro-inflammatory activities. Front. Cell Dev. Biol. 2025, 13, 1419651. [Google Scholar] [CrossRef] [PubMed]
- Sczepanik, F.S.C.; Grossi, M.L.; Casati, M.; Goldberg, M.; Glogauer, M.; Fine, N.; Tenenbaum, H.C. Periodontitis is an inflammatory disease of oxidative stress: We should treat it that way. Periodontol. 2000 2020, 84, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Aitken, S.; Sodek, J.; McCulloch, C.A. Evidence of a direct relationship between neutrophil collagenase activity and periodontal tissue destruction in vivo: Role of active enzyme in human periodontitis. J. Periodontal Res. 1995, 30, 23–33. [Google Scholar] [CrossRef]
- Kim, T.S.; Silva, L.M.; Theofilou, V.I.; Greenwell-Wild, T.; Li, L.; Williams, D.W.; Ikeuchi, T.; Brenchley, L.; Bugge, T.H.; Diaz, P.I.; et al. Neutrophil extracellular traps and extracellular histones potentiate IL-17 inflammation in periodontitis. J. Exp. Med. 2023, 220, e20221751. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Ren, B.; Zou, L.; He, B.; Li, M. The Role of Neutrophil Extracellular Traps in Periodontitis. Front. Cell Infect. Microbiol. 2021, 11, 639144. [Google Scholar] [CrossRef]
- Qiu, W.; Guo, R.; Yu, H.; Chen, X.; Chen, Z.; Ding, D.; Zhong, J.; Yang, Y.; Fang, F. Single-cell atlas of human gingiva unveils a NETs-related neutrophil subpopulation regulating periodontal immunity. J. Adv. Res. 2024, 72, 287–301. [Google Scholar] [CrossRef]
- Sansores-España, L.D.; Melgar-Rodríguez, S.; Vernal, R.; Carrillo-Ávila, B.A.; Martínez-Aguilar, V.M.; Díaz-Zúñiga, J. Neutrophil N1 and N2 Subsets and Their Possible Association with Periodontitis: A Scoping Review. Int. J. Mol. Sci. 2022, 23, 12068. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Nicu, E.A.; Rijkschroeff, P.; Wartewig, E.; Nazmi, K.; Loos, B.G. Characterization of oral polymorphonuclear neutrophils in periodontitis patients: A case-control study. BMC Oral. Health 2018, 18, 149. [Google Scholar] [CrossRef]
- Westerlund, U.; Ingman, T.; Lukinmaa, P.L.; Salo, T.; Kjeldsen, L.; Borregaard, N.; Tjäderhane, L.; Konttinen, Y.T.; Sorsa, T. Human neutrophil gelatinase and associated lipocalin in adult and localized juvenile periodontitis. J. Dent. Res. 1996, 75, 1553–1563. [Google Scholar] [CrossRef] [PubMed]
- Klebanoff, S.J. Myeloperoxidase. Proc. Assoc. Am. Physicians 1999, 111, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Pöllänen, M.T.; Häkkinen, L.; Overman, D.O.; Salonen, J.I. Lactoferrin impedes epithelial cell adhesion in vitro. J. Periodontal Res. 1998, 33, 8–16. [Google Scholar] [CrossRef]
- Ujiie, Y.; Oida, S.; Gomi, K.; Arai, T.; Fukae, M. Neutrophil elastase is involved in the initial destruction of human periodontal ligament. J. Periodontal Res. 2007, 42, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Trocme, C.; Deffert, C.; Cachat, J.; Donati, Y.; Tissot, C.; Papacatzis, S.; Braunersreuther, V.; Pache, J.C.; Krause, K.H.; Holmdahl, R.; et al. Macrophage-specific NOX2 contributes to the development of lung emphysema through modulation of SIRT1/MMP-9 pathways. J. Pathol. 2015, 235, 65–78. [Google Scholar] [CrossRef]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef]
- Ye, X.; Bai, Y.; Li, M.; Ye, Y.; Chen, Y.; Liu, B.; Dai, Y.; Wang, S.; Pan, W.; Wang, Z.; et al. Genetic associations between circulating immune cells and periodontitis highlight the prospect of systemic immunoregulation in periodontal care. Elife 2024, 12, RP92895. [Google Scholar] [CrossRef]
- Huoshen, W.; Zhu, H.; Xiong, J.; Chen, X.; Mou, Y.; Hou, S.; Yang, B.; Yi, S.; He, Y.; Huang, H.; et al. Identification of Potential Biomarkers and Therapeutic Targets for Periodontitis. Int. Dent. J. 2025, 75, 1370–1383. [Google Scholar] [CrossRef]
- Kim, H.D.; Karna, S.; Shin, Y.; Vu, H.; Cho, H.J.; Kim, S. S100A8 and S100A9 in saliva, blood and gingival crevicular fluid for screening established periodontitis: A cross-sectional study. BMC Oral. Health 2021, 21, 388. [Google Scholar] [CrossRef] [PubMed]
- Kurniyati, K.; Clark, N.D.; Wang, H.; Deng, Y.; Sze, C.W.; Visser, M.B.; Malkowski, M.G.; Li, C. A bipartite bacterial virulence factor targets the complement system and neutrophil activation. Embo J. 2025, 44, 1154–1184. [Google Scholar] [CrossRef] [PubMed]
- Turkmen, M.; Firatli, E. The study of genetic predisposition on periodontitis and peri-implantitis. Niger. J. Clin. Pract. 2022, 25, 1799–1804. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Hu, S.; Yang, R.; Lin, L.; Mao, C.; Jin, M.; Gu, Y.; Li, G.; Jiang, B.; Gong, Y.; et al. Upregulated Vanins and their potential contribution to periodontitis. BMC Oral. Health 2022, 22, 614. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Huang, L.; Cai, S.; Xiong, X.; He, Y. Diagnosis of Periodontitis via Neutrophil Degranulation Signatures Identified by Integrated scRNA-Seq and Deep Learning. Genes 2025, 16, 1005. https://doi.org/10.3390/genes16091005
Wu H, Huang L, Cai S, Xiong X, He Y. Diagnosis of Periodontitis via Neutrophil Degranulation Signatures Identified by Integrated scRNA-Seq and Deep Learning. Genes. 2025; 16(9):1005. https://doi.org/10.3390/genes16091005
Chicago/Turabian StyleWu, Huijian, Linqing Huang, Shuting Cai, Xiaoming Xiong, and Yan He. 2025. "Diagnosis of Periodontitis via Neutrophil Degranulation Signatures Identified by Integrated scRNA-Seq and Deep Learning" Genes 16, no. 9: 1005. https://doi.org/10.3390/genes16091005
APA StyleWu, H., Huang, L., Cai, S., Xiong, X., & He, Y. (2025). Diagnosis of Periodontitis via Neutrophil Degranulation Signatures Identified by Integrated scRNA-Seq and Deep Learning. Genes, 16(9), 1005. https://doi.org/10.3390/genes16091005





