Intron Retention: A Reemerging Paradigm in RNA Biology and Post-Transcriptional Gene Regulation
Abstract
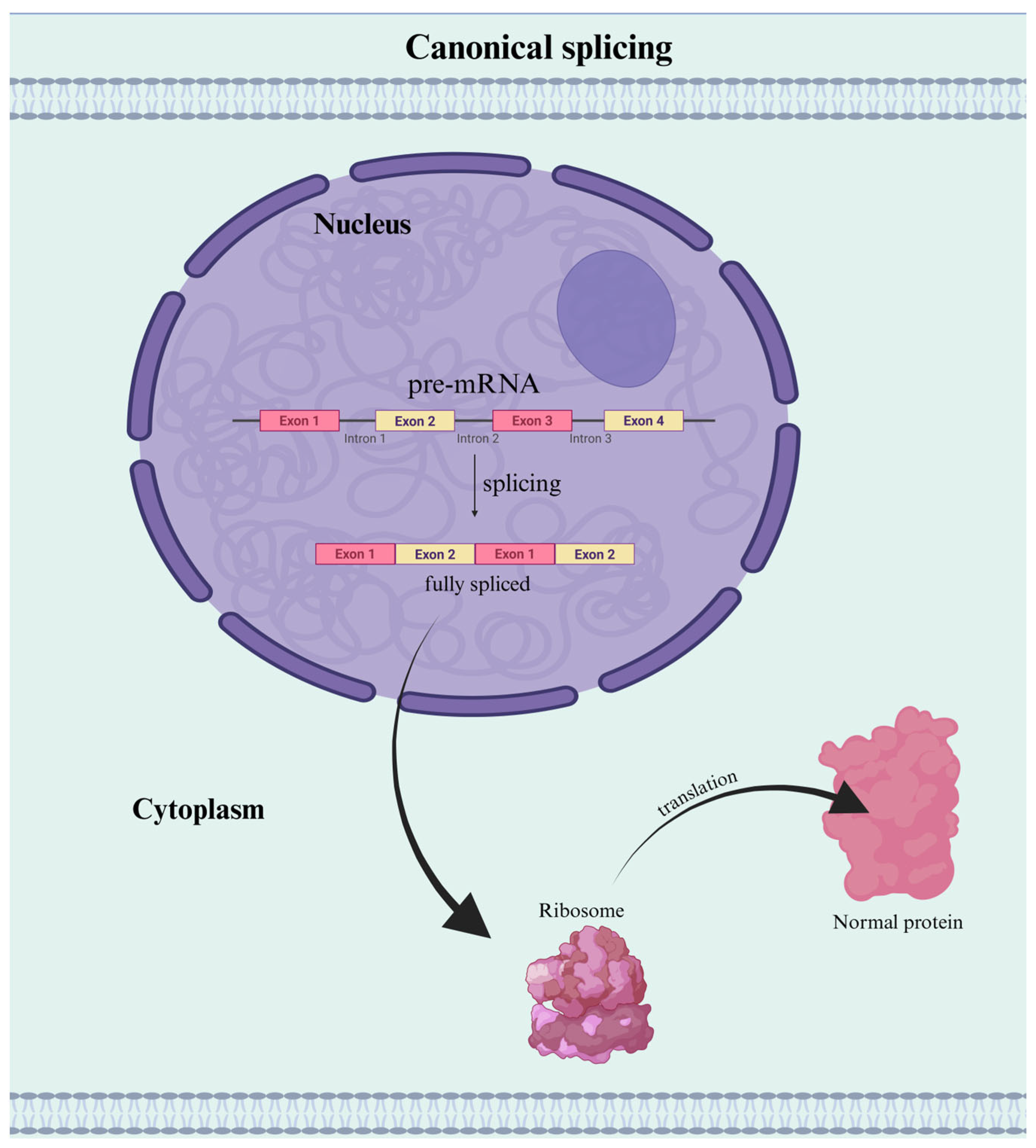
1. Introduction
2. IR Regulation: A Complex, Multifactorial Process
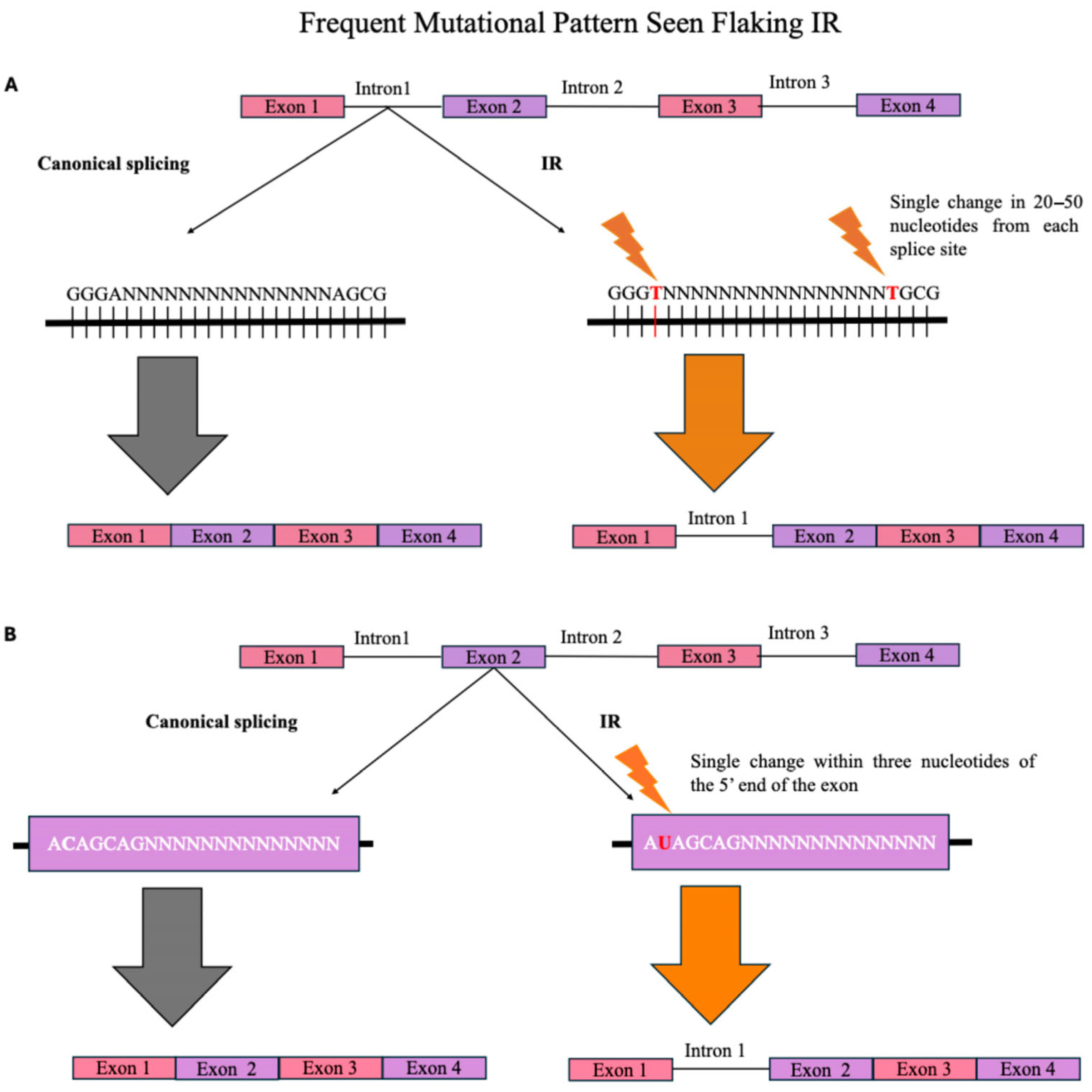
2.1. Sequence-Dependent IR Regulation
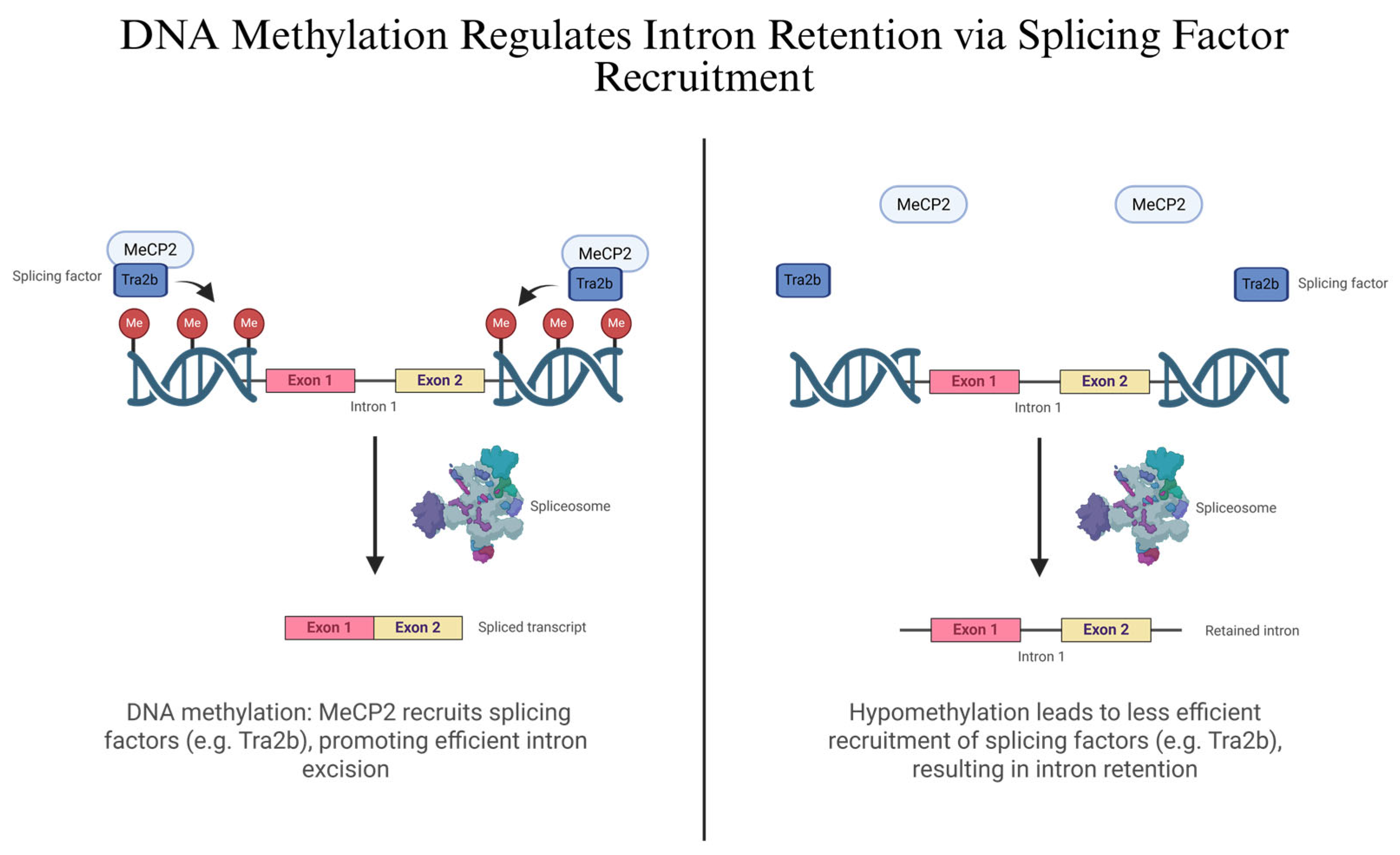
2.2. Epigenetic Regulation
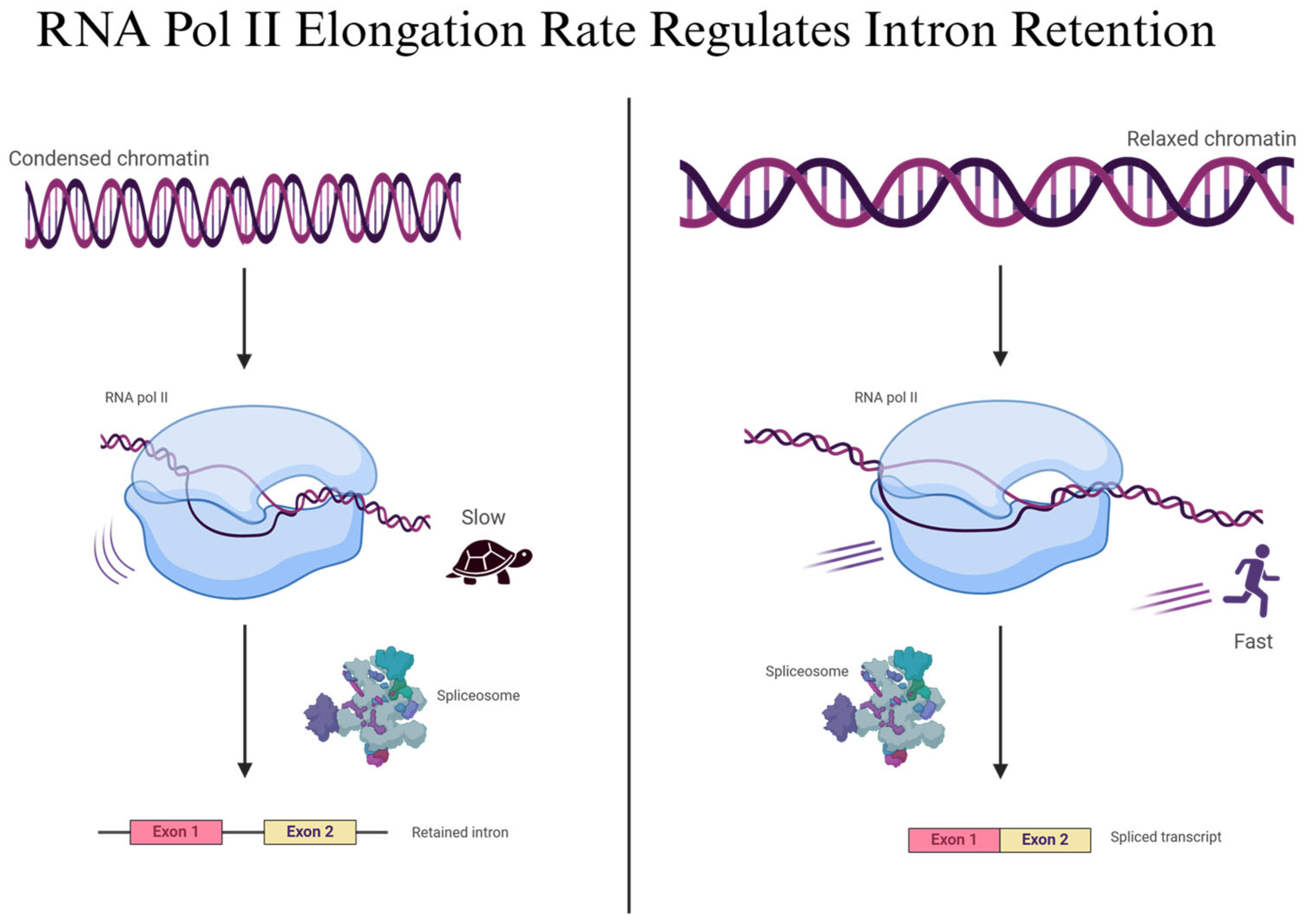
2.3. Kinetic Coupling
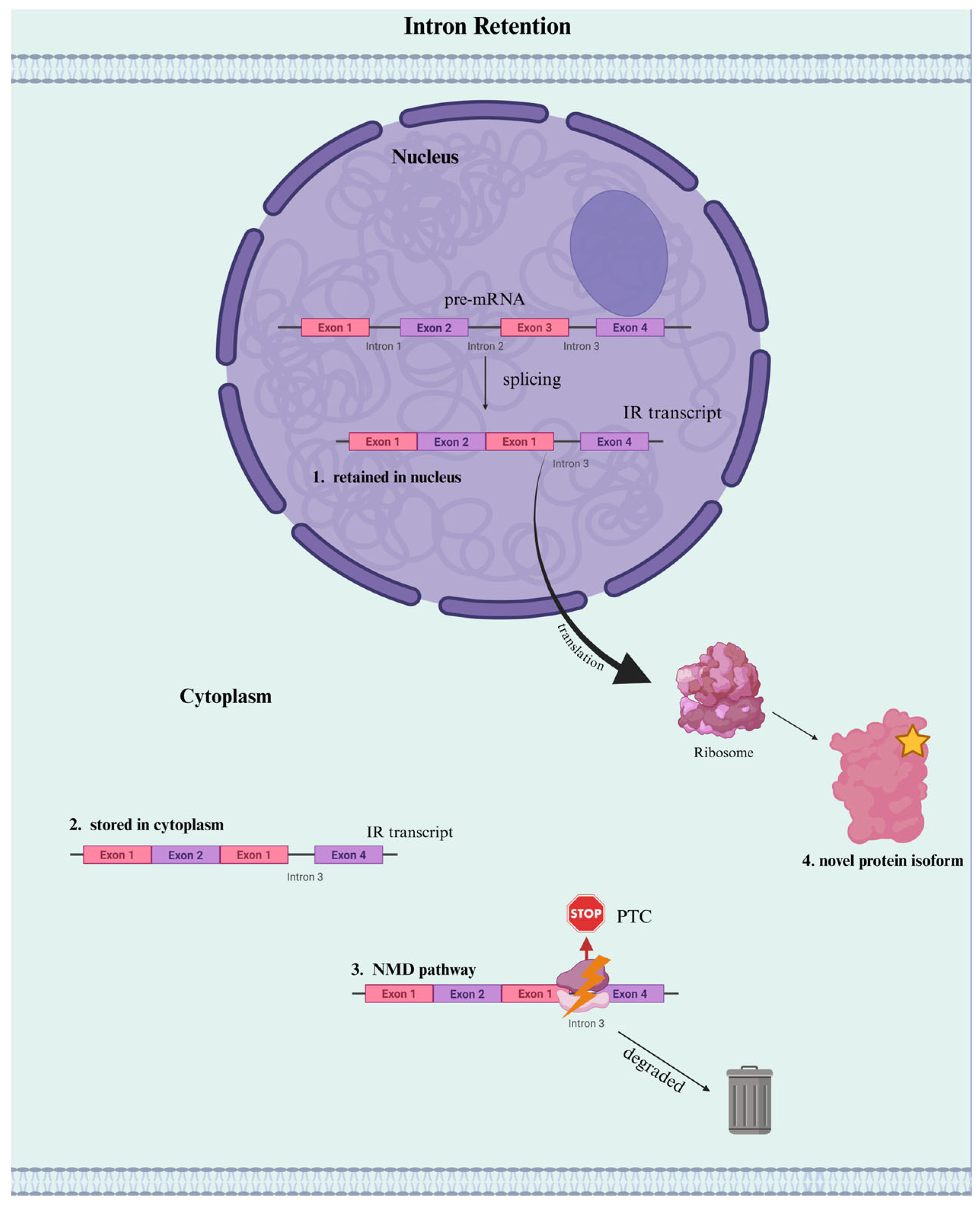
3. IR Functions as a Regulator of Gene Expression
3.1. Mechanisms of IR as a Regulator of Gene Expression
3.2. Dynamic Role of IR in Cell Differentiation
3.3. IR Role in Stress and Stimulus Response
3.4. Role of IR in Cell-Type-Specific Gene Expression Programs
3.4.1. Neurons
3.4.2. Immune System
3.4.3. Heart
4. Detection and Quantification of IR
4.1. Sequencing Technologies: Short- vs. Long-Read Approaches
4.1.1. Short-Read Sequencing
4.1.2. Long-Read Sequencing
4.2. Computational Tools
4.3. Quantitative Metrics for IR
4.3.1. IR Ratio
4.3.2. Percent Spliced-In (PSI)
4.3.3. PSI vs. IR Ratio
5. Diseases Associated with IR
5.1. Aging
- Spliceosomal Decline and IR Accumulation: Aging alters the expression and localization of core splicing factors such as SF3B1, SRSF proteins, PUF60, and ASF/SF2, compromising spliceosome fidelity. This leads to increased IR in genes like POLR2A, impairing global transcription and promoting cellular senescence. XAB2 depletion further induces IR in POLR2A, linking splicing failure to aging processes [7,16,101,102].
- Sensor Gene IR in Pre-Symptomatic Aging: In klotho and SAMP8 mouse models, IR accumulates in stress-responsive genes such as Nr1h2 and Slc16a3 (MCT4) during early aging stages, particularly in the hippocampus. These events precede neurodegeneration and may serve as early biomarkers of age-related brain dysfunction [86,87].
- Transcriptional Readthrough and IR Coupling: Aging and senescence are associated with increased transcriptional readthrough and IR, specifically in the mouse hippocampus and human prefrontal cortex. These defects are linked to altered RNA polymerase II dynamics and elevated transposon expression, indicating transcriptional dysfunction [38,88].
- Therapeutic Reversal by Kampo Medicine: The traditional Japanese herbal medicine Juzentaihoto (JTT) restores normal splicing patterns in aging models by reversing IR in sensor genes. This intervention improves metabolic regulation and may serve as a functional marker for anti-aging therapies [16,87].
5.2. Neurodegenerative Disorders
Alzheimer’s Disease (AD)
- DDIT4L IR (DIR): Retention of an intron in the DDIT4L gene produces a toxic isoform known as DIR. This protein promotes amyloid β aggregation when interacted with gelsolin, accelerating plaque formation. When bonded with GluA1, an AMPA receptor subunit, it leads to synaptic dysfunction, which contributes to cognitive decline. Hypoxia was shown to enhance DIR expression, suggesting that environmental stressors can trigger or worsen IR-mediated AD pathology [98].
- Sex-Biased IR Patterns: Females with AD exhibit 1645 elevated IR events compared to 80 in males, particularly affecting genes involved in ubiquitin signaling and Tau protein binding, which are critical in AD pathology. These events are associated with lower mRNA levels via NMD and are regulated by epigenetic markers such as H3K27ac and CTCF near splice sites, implicating at the chromatin level control of IR [103].
- IR in gene Slc16a3 (MCT4): In the AD mouse model and aged brains, there was found a retention of intron 2 between exons 2 and 3 in the Slc16a3 gene in astrocytes and endothelial cells, especially under inflammatory stimulation. This variant may indicate disruption of both the lactate transport and metabolic regulation [86].
- Tau11i from IR in MAPT: Retention of intron 11 in the MAPT gene is associated with the synthesis of a truncated protein isoform, Tau11i, that accumulates in AD brain regions. Tau11i displays features shared with full-length Tau441: altered aggregation, higher stability, and reduced microtubule binding. However, Tau11i weakly co-localizes with α-tubulin and Tau fibrils, which drives early neurodegenerative progression [99].
5.3. Oncogenic Mechanisms and Tumor Immunogenicity
5.3.1. Ovarian Cancer (OC)
- SMARCA4 Loss in SCCOHT: In SCCOHT, the loss of gene SMARCA4 is seen to induce widespread IR and exhibit tumor-specific splicing patterns. Mass spectrometry confirmed IR-derived peptides capable of MHC-I binding and T cell activation, suggesting that IR could be a source of neoantigens and a promising target for immunotherapy [89].
- WBP11 and MCM7 IR Suppression: In OC, overexpression of the splicing factor WBP11 prevents the retention of intron 4 in the MCM7 gene, leading to increased cancer cell proliferation. When WBP11 is silenced, IR is restored, MCM7 expression is reduced, and tumor growth is suppressed, making WBP11 a potential therapeutic target [104]. This mechanism reveals how splicing factor dysregulation can drive oncogenesis by repressing IR.
- CD44 Intron 9 Retention as a Biomarker: Abnormal retention of intron 9 in CD44 mRNA is seen in 60% of OC cell lines, but not in normal ovarian tissue. This intron retention may disrupt CD44 isoform expression and contribute to tumor development, making it a potential diagnostic marker for OC [105].
5.3.2. Breast Cancer Paradox
- High Baseline IR in Normal Breast Tissue: Pan-cancer analyses have shown that normal breast tissue exhibits the highest IR levels among all tissue types. This elevated baseline explains why breast tumors show reduced IR, despite IR being generally increased in other cancers [90].
- Prognostic Implications in the Luminal B Subtype: Within breast cancer subtypes, the luminal B subtype shows a link between IR levels and poor prognosis. Lower IR is associated with higher cell proliferation, suggesting that rapidly dividing tumor cells may suppress IR to streamline gene expression and support faster growth [90].
- Epigenetic Influence and Population Differences: In breast cancer, DNA hypomethylation of introns is linked to increased IR. Comparative analysis shows that African-American patients have lower intronic methylation and higher levels of retained intron expression compared to European-American patients, suggesting that the epigenetic regulation of IR may vary across populations [26].
5.4. Developmental and Sex Differentiation Disorders
46,XX DSD
- WT1 Intron 9 Retention in 46,XX DSD: A novel splice-site mutation (c.1437A > G) in the WT1 gene causes retention of intron 9, which produces a truncated +KTS isoform lacking zinc finger 4 (ZnF4) and fails to express the production of the -KTS isoform. This imbalance interferes with WT1’s role in controlling gene activity and disrupts normal sex determination, resulting in testicular or ovotesticular development in a genetically 46,XX individual [94].
5.5. Fibrotic Disorders
Renal Fibrosis
- Renal Fibrosis and TGF-β co-receptor endoglin (ENG) IR Modulation: In chronic kidney disease, ENG exists in two isoforms. One promotes fibrosis and a shorter one that protects against it. This study used antisense oligonucleotides to trigger terminal intron retention in ENG pre-mRNA, shifting splicing toward the anti-fibrotic short isoform. Patient biopsies showed that overall ENG levels were high, but the protective short isoform was reduced. Antisense oligonucleotide treatment restored short-ENG expression and reduced TGF-β1-driven pro-fibrotic genes and proteins such as ACTA2, COL1A1, and FN1. These results suggest that modulating IR can be a promising strategy to treat renal fibrosis by enhancing protective splicing outcomes [95].
6. Discussion
7. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wong, J.J.-L.; Schmitz, U. Intron retention: Importance, challenges, and opportunities. Trends Genet. 2022, 38, 789–792. [Google Scholar] [CrossRef]
- Rekosh, D.; Hammarskjold, M.L. Intron retention in viruses and cellular genes: Detention, border controls and passports. Wiley Interdiscip. Rev. RNA 2018, 9, e1470. [Google Scholar] [CrossRef]
- Ullah, F.; Hamilton, M.; Reddy, A.S.; Ben-Hur, A. Exploring the relationship between intron retention and chromatin accessibility in plants. BMC Genom. 2018, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Vanichkina, D.P.; Schmitz, U.; Wong, J.J.-L.; Rasko, J.E. Challenges in defining the role of intron retention in normal biology and disease. Semin. Cell Dev. Biol. 2018, 75, 40–49. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Argandoña-Picazo, J.; Castillo, R.; Gómez-Gómez, L. Intron retention and rhythmic diel pattern regulation of carotenoid cleavage dioxygenase 2 during crocetin biosynthesis in saffron. Plant Mol. Biol. 2016, 91, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef]
- Blanco, F.J.; Bernabeu, C. Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence. Aging Cell 2011, 10, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, N.J.; de Souza, S.J. Sequence features responsible for intron retention in human. BMC Genom. 2007, 8, 59. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Schmitz, U.; Pinello, N.; Jia, F.; Alasmari, S.; Ritchie, W.; Keightley, M.-C.; Shini, S.; Lieschke, G.J.; Wong, J.J.; Rasko, J.E.J. Intron retention enhances gene regulatory complexity in vertebrates. Genome Biol. 2017, 18, 216. [Google Scholar] [CrossRef]
- Edwards, C.R.; Ritchie, W.; Wong, J.J.-L.; Schmitz, U.; Middleton, R.; An, X.; Mohandas, N.; Rasko, J.E.J.; Blobel, G.A. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood 2016, 127, e24–e34. [Google Scholar] [CrossRef]
- Gómez-Montalvo, J.; Valle, A.d.O.F.d.; Gutiérrez, L.F.D.l.C.; Gonzalez-Meljem, J.M.; Scheckhuber, C.Q. Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination. Microb. Cell 2024, 11, 69–78. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef]
- Green, I.D.; Pinello, N.; Song, R.; Lee, Q.; Halstead, J.M.; Kwok, C.-T.; Wong, A.C.H.; Nair, S.S.; Clark, S.J.; Roediger, B.; et al. Macrophage development and activation involve coordinated intron retention in key inflammatory regulators. Nucleic Acids Res. 2020, 48, 6513–6529. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Allu, A.D.; Li, W.; Lohani, N.; Singh, M.B.; Bhalla, P.L. A dynamic intron retention program regulates the expression of several hundred genes during pollen meiosis. Plant Reprod. 2021, 34, 225–242. [Google Scholar] [CrossRef]
- Okada, N.; Oshima, K.; Iwasaki, Y.; Maruko, A.; Matsumura, K.; Iioka, E.; Vu, T.-D.; Fujitsuka, N.; Nishi, A.; Sugiyama, A.; et al. Intron retention as a new pre-symptomatic marker of aging and its recovery to the normal state by a traditional Japanese multi-herbal medicine. Gene 2021, 794, 145752. [Google Scholar] [CrossRef] [PubMed]
- Flynn, L.L.; Mitrpant, C.; Pitout, I.L.; Fletcher, S.; Wilton, S.D. Antisense Oligonucleotide-Mediated Terminal Intron Retention of the SMN2 Transcript. Mol. Ther. Nucleic Acids 2018, 11, 91–102. [Google Scholar] [CrossRef]
- Frankiw, L.; Majumdar, D.; Burns, C.; Vlach, L.; Moradian, A.; Sweredoski, M.J.; Baltimore, D. BUD13 Promotes a Type I Interferon Response by Countering Intron Retention in Irf7. Mol. Cell 2019, 73, 803–814.e6. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Cai, M. Prediction and feature analysis of intron retention events in plant genome. Comput. Biol. Chem. 2017, 68, 219–223. [Google Scholar] [CrossRef]
- Naro, C.; Jolly, A.; Persio, S.D.; Bielli, P.; Setterblad, N.; Alberdi, A.J.; Vicini, E.; Geremia, R.; la Grange, P.D.; Sette, C. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev. Cell 2017, 41, 82–93. [Google Scholar] [CrossRef]
- Ma, L.; Tan, Z.; Teng, Y.; Hoersch, S.; Horvitz, H.R. In vivo effects on intron retention and exon skipping by the U2AF large subunit and SF1/BBP in the nematode Caenorhabditis elegans. RNA 2011, 17, 2201–2211. [Google Scholar] [CrossRef]
- Ding, W.; Lin, L.; Xiao, Z.; Zou, H.; Duan, Z.; Dai, J. Multiple sequence elements determine the intron retention in histamine H3 receptors in rats and mice. Int. J. Biochem. Cell Biol. 2009, 41, 2281–2286. [Google Scholar] [CrossRef]
- Dirksen, W.P.; Sun, Q.; Rottman, F.M. Multiple splicing signals control alternative intron retention of bovine growth hormone pre-mRNA. J. Biol. Chem. 1995, 270, 5346–5352. [Google Scholar] [CrossRef]
- Hu, H.-J.; Goh, S.-H.; Lee, Y.-S. Association pattern mining of intron retention events in human based on hybrid learning machine. Genes. Genet. Syst. 2010, 85, 383–394. [Google Scholar] [CrossRef]
- Ullah, F.; Jabeen, S.; Salton, M.; Reddy, A.S.N.; Ben-Hur, A. Evidence for the role of transcription factors in the co-transcriptional regulation of intron retention. Genome Biol. 2023, 24, 53. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Shivakumar, M.; Han, S.; Sinclair, M.S.; Lee, Y.-J.; Zheng, Y.; Olopade, O.I.; Kim, D.; Lee, Y. Population-dependent Intron Retention and DNA Methylation in Breast Cancer. Mol. Cancer Res. 2018, 16, 461–469. [Google Scholar] [CrossRef]
- Bergeron, D.; Pal, G.; Beaulieu, Y.B.; Chabot, B.; Bachand, F. Regulated Intron Retention and Nuclear Pre-mRNA Decay Contribute to PABPN1 Autoregulation. Mol. Cell Biol. 2015, 35, 2503–2517. [Google Scholar] [CrossRef]
- Cho, V.; Mei, Y.; Sanny, A.; Chan, S.; Enders, A.; Bertram, E.M.; Tan, A.; Goodnow, C.C.; Andrews, T.D. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 2014, 15, R26. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Lages, A.; Patel, A.; Dhir, A.; Buratti, E.; Searle, M.; Vorechovsky, I. Optimal antisense target reducing INS intron 1 retention is adjacent to a parallel G quadruplex. Nucleic Acids Res. 2014, 42, 8161–8173. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.; Lim, Z.Q.; Khandelia, P.; Friedman, B.; Makeyev, E.V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes. Dev. 2012, 26, 1209–1223. [Google Scholar] [CrossRef]
- Marinescu, V.; Loomis, P.A.; Ehmann, S.; Beales, M.; Potashkin, J.A.; Valcarcel, J. Regulation of Retention of FosB Intron 4 by PTB. PLoS ONE 2007, 2, e828. [Google Scholar] [CrossRef] [PubMed]
- Unsworth, B.R.; Hayman, G.T.; Carroll, A.; Lelkes, P.I. Tissue-specific alternative mRNA splicing of phenylethanolamine N-methyltransferase (PNMT) during development by intron retention. Int. J. Dev. Neurosci. 1999, 17, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Luisier, R.; Tyzack, G.E.; Hall, C.E.; Mitchell, J.S.; Devine, H.; Taha, D.M.; Malik, B.; Meyer, I.; Greensmith, L.; Newcombe, J.; et al. Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat. Commun. 2018, 9, 2010. [Google Scholar] [CrossRef]
- Izumikawa, K.; Yoshikawa, H.; Ishikawa, H.; Nobe, Y.; Yamauchi, Y.; Philipsen, S.; Simpson, R.J.; Isobe, T.; Takahashi, N. Chtop (Chromatin target of Prmt1) auto-regulates its expression level via intron retention and nonsense-mediated decay of its own mRNA. Nucleic Acids Res. 2016, 44, 9847–9859. [Google Scholar] [CrossRef]
- Yeom, K.-H.; Pan, Z.; Lin, C.-H.; Lim, H.Y.; Xiao, W.; Xing, Y.; Black, D.L. Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring. Genome Res. 2021, 31, 1106–1119. [Google Scholar] [CrossRef]
- Smith, J.E.; Baker, K.E. Nonsense-mediated RNA decay—A switch and dial for regulating gene expression. Bioessays 2015, 37, 612–623. [Google Scholar] [CrossRef]
- Tan, K.; Stupack, D.G.; Wilkinson, M.F. Nonsense-mediated RNA decay: An emerging modulator of malignancy. Nat. Rev. Cancer 2022, 22, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Ding, D.; Li, X.; Shen, T.; Fu, H.; Zhong, H.; Wei, G.; Ni, T. Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence. Aging Cell 2020, 19, e13276. [Google Scholar] [CrossRef]
- Kaur, G.; Helmer, R.A.; Smith, L.A.; Martinez-Zaguilan, R.; Dufour, J.M.; Chilton, B.S.; Afink, G.B. Alternative splicing of helicase-like transcription factor (Hltf): Intron retention-dependent activation of immune tolerance at the feto-maternal interface. PLoS ONE 2018, 13, e0200211. [Google Scholar] [CrossRef]
- Mastrangelo, A.M.; Belloni, S.; Barilli, S.; Ruperti, B.; Di Fonzo, N.; Stanca, A.M.; Cattivelli, L. Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 2005, 221, 705–715. [Google Scholar] [CrossRef]
- Daoud, A.; Ben-Hur, A.; Bader, J.S. The role of chromatin state in intron retention: A case study in leveraging large scale deep learning models. PLoS Comput. Biol. 2025, 21, e1012755. [Google Scholar] [CrossRef]
- Dey, P.; Mattick, J.S. High frequency of intron retention and clustered H3K4me3-marked nucleosomes in short first introns of human long non-coding RNAs. Epigenetics Chromatin 2021, 14, 45. [Google Scholar] [CrossRef]
- Petrova, V.; Song, R.; DEEP Consortium; Nordström, K.J.V.; Walter, J.; Wong, J.J.L.; Armstrong, N.J.; Rasko, J.E.J.; Schmitz, U. Increased chromatin accessibility facilitates intron retention in specific cell differentiation states. Nucleic Acids Res. 2022, 50, 11563–11579. [Google Scholar] [CrossRef]
- Wei, G.; Liu, K.; Shen, T.; Shi, J.; Liu, B.; Han, M.; Peng, M.; Fu, H.; Song, Y.; Zhu, J.; et al. Position-specific intron retention is mediated by the histone methyltransferase SDG725. BMC Biol. 2018, 16, 44. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.-L.; Gao, D.; Nguyen, T.V.; Kwok, C.-T.; van Geldermalsen, M.; Middleton, R.; Pinello, N.; Thoeng, A.; Nagarajah, R.; Holst, J.; et al. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 2017, 8, 15134. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m 6 A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, J.; Song, Y.; Qin, X.; Lu, X.; Huang, W.; Peng, C.; Wei, J.; Huang, D.; Wang, W. CLK2 Condensates Reorganize Nuclear Speckles and Induce Intron Retention. Adv. Sci. 2024, 11, e2309588. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sengupta, S.; Mukherjee, A.; Basak, P.; Majumder, A.L. Abiotic stress regulates expression of galactinol synthase genes post-transcriptionally through intron retention in rice. Planta 2019, 249, 891–912. [Google Scholar] [CrossRef]
- Mauger, O.; Lemoine, F.; Scheiffele, P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 2016, 92, 1266–1278. [Google Scholar] [CrossRef]
- Ni, T.; Yang, W.; Han, M.; Zhang, Y.; Shen, T.; Nie, H.; Zhou, Z.; Dai, Y.; Yang, Y.; Liu, P.; et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016, 44, 6817–6829. [Google Scholar] [CrossRef]
- Pimentel, H.; Parra, M.; Gee, S.L.; Mohandas, N.; Pachter, L.; Conboy, J.G. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2015, 44, 838–851. [Google Scholar] [CrossRef]
- Uzor, S.; Zorzou, P.; Bowler, E.; Porazinski, S.; Wilson, I.; Ladomery, M. Autoregulation of the human splice factor kinase CLK1 through exon skipping and intron retention. Gene 2018, 670, 46–54. [Google Scholar] [CrossRef]
- Oghabian, A.; Greco, D.; Frilander, M.J. IntEREst: Intron-exon retention estimator. BMC Bioinform. 2018, 19, 130. [Google Scholar] [CrossRef] [PubMed]
- Dumbović, G.; Braunschweig, U.; Langner, H.K.; Smallegan, M.; Biayna, J.; Hass, E.P.; Jastrzebska, K.; Blencowe, B.; Cech, T.R.; Caruthers, M.H.; et al. Publisher Correction: Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. Nat. Commun. 2021, 12, 6189. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Wan, R.; Luan, S.; Zeng, W.; Cheung, T.H. Dek Modulates Global Intron Retention during Muscle Stem Cells Quiescence Exit. Dev. Cell 2020, 53, 661–676. [Google Scholar] [CrossRef]
- Buckley, P.T.; Khaladkar, M.; Kim, J.; Eberwine, J. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip. Rev. RNA 2014, 5, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Brady, L.K.; Wang, H.; Radens, C.M.; Bi, Y.; Radovich, M.; Maity, A.; Ivan, C.; Ivan, M.; Barash, Y.; Koumenis, C. Transcriptome analysis of hypoxic cancer cells uncovers intron retention in EIF2B5 as a mechanism to inhibit translation. PLoS Biol. 2017, 15, e2002623. [Google Scholar] [CrossRef]
- Adusumalli, S.; Ngian, Z.; Lin, W.; Benoukraf, T.; Ong, C. Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell 2019, 18, e12928. [Google Scholar] [CrossRef]
- Blanco, E.; Rojas, R.; Haeger, P.; Cuevas, R.; Perez, C.; Munita, R.; Quiroz, G.; Andrés, M.E.; Forray, M.; Gysling, K. Intron retention as an alternative splice variant of the rat urocortin 1 gene. Neuroscience 2006, 140, 1245–1252. [Google Scholar] [CrossRef]
- Song, R.; Tikoo, S.; Jain, R.; Pinello, N.; Au, A.Y.M.; Nagarajah, R.; Porse, B.; Rasko, J.E.J.; Wong, J.J. Dynamic intron retention modulates gene expression in the monocytic differentiation pathway. Immunology 2022, 165, 274–286. [Google Scholar] [CrossRef]
- Wong, J.J.; Ritchie, W.; Ebner, O.A.; Selbach, M.; Wong, J.W.; Huang, Y.; Gao, D.; Pinello, N.; Gonzalez, M.; Baidya, K.; et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 2013, 154, 583–595. [Google Scholar] [CrossRef]
- Marroquín-Flores, R.A.; Bowden, R.M.; Paitz, R.T. Brief exposure to warm temperatures reduces intron retention in Kdm6b in a species with temperature-dependent sex determination. Biol. Lett. 2021, 17, 20210167. [Google Scholar] [CrossRef]
- Wang, S.; Wu, H.; Zhao, Y.; Wang, L.; Guan, X.; Zhao, T. Mapping intron retention events contributing to complex traits using splice quantitative trait locus. Plant Methods 2023, 19, 72. [Google Scholar] [CrossRef]
- Wang, M.; Branco, A.T.; Lemos, B. The Y Chromosome Modulates Splicing and Sex-Biased Intron Retention Rates in Drosophila. Genetics 2018, 208, 1057–1067. [Google Scholar] [CrossRef]
- Lunghi, M.; Galizi, R.; Magini, A.; Carruthers, V.B.; Cristina, M.D. Expression of the glycolytic enzymes enolase and lactate dehydrogenase during the early phase of Toxoplasma differentiation is regulated by an intron retention mechanism. Mol. Microbiol. 2015, 96, 1159–1175. [Google Scholar] [CrossRef]
- Gao, Y.; Pang, A.-P.; Ma, L.; Wang, H.; Durrani, S.; Li, B.; Wu, F.-G.; Lin, F. Intron retention coupled with nonsense-mediated decay is involved in cellulase biosynthesis in cellulolytic fungi. Biotechnol. Biofuels Bioprod. 2022, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Hernández, L. The mRNA puzzle: Intron retention under stress. Plant Cell 2024, 36, 2057–2058. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, A.B.; Schmitz, U.; Marshall, A.D.; Vanichkina, D.; Nagarajah, R.; Vellozzi, M.; Wong, J.J.; Bailey, C.G.; Rasko, J.E. Ctcf haploinsufficiency mediates intron retention in a tissue-specific manner. RNA Biol. 2021, 18, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Walker, D.; Bernardo, A.; Brodbeck, J.; Balestra, M.E.; Huang, Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 2008, 28, 1452–1459. [Google Scholar] [CrossRef]
- Jangi, M.; Fleet, C.; Cullen, P.; Gupta, S.V.; Mekhoubad, S.; Chiao, E.; Allaire, N.; Bennett, C.F.; Rigo, F.; Krainer, A.R.; et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc. Natl. Acad. Sci. USA 2017, 114, E2347–E2356. [Google Scholar] [CrossRef]
- Ziff, O.J.; Taha, D.M.; Crerar, H.; Clarke, B.E.; Chakrabarti, A.M.; Kelly, G.; Neeves, J.; Tyzack, G.E.; Luscombe, N.M.; Patani, R. Reactive astrocytes in ALS display diminished intron retention. Nucleic Acids Res. 2021, 49, 3168–3184. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Özçam, M.; Abasht, B.; Balasubramanian, B. 3’UTR-Seq analysis of chicken abdominal adipose tissue reveals widespread intron retention in 3’UTR and provides insight into molecular basis of feed efficiency. PLoS ONE 2022, 17, e0269534. [Google Scholar] [CrossRef]
- Li, C.; Feng, L.; Luo, W.-W.; Lei, C.-Q.; Li, M.; Shu, H.-B. The RNA-binding protein LUC7L2 mediates MITA/STING intron retention to negatively regulate innate antiviral response. Cell Discov. 2021, 7, 46. [Google Scholar] [CrossRef]
- Sleiman, M.B.; Frochaux, M.V.; Andreani, T.; Osman, D.; Guigo, R.; Deplancke, B. Enteric infection induces Lark-mediated intron retention at the 5′ end of Drosophila genes. Genome Biol. 2020, 21, 4. [Google Scholar] [CrossRef]
- Torrado, M.; Iglesias, R.; Nespereira, B.; Centeno, A.; López, E.; Mikhailov, A.T. Intron retention generates ANKRD1 splice variants that are co-regulated with the main transcript in normal and failing myocardium. Gene 2009, 440, 28–41. [Google Scholar] [CrossRef]
- Gualandi, N.; Iperi, C.; Esposito, M.; Ansaloni, F.; Gustincich, S.; Sanges, R. Meta-Analysis Suggests That Intron Retention Can Affect Quantification of Transposable Elements from RNA-Seq Data. Biology 2022, 11, 826. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Oshima, K.; Maruko, A.; Sekine, M.; Ito, N.; Wakasugi, A.; Mori, E.; Odaguchi, H.; Kobayashi, Y. Intron retention as an excellent marker for diagnosing depression and for discovering new potential pathways for drug intervention. Front. Psychiatry 2024, 15, 1450708. [Google Scholar] [CrossRef]
- Broseus, L.; Ritchie, W. Challenges in detecting and quantifying intron retention from next generation sequencing data. Comput. Struct. Biotechnol. J. 2020, 18, 501–508. [Google Scholar] [CrossRef]
- Ura, H.; Togi, S.; Niida, Y. Target-capture full-length double-stranded cDNA long-read sequencing through Nanopore revealed novel intron retention in patient with tuberous sclerosis complex. Front. Genet. 2023, 14, 1256064. [Google Scholar] [CrossRef] [PubMed]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.-L.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.J.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef]
- Lorenzi, C.; Barriere, S.; Arnold, K.; Luco, R.F.; Oldfield, A.J.; Ritchie, W. IRFinder-S: A comprehensive suite to discover and explore intron retention. Genome Biol. 2021, 22, 307. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-D.; Funk, C.C.; Price, N.D. iREAD: A tool for intron retention detection from RNA-seq data. BMC Genom. 2020, 21, 128. [Google Scholar] [CrossRef]
- Shiraishi, Y.; Okada, A.; Chiba, K.; Kawachi, A.; Omori, I.; Mateos, R.N.; Iida, N.; Yamauchi, H.; Kosaki, K.; Yoshimi, A. Systematic identification of intron retention associated variants from massive publicly available transcriptome sequencing data. Nat. Commun. 2022, 13, 5357. [Google Scholar] [CrossRef]
- Canson, D.M.; Davidson, A.L.; de la Hoya, M.; Parsons, M.T.; Glubb, D.M.; Kondrashova, O.; Spurdle, A.B.; Kendziorski, C. SpliceAI-10k calculator for the prediction of pseudoexonization, intron retention, and exon deletion. Bioinformatics 2023, 39, btad179. [Google Scholar] [CrossRef]
- Zheng, J.-T.; Lin, C.-X.; Fang, Z.-Y.; Li, H.-D. Intron Retention as a Mode for RNA-Seq Data Analysis. Front. Genet. 2020, 11, 586. [Google Scholar] [CrossRef]
- El-Seedy, A.; Pellerin, L.; Page, G.; Ladeveze, V. Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice. Genes. 2023, 14, 1949. [Google Scholar] [CrossRef]
- Vu, T.-D.; Ito, N.; Oshima, K.; Maruko, A.; Nishi, A.; Mizoguchi, K.; Odaguchi, H.; Kobayashi, Y.; Okada, N. Intron retention is a stress response in sensor genes and is restored by Japanese herbal medicines: A basis for future clinical applications. Gene 2022, 830, 146496. [Google Scholar] [CrossRef] [PubMed]
- Pabis, K.; Barardo, D.; Sirbu, O.; Selvarajoo, K.; Gruber, J.; Kennedy, B.K. A concerted increase in readthrough and intron retention drives transposon expression during aging and senescence. eLife 2024, 12, RP87811. [Google Scholar] [CrossRef] [PubMed]
- Raupach, E.A.; Hegde, A.M.; Garcia-Mansfield, K.; Alcantara, M.; Rose, D.L.; Halperin, R.F.; Orlando, K.A.; Lang, J.D.; Sharma, R.; David-Dirgo, V.; et al. Loss of SMARCA4 Leads to Intron Retention and Generation of Tumor-Associated Antigens in Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Cancer Res. 2025, 85, 2626–2642. [Google Scholar] [CrossRef]
- Shah, J.S.; Milevskiy, M.J.G.; Petrova, V.; Au, A.Y.M.; Wong, J.J.L.; Visvader, J.E.; Schmitz, U.; Rasko, J.E.J. Towards resolution of the intron retention paradox in breast cancer. Breast Cancer Res. 2022, 24, 100. [Google Scholar] [CrossRef]
- Zhou, J.; Lai, P.B.-S.; Tsui, S.K.-W. Identification of a non-coding KLF4 transcript generated from intron retention and downregulated in human hepatocellular carcinoma. Int. J. Oncol. 2015, 47, 1554–1562. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Cesarano, A.; Bombaci, G.; Reiter, J.L.; Yu, C.Y.; Wang, Y.; Jiang, Z.; Abu Zaid, M.; Huang, K.; Lu, X.; et al. Intron retention-induced neoantigen load correlates with unfavorable prognosis in multiple myeloma. Oncogene 2021, 40, 6130–6138. [Google Scholar] [CrossRef]
- Kanagasabai, R.; Serdar, L.; Karmahapatra, S.; Kientz, C.A.; Ellis, J.; Ritke, M.K.; Elton, T.S.; Yalowich, J.C. Alternative RNA Processing of Topoisomerase IIα in Etoposide-Resistant Human Leukemia K562 Cells: Intron Retention Results in a Novel C-Terminal Truncated 90-kDa Isoform. J. Pharmacol. Exp. Ther. 2017, 360, 152–163. [Google Scholar] [CrossRef]
- Sirokha, D.; Gorodna, O.; Vitrenko, Y.; Zelinska, N.; Ploski, R.; Nef, S.; Jaruzelska, J.; Kusz-Zamelczyk, K.; Livshits, L. A Novel WT1 Mutation Identified in a 46,XX Testicular/Ovotesticular DSD Patient Results in the Retention of Intron 9. Biology 2021, 10, 1248. [Google Scholar] [CrossRef]
- Gerrits, T.; Dijkstra, K.L.; Bruijn, J.A.; Scharpfenecker, M.; Bijkerk, R.; Baelde, H.J. Antisense oligonucleotide-mediated terminal intron retention of endoglin: A potential strategy to inhibit renal interstitial fibrosis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2024, 1870, 167186. [Google Scholar] [CrossRef]
- Yin, J.; Zhou, J.; Chen, J.; Xu, T.; Zhang, Z.; Zhang, H.; Yuan, C.; Cheng, X.; Qin, Y.; Zheng, B.; et al. Case Report: A Novel Variant c.2262+3A>T of the SCN5A Gene Results in Intron Retention Associated with Incessant Ventricular Tachycardias. Front. Med. 2021, 8, 659119. [Google Scholar] [CrossRef] [PubMed]
- Kallabi, F.; Ben Rhouma, B.; Baklouti, S.; Ghorbel, R.; Felhi, R.; Keskes, L.; Kamoun, H. Splicing Defects in the AAAS Gene Leading to both Exon Skipping and Partial Intron Retention in a Tunisian Patient with Allgrove Syndrome. Horm. Res. Paediatr. 2016, 86, 90–93. [Google Scholar] [CrossRef]
- Li, K.-C.; Shi, H.-X.; Li, Z.; You, P.; Pan, J.; Cai, Y.-C.; Li, J.-W.; Ma, X.-F.; Zhang, S.; Diao, L.; et al. Human DDIT4L intron retention contributes to cognitive impairment and amyloid plaque formation. Cell Discov. 2025, 11, 12. [Google Scholar] [CrossRef]
- Ngian, Z.-K.; Tan, Y.-Y.; Choo, C.-T.; Lin, W.-Q.; Leow, C.-Y.; Mah, S.-J.; Lai, M.K.-P.; Chen, C.L.-H.; Ong, C.-T. Truncated Tau caused by intron retention is enriched in Alzheimer’s disease cortex and exhibits altered biochemical properties. Proc. Natl. Acad. Sci. USA 2022, 119, e2204179119. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, Y.; Zhang, H.; Wright, J.T.; Simmer, J.P.; Hu, J.C.-C.; Kim, J.-W. Translational Attenuation by an Intron Retention in the 5′ UTR of ENAM Causes Amelogenesis Imperfecta. Biomedicines 2021, 9, 456. [Google Scholar] [CrossRef]
- Hou, S.; Qu, D.; Li, Y.; Zhu, B.; Liang, D.; Wei, X.; Tang, W.; Zhang, Q.; Hao, J.; Guo, W.; et al. XAB2 depletion induces intron retention in POLR2A to impair global transcription and promote cellular senescence. Nucleic Acids Res. 2019, 47, 8239–8254. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Kew, C.; Fernandes, S.d.A.; Löhrke, A.; Han, L.; Demetriades, C.; Antebi, A. Decreased spliceosome fidelity and egl-8 intron retention inhibit mTORC1 signaling to promote longevity. Nat. Aging 2022, 2, 796–808. [Google Scholar] [CrossRef]
- Choo, C.; Leow, C.; Ong, C. Higher Intron Retention Levels in Female Alzheimer’s Brains May Be Linked to Disease Prevalence. Aging Cell 2025, 24, e14457. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, Z.; Li, Y.; Song, K. The splicing factor WBP11 mediates MCM7 intron retention to promote the malignant progression of ovarian cancer. Oncogene 2024, 43, 1565–1578. [Google Scholar] [CrossRef]
- Stickeler, E.; Möbus, V.J.; Kieback, D.G.; Kohlberger, P.; Runnebaum, I.B.; Kreienberg, R. Intron 9 retention in gene transcripts suggests involvement of CD44 in the tumorigenesis of ovarian cancer. Anticancer Res. 1997, 17, 4395–4398. [Google Scholar]
- Jung, H.; Lee, D.; Lee, J.; Park, D.; Kim, Y.J.; Park, W.-Y.; Hong, D.; Park, P.J.; Lee, E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015, 47, 1242–1248. [Google Scholar] [CrossRef]
- Moran, Y.; Weinberger, H.; Reitzel, A.M.; Sullivan, J.C.; Kahn, R.; Gordon, D.; Finnerty, J.R.; Gurevitz, M. Intron retention as a posttranscriptional regulatory mechanism of neurotoxin expression at early life stages of the starlet anemone Nematostella vectensis. J. Mol. Biol. 2008, 380, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hilarion, S.; Paulet, D.; Lee, K.-T.; Hon, C.-C.; Lechat, P.; Mogensen, E.; Moyrand, F.; Proux, C.; Barboux, R.; Bussotti, G.; et al. Intron retention-dependent gene regulation in Cryptococcus neoformans. Sci. Rep. 2016, 6, 32252. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, V.; Mallya, M.; Campbell, R.D.; Aguado, B. Regulation of expression of two LY-6 family genes by intron retention and transcription induced chimerism. BMC Mol. Biol. 2008, 9, 81. [Google Scholar] [CrossRef]
- Lee, S.; Jung, H.; Choi, S.; Cho, N.; Kim, E.-M.; Kim, K.K. Intron retention decreases METTL3 expression by inhibiting mRNA export to the cytoplasm. BMB Rep. 2023, 56, 514–519. [Google Scholar] [CrossRef]
- Erhardt, S.; Stoecklin, G. The heat’s on: Nuclear stress bodies signal intron retention. EMBO J. 2020, 39, e104154. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, L.; Landry-Voyer, A.-M.; Latour, M.; Yague-Sanz, C.; Bachand, F. PABPN1 prevents the nuclear export of an unspliced RNA with a constitutive transport element and controls human gene expression via intron retention. RNA 2023, 29, 644–662. [Google Scholar] [CrossRef] [PubMed]
- Hadar, S.; Meller, A.; Saida, N.; Shalgi, R. Stress-induced transcriptional readthrough into neighboring genes is linked to intron retention. iScience 2022, 25, 105543. [Google Scholar] [CrossRef]
- Lunghi, M.; Spano, F.; Magini, A.; Emiliani, C.; Carruthers, V.B.; Di Cristina, M. Alternative splicing mechanisms orchestrating post-transcriptional gene expression: Intron retention and the intron-rich genome of apicomplexan parasites. Curr. Genet. 2016, 62, 31–38. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic Stresses Modulate Landscape of Poplar Transcriptome via Alternative Splicing, Differential Intron Retention, and Isoform Ratio Switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Bourdin, C.M.; Moignot, B.; Wang, L.; Murillo, L.; Juchaux, M.; Quinchard, S.; Lapied, B.; Guérineau, N.C.; Dong, K.; Legros, C.; et al. Intron Retention in mRNA encoding ancillary subunit of insect voltage-gated sodium channel modulates channel expression, gating regulation and drug sensitivity. PLoS ONE 2013, 8, e67290. [Google Scholar] [CrossRef]
- Rocchi, V.; Janni, M.; Bellincampi, D.; Giardina, T.; D’oVidio, R. Intron retention regulates the expression of pectin methyl esterase inhibitor (Pmei) genes during wheat growth and development. Plant Biol. 2012, 14, 365–373. [Google Scholar] [CrossRef]
- Auffray, C.; Gayon, R.; Benraiss, A.; Martin, N.; Laurendeau, I.; Garaud, J.; Lucas, B.; Boitard, C.; Krief, P. An 8-bp deletion in mNOTCH4 intron 10 leads to its retention in mRNA and to synthesis of a truncated protein. Exp. Cell Res. 2006, 312, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef]
- Smith, L.D.; Lucas, C.M.; Eperon, I.C.; Buratti, E. Intron retention in the alternatively spliced region of RON results from weak 3’ splice site recognition. PLoS ONE 2013, 8, e77208. [Google Scholar] [CrossRef]
- Brown, P.J.; Kagaya, R.; Banham, A.H. Characterization of human FOXP1 isoform 2, using monoclonal antibody 4E3-G11, and intron retention as a tissue-specific mechanism generating a novel FOXP1 isoform. Histopathology 2008, 52, 632–637. [Google Scholar] [CrossRef]
- Flodrops, M.; Dujardin, G.; Busson, A.; Trouvé, P.; Ka, C.; Simon, B.; Arzur, D.; Le Jossic-Corcos, C.; Corcos, L. TIMP1 intron 3 retention is a marker of colon cancer progression controlled by hnRNPA1. Mol. Biol. Rep. 2020, 47, 3031–3040. [Google Scholar] [CrossRef]
- Niba, E.T.E.; Yamanaka, R.; Rani, A.Q.M.; Awano, H.; Matsumoto, M.; Nishio, H.; Matsuo, M. DMD transcripts in CRL-2061 rhabdomyosarcoma cells show high levels of intron retention by intron-specific PCR amplification. Cancer Cell Int. 2017, 17, 58. [Google Scholar] [CrossRef]
- Chen, S.X.; Simpson, E.; Reiter, J.L.; Liu, Y. Bioinformatics detection of modulators controlling splicing factor-dependent intron retention in the human brain. Hum. Mutat. 2022, 43, 1629–1641. [Google Scholar] [CrossRef]
- Dvinge, H.; Bradley, R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015, 7, 45. [Google Scholar] [CrossRef]
- Asakawa, T.; Esumi, M.; Endo, S.; Kida, A.; Ikeda, M. A mutation at IVS1 + 5 of the von Hippel-Lindau gene resulting in intron retention in transcripts is not pathogenic in a patient with a tongue cancer? Case report. BMC Med. Genet. 2012, 13, 23. [Google Scholar] [CrossRef]
| RNA Binding Protein | Key Function | Binding to Specific RNA Sequences | IR Highlights | References |
|---|---|---|---|---|
| Polypyrimidine Tract-Binding Protein 1 (PTBP1) | Regulator of alternative splicing | Pyrimidine-rich RNA sequences, particularly those rich in cytosine (C) and uracil (U) bases near splice junctions | Represses splicing of terminal introns to retain transcripts in the nucleus | [31] |
| Splicing Factor Proline and Glutamine Rich (SFPQ) | Splicing factor, particularly important for splicing long genes, and regulates the formation of circular RNAs (circRNAs) | RNA sequences, particularly those surrounding cryptic last exons and in long introns. | SFPQ exports IR transcripts to the cytoplasm that is associated with ALS | [33] |
| Heterogeneous Nuclear Ribonucleoprotein L Like (hnRNPLL) | Regulates alternative splicing | 5′-YCAY-3′ | Regulates lineage-specific IR during T cell development | [28] |
| Chromatin Target of PRMT1 (Chtop) | Component of the TREX (TRanscription-EXport) complex, which links transcription to mRNA export | Interacts with RNA through its arginine-glycine-rich (RG) domain and its N-terminal (N1) domain | Regulates Chtop mRNA expression antagonistically under specific stimuli | [34] |
| Poly(A) Binding Protein Nuclear 1 (PABPN1) | mRNA processing and export | Poly(A) tails of mRNA molecules | PABPN1 gene mutation results in oculopharyngeal muscular dystrophy | [27] |
| NOVA Alternative Splicing Regulator 1 (Nova-1) | Regulates alternative splicing | 5′-YCAY-3′ | Regulators in certain mammalian neurons | [33] |
| ASF/SF2 (SRSF1) | Constitutive and alternative pre-mRNA splicing | Exonic splicing enhancers and 5’ splice sites | Specifically involved in the synthesis of endoglin | [7] |
| Tool/Approach | Highlights | References |
|---|---|---|
| IRFinder/IRFinder-S | Benchmark IR detection tool; IRFinder-S adds CNN filtering, long-read support, and integrated differential IR analysis | [78,80,81] |
| iREAD | Uses entropy scoring across independent introns to detect flat read distributions; avoids exon overlap | [82] |
| KMA | Involves transcript quantification; suitable for differential analysis with minimal artifacts | [78,82] |
| IntEREst | Supports non-annotated introns; integrates statistical tests for intra- and inter- sample comparisons | [53,78] |
| IRAVNet | Identifies IR-causing variants directly from transcriptomic data; assesses the connection between genomic variant status and the amount of splicing changes | [83] |
| SpliceAI/SpliceAI-10k | Deep learning model for splicing prediction; identifies partial IR and spliceogenic variants with high sensitivity | [84,85] |
| Disease | Key IR Feature | Gene(s) Involved | Functional Consequences | References |
|---|---|---|---|---|
| Aging | Increased IR in genes | Various | Impaired metabolic homeostasis; reversible by JTT, which may serve as a functional marker for anti-aging therapies | [16,38,86,87,88] |
| SCCOHT (Ovarian Cancer) | SMARCA4 loss triggers IR → neoantigens | SMARCA4, MHC-I pathway | IR-derived peptides activate T cells; opens immunotherapy avenues in chromatin-remodeling cancers | [89] |
| Breast Cancer | IR globally decreased; low-IR tumors linked to poor prognosis | Various | Contrasts most cancers; IR levels correlate with proliferation and prognosis | [90] |
| Hepatocellular Carcinoma | IR generates a non-coding isoform (KLF-003) that is downregulated | KLF4 | Epigenetic silencing of IR transcript via CpG methylation; loss may contribute to recurrence and could serve as a diagnostic biomarker | [91] |
| Multiple Myeloma | Elevated IR events → generation of IR-neoantigens | RNA splicing machinery; immune checkpoint pathways | High IR-neoAg load correlates with poor survival, immune suppression via co-inhibitory molecules, reduced MHC-II, and immune escape | [92] |
| Etoposide-Resistant Leukemia | Retention of intron 19 in TOP2α mRNA | TOP2α gene (DNA topoisomerase Iiα) | Production of truncated TOP2α/90 isoform, reduced drug induced DNA damage; chemoresistance | [93] |
| 46,XX DSD | IR in WT1 alters isoform ratio → sex development disruption | WT1 | First documented IR-induced mechanism affecting human sexual differentiation | [94] |
| Renal Fibrosis | ASO-induced IR in ENG shifts isoform balance to anti-fibrotic variant | ENG | Demonstrates therapeutic modulation of IR to treat fibrosis via isoform control | [95] |
| Incessant Ventricular Tachycardia | Retention of 79 bp from intron 14 | SCN5A (Nav1.5 sodium channel) | PTC (p.R818 *) → truncated protein → impaired sodium channel function → arrhythmia | [96] |
| Allgrove Syndrome | Partial retention of 99 bp of intron 14 plus exon 14 skipping | AAAS | Frameshift mutations causing PTCs, likely resulting in non-functional protein and disease phenotype | [97] |
| Alzheimer’s Disease | IR in DDIT4L and MAPT produces toxic isoforms (DIR and Tau11i) | DDIT4L, MAPT | Links IR to neurodegeneration, synaptic dysfunction, and cognitive decline | [98,99] |
| Hypoplastic Amelogenesis Imperfecta | Retention of intron 1 and normally skipped exon 2 → elongated 5’ UTR | ENAM (enamelin) | Complex 5’ UTR secondary structure attenuates translation → reduced enamelin → defective enamel | [100] |
| ALS | Premature and increased IR during motor neuron differentiation; prominent IR in SFPQ transcript | SFPQ | Reduced nuclear SFPQ protein; disrupted RNA metabolism; ALS pathogenesis marker | [33] |
| SMA | Intron 7 retention in SMN2 transcripts | SMN2 | Longer 3’ UTR → translational repression of SMN protein | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porras-Tobias, A.L.; Caldera, A.; Castro-Piedras, I. Intron Retention: A Reemerging Paradigm in RNA Biology and Post-Transcriptional Gene Regulation. Genes 2025, 16, 986. https://doi.org/10.3390/genes16080986
Porras-Tobias AL, Caldera A, Castro-Piedras I. Intron Retention: A Reemerging Paradigm in RNA Biology and Post-Transcriptional Gene Regulation. Genes. 2025; 16(8):986. https://doi.org/10.3390/genes16080986
Chicago/Turabian StylePorras-Tobias, Ana L., Abigail Caldera, and Isabel Castro-Piedras. 2025. "Intron Retention: A Reemerging Paradigm in RNA Biology and Post-Transcriptional Gene Regulation" Genes 16, no. 8: 986. https://doi.org/10.3390/genes16080986
APA StylePorras-Tobias, A. L., Caldera, A., & Castro-Piedras, I. (2025). Intron Retention: A Reemerging Paradigm in RNA Biology and Post-Transcriptional Gene Regulation. Genes, 16(8), 986. https://doi.org/10.3390/genes16080986






