Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and Skeletal Muscle Sampling
2.3. RNA Extraction and Sequencing
2.4. RNA-Seq Data Processing and Analysis
2.5. Differentially Expressed Genes
2.6. Protein–Protein Interaction (PPI) Network
2.7. Functional Analysis (Gene Ontology and Pathway Enrichment Analyses)
3. Results
3.1. Measuring Data Quality
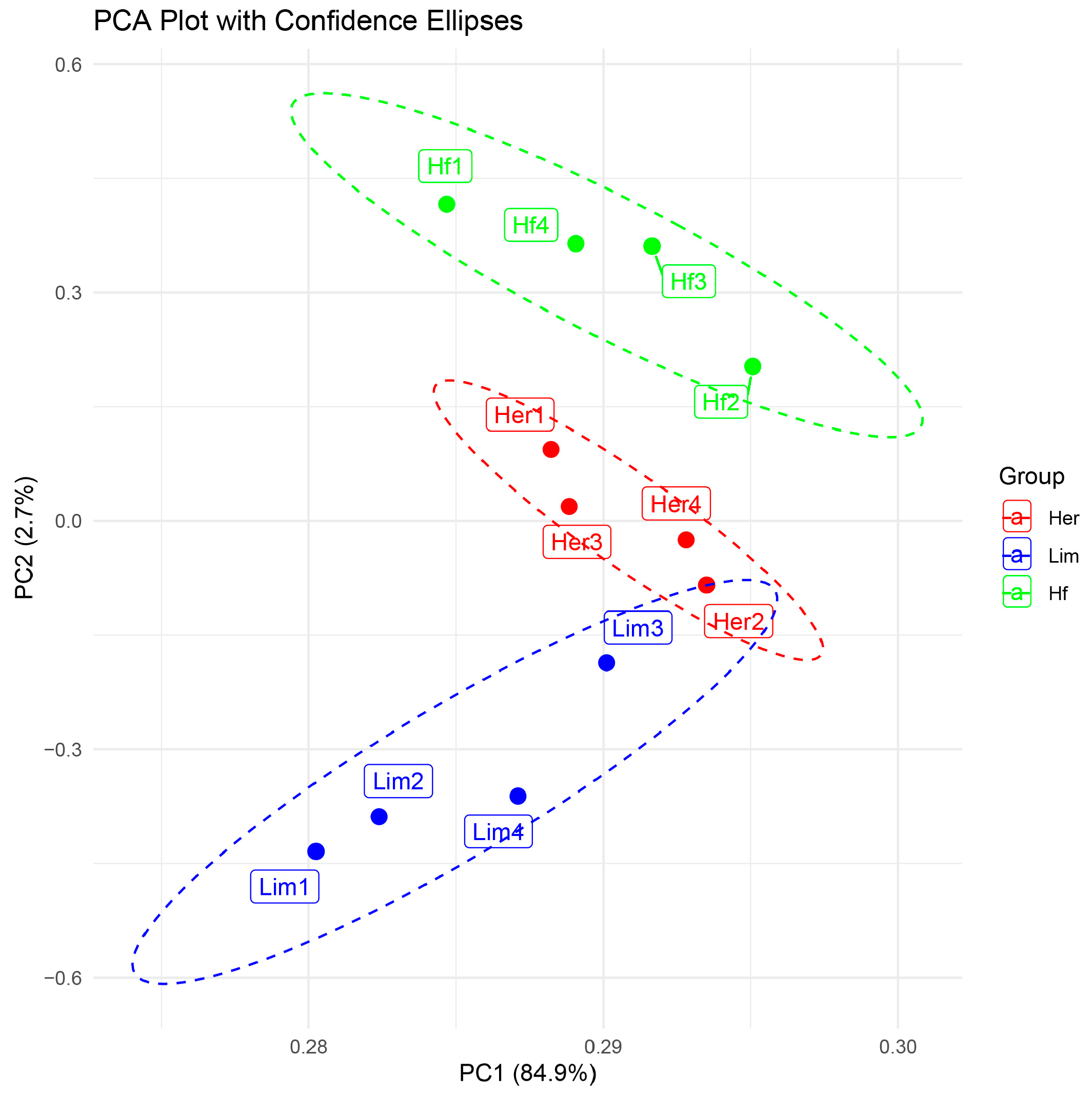
3.2. RNA-Sequencing Data Processing
3.3. Analysis of Differential Gene Expression
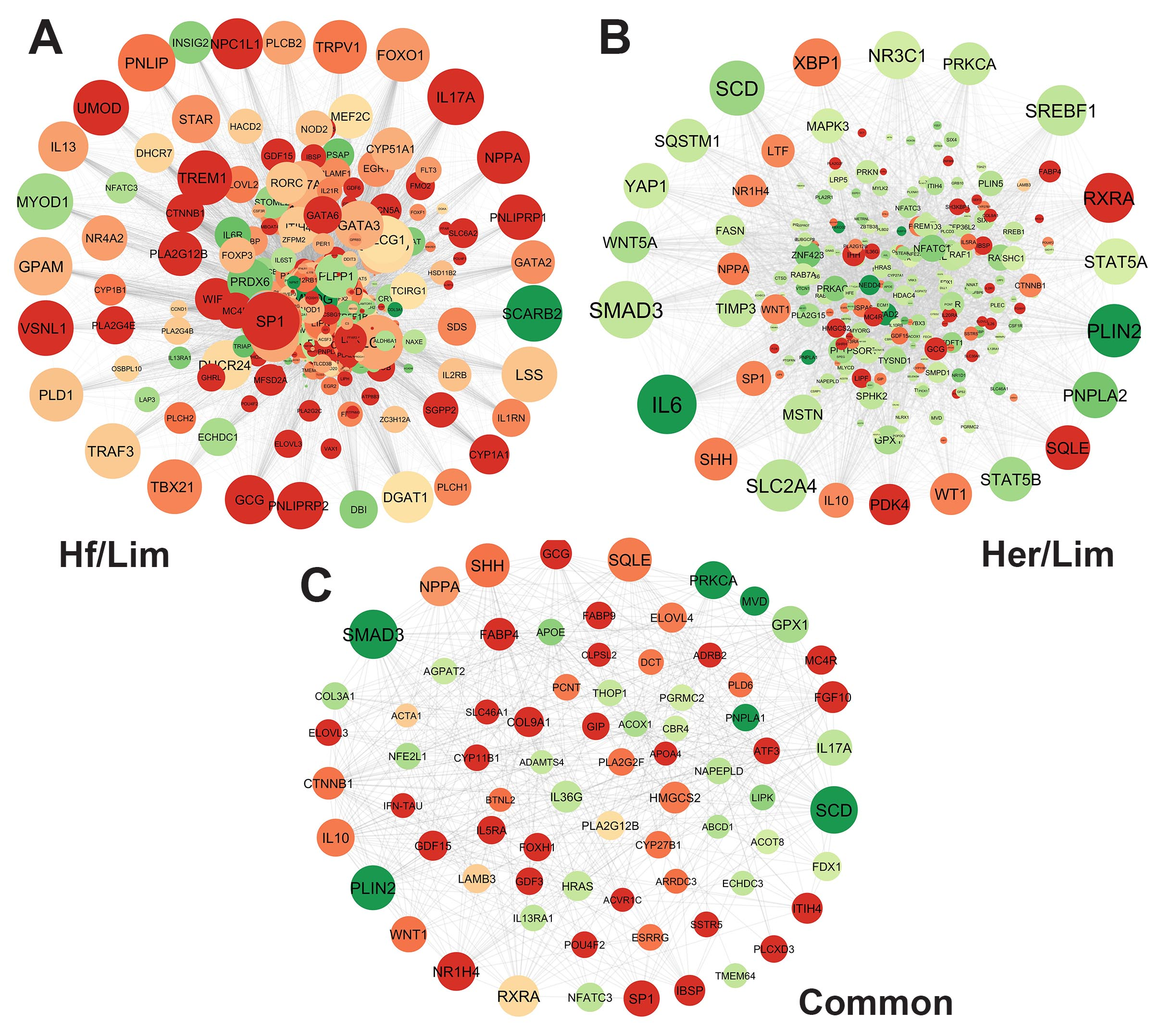
3.4. PPI Network Analysis
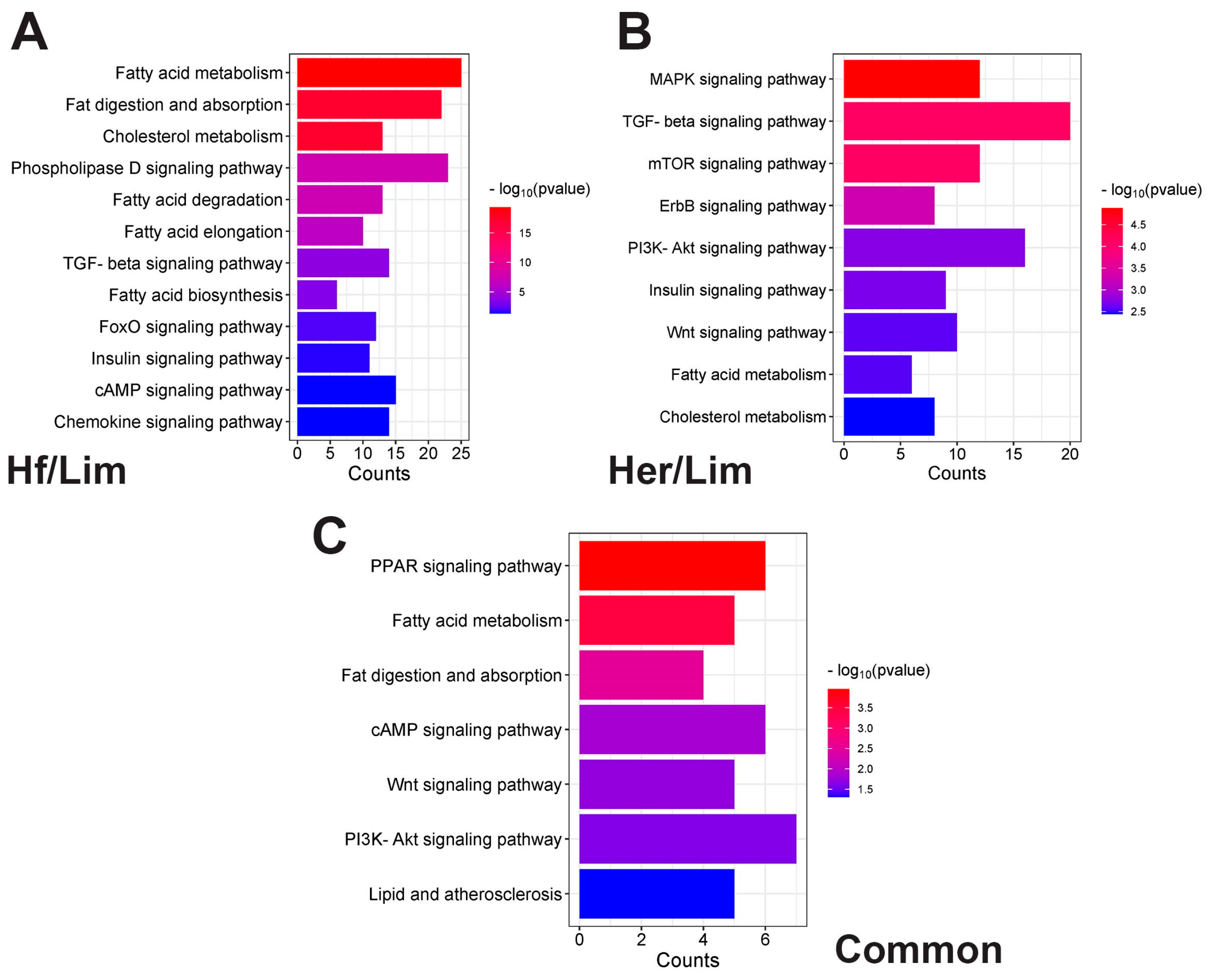
3.5. Signaling Pathway
3.6. Gene Ontology
4. Discussion
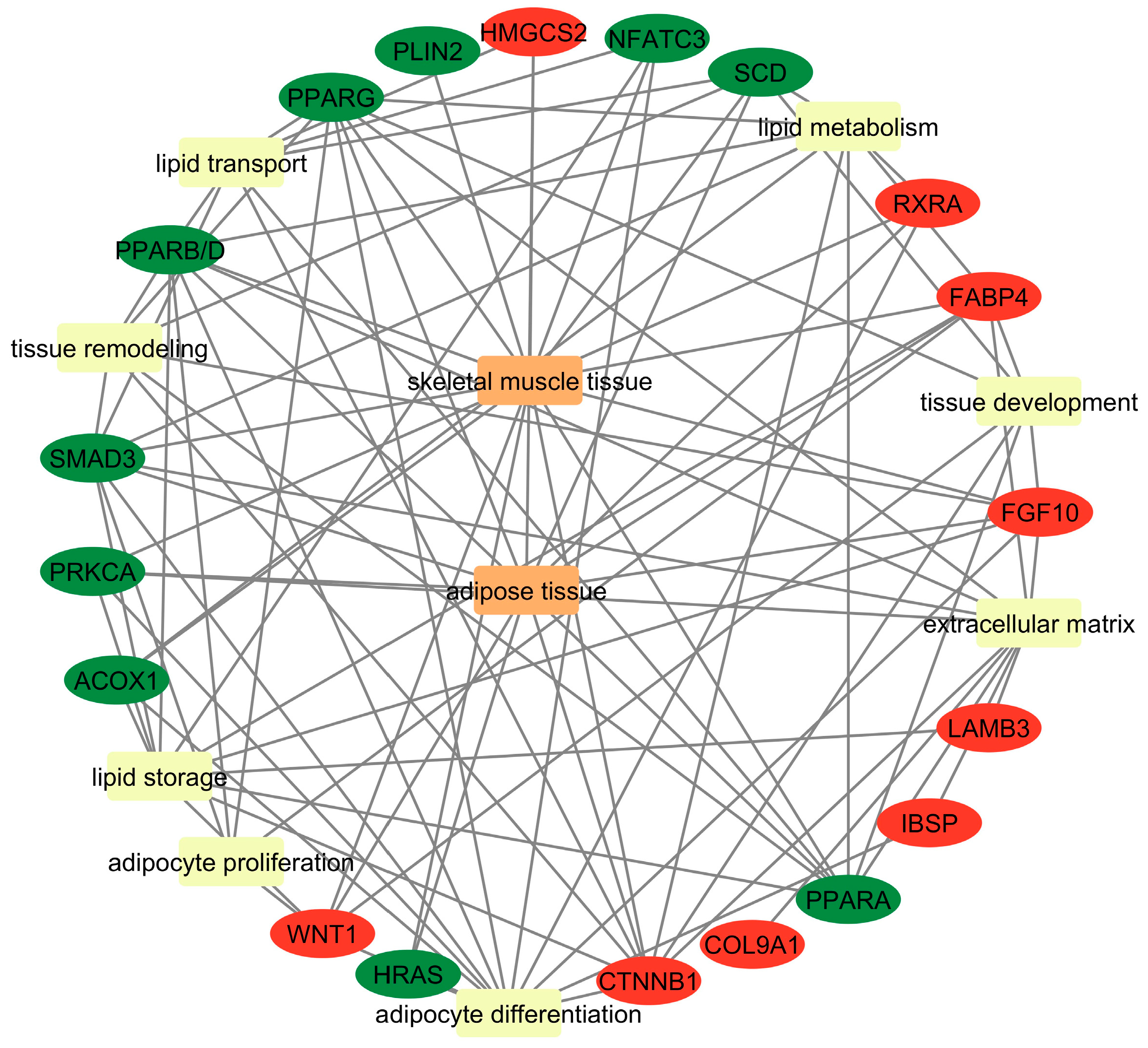
4.1. Common Genes Related to Adipogenesis
4.2. Common Genes Related to Lipid Metabolism (Breakdown, Synthesis, and Regulation of Lipids)
4.3. Common Genes Related to Lipid Storage (Lipid Accumulation and Fat Droplet Formation)
4.4. Common Genes Related to Skeletal Muscle Remodeling During IMF Deposition
4.5. Common Signaling Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leroy, F.; Smith, N.W.; Adesogan, A.T.; Beal, T.; Iannotti, L.; Moughan, P.J.; Mann, N. The Role of Meat in the Human Diet: Evolutionary Aspects and Nutritional Value. Anim. Front. 2023, 13, 11–18. [Google Scholar] [CrossRef]
- Fortova, J.; del Mar Campo, M.; Valenta, J.; Needham, T.; Rehak, D.; Lebedova, N.; Barton, L.; Kloucek, P.; Bureš, D. Preferences and Acceptance of Czech and Spanish Consumers Regarding Beef with Varying Intramuscular Fat Content. Meat Sci. 2022, 192, 108912. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.V.; Nguyen, O.C.; Malau-Aduli, A.E.O. Main Regulatory Factors of Marbling Level in Beef Cattle. Vet. Anim. Sci. 2021, 14, 100219. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.L.; Boler, D.D.; Kutzler, L.W.; Jones, K.A.; McKeith, F.K.; Killefer, J.; Carr, T.R.; Dilger, A.C. Muscle Gene Expression Associated with Increased Marbling in Beef Cattle. Anim. Biotechnol. 2011, 22, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.L.; Wang, B.; Deavila, J.M.; Busboom, J.R.; Maquivar, M.; Parish, S.M.; McCann, B.; Nelson, M.L.; Du, M. Vitamin A Administration at Birth Promotes Calf Growth and Intramuscular Fat Development in Angus Beef Cattle. J. Anim. Sci. Biotechnol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.; Fan, J.; Yu, J.; Li, K.; Bai, J. Lipidomics Reveals Alterations of Lipid Composition and Molecular Nutrition in Irradiated Marble Beef. Food Chem. X 2023, 17, 100617. [Google Scholar] [CrossRef]
- Nogoy, K.M.C.; Sun, B.; Shin, S.; Lee, Y.; Li, X.Z.; Choi, S.H.; Park, S. Fatty Acid Composition of Grain- and Grass-Fed Beef and Their Nutritional Value and Health Implication. Food Sci. Anim. Resour. 2022, 42, 18. [Google Scholar] [CrossRef]
- Bolormaa, S.; Neto, L.R.P.; Zhang, Y.D.; Bunch, R.J.; Harrison, B.E.; Goddard, M.E.; Barendse, W. A Genome-Wide Association Study of Meat and Carcass Traits in Australian Cattle. J. Anim. Sci. 2011, 89, 2297–2309. [Google Scholar] [CrossRef]
- Hongfang, G.; Khan, R.; Raza, S.H.A.; Nurgulsim, K.; Suhail, S.M.; Rahman, A.; Ahmed, I.; Ijaz, A.; Ahmad, I.; Linsen, Z. Transcriptional Regulation of Adipogenic Marker Genes for the Improvement of Intramuscular Fat in Qinchuan Beef Cattle. Anim. Biotechnol. 2022, 33, 776–795. [Google Scholar] [CrossRef]
- Santiago, B.M.; Baldassini, W.A.; Chiaratti, M.R.; Pandey, A.K.; Torrecilhas, J.A.; Torres, R.N.S.; Ribeiro, R.V.; Lanna, D.P.D.; Pereira, G.L.; Curi, R.A. Skeletal Muscle Gene Expression and Meat Quality of F1 Angus–Nellore Young Steers and Bulls Feedlot Finished. Livest. Sci. 2023, 268, 105151. [Google Scholar] [CrossRef]
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, Acquisition, and Functional Impact of Sex-Biased Gene Expression in Mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef]
- Naldurtiker, A.; Batchu, P.; Kouakou, B.; Terrill, T.H.; Shaik, A.; Kannan, G. RNA-Seq Exploration of the Influence of Stress on Meat Quality in Spanish Goats. Sci. Rep. 2022, 12, 20573. [Google Scholar] [CrossRef]
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. MEATabolomics: Muscle and Meat Metabolomics in Domestic Animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef]
- Hocquette, J.-F.; Ellies-Oury, M.-P.; Lherm, M.; Pineau, C.; Deblitz, C.; Farmer, L. Current Situation and Future Prospects for Beef Production in Europe—A Review. Asian-Australas. J. Anim. Sci. 2018, 31, 1017. [Google Scholar] [CrossRef] [PubMed]
- Sadkowski, T.; Ciecierska, A.; Majewska, A.; Oprządek, J.; Dasiewicz, K.; Ollik, M.; Wicik, Z.; Motyl, T. Transcriptional Background of Beef Marbling—Novel Genes Implicated in Intramuscular Fat Deposition. Meat Sci. 2014, 97, 32–41. [Google Scholar] [CrossRef]
- PN ISO 1444:2000; Mrat and Meta Products-Determination of Free Fat Content. Polish Committee for Standardization: Warsaw, Poland, 2013.
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.; Ono, K.; Ideker, T.; Maere, S. PiNGO: A Cytoscape Plugin to Find Candidate Genes in Biological Networks. Bioinformatics 2011, 27, 1030–1031. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Yu, H.; Greenbaum, D.; Kluger, Y.; Krogan, N.J.; Chung, S.; Emili, A.; Snyder, M.; Greenblatt, J.F.; Gerstein, M. A Bayesian Networks Approach for Predicting Protein-Protein Interactions from Genomic Data. Science 2003, 302, 449–453. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Liu, J.; Farmer, S.R. Regulating the Balance between Peroxisome Proliferator-Activated Receptor γ and β-Catenin Signaling during Adipogenesis: A Glycogen Synthase Kinase 3β Phosphorylation-Defective Mutant of β-Catenin Inhibits Expression of a Subset of Adipogenic Genes. J. Biol. Chem. 2004, 279, 45020–45027. [Google Scholar] [CrossRef]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and Characterization of the Promoter Region of Perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in Bovine Adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef]
- Liu, D.; Black, B.L.; Derynck, R. TGF-β Inhibits Muscle Differentiation through Functional Repression of Myogenic Transcription Factors by Smad3. Genes Dev. 2001, 15, 2950–2966. [Google Scholar] [CrossRef]
- Tsurutani, Y.; Fujimoto, M.; Takemoto, M.; Irisuna, H.; Koshizaka, M.; Onishi, S.; Ishikawa, T.; Mezawa, M.; He, P.; Honjo, S. The Roles of Transforming Growth Factor-β and Smad3 Signaling in Adipocyte Differentiation and Obesity. Biochem. Biophys. Res. Commun. 2011, 407, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-X.; Luo, X.; Liu, R.-X.; Yang, Y.-J.; Yang, G.-S. Roles of Wnt/β-Catenin Signaling in Adipogenic Differentiation Potential of Adipose-Derived Mesenchymal Stem Cells. Mol. Cell. Endocrinol. 2008, 291, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Prestwich, T.C.; MacDougald, O.A. Wnt/β-Catenin Signaling in Adipogenesis and Metabolism. Curr. Opin. Cell Biol. 2007, 19, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Andrzejewska, A.; Lukomska, B.; Janowski, M. Concise Review: Mesenchymal Stem Cells: From Roots to Boost. Stem Cells 2019, 37, 855–864. [Google Scholar] [CrossRef]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and WNT Signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef]
- Kang, S.; Bennett, C.N.; Gerin, I.; Rapp, L.A.; Hankenson, K.D.; MacDougald, O.A. Wnt Signaling Stimulates Osteoblastogenesis of Mesenchymal Precursors by Suppressing CCAAT/Enhancer-Binding Protein α and Peroxisome Proliferator-Activated Receptor γ. J. Biol. Chem. 2007, 282, 14515–14524. [Google Scholar] [CrossRef]
- Lim, K.S.; Lee, K.T.; Park, J.E.; Chung, W.H.; Jang, G.W.; Choi, B.H.; Hong, K.C.; Kim, T.H. Identification of Differentially Expressed Genes in Longissimus Muscle of Pigs with High and Low Intramuscular Fat Content Using RNA Sequencing. Anim. Genet. 2017, 48, 166–174. [Google Scholar] [CrossRef]
- Renaville, B.; Prandi, A.; Fan, B.; Sepulcri, A.; Rothschild, M.F.; Piasentier, E. Candidate Gene Marker Associations with Fatty Acid Profiles in Heavy Pigs. Meat Sci. 2013, 93, 495–500. [Google Scholar] [CrossRef]
- Doran, O.; Moule, S.K.; Teye, G.A.; Whittington, F.M.; Hallett, K.G.; Wood, J.D. A Reduced Protein Diet Induces Stearoyl-CoA Desaturase Protein Expression in Pig Muscle but Not in Subcutaneous Adipose Tissue: Relationship with Intramuscular Lipid Formation. Br. J. Nutr. 2006, 95, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Enoch, H.G.; Catala, A.; Strittmatter, P. Mechanism of Rat Liver Microsomal Stearyl-CoA Desaturase. Studies of the Substrate Specificity, Enzyme-Substrate Interactions, and the Function of Lipid. J. Biol. Chem. 1976, 251, 5095–5103. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, R.; Joshi, V.C. Hormonal Regulation of Stearoyl Coenzyme A Desaturase Activity and Lipogenesis during Adipose Conversion of 3T3-L1 Cells. J. Biol. Chem. 1982, 257, 12224–12230. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Lee, Y.; Yeo, J. Identification of the SNP (Single Necleotide Polymorphism) of the Stearoyl-CoA Desaturase (SCD) Associated with Unsaturated Fatty Acid in Hanwoo (Korean Cattle). Asian-Australas. J. Anim. Sci. 2011, 24, 757–765. [Google Scholar] [CrossRef]
- Gerhard, G.S.; Styer, A.M.; Strodel, W.E.; Roesch, S.L.; Yavorek, A.; Carey, D.J.; Wood, G.C.; Petrick, A.T.; Gabrielsen, J.; Ibele, A. Gene Expression Profiling in Subcutaneous, Visceral and Epigastric Adipose Tissues of Patients with Extreme Obesity. Int. J. Obes. 2014, 38, 371–378. [Google Scholar] [CrossRef]
- Sakaue, H.; Konishi, M.; Ogawa, W.; Asaki, T.; Mori, T.; Yamasaki, M.; Takata, M.; Ueno, H.; Kato, S.; Kasuga, M. Requirement of Fibroblast Growth Factor 10 in Development of White Adipose Tissue. Genes Dev. 2002, 16, 908–912. [Google Scholar] [CrossRef]
- Ohta, H.; Konishi, M.; Itoh, N. FGF10 and FGF21 as Regulators in Adipocyte Development and Metabolism. Endocr. Metab. Immune Disord. Targets Former. Curr. Drug Targets–Immune Endocr. Metab. Disord. 2011, 11, 302–309. [Google Scholar] [CrossRef]
- Metzger, D.; Imai, T.; Jiang, M.; Takukawa, R.; Desvergne, B.; Wahli, W.; Chambon, P. Functional Role of RXRs and PPARγ in Mature Adipocytes. Prostaglandins Leukot. Essent. Fat. acids 2005, 73, 51–58. [Google Scholar] [CrossRef]
- Kosters, A.; Sun, D.; Wu, H.; Tian, F.; Felix, J.C.; Li, W.; Karpen, S.J. Sexually Dimorphic Genome-Wide Binding of Retinoid X Receptor Alpha (RXRα) Determines Male-Female Differences in the Expression of Hepatic Lipid Processing Genes in Mice. PLoS ONE 2013, 8, e71538. [Google Scholar] [CrossRef]
- Lim, D.; Chai, H.-H.; Lee, S.-H.; Cho, Y.-M.; Choi, J.-W.; Kim, N.-K. Gene Expression Patterns Associated with Peroxisome Proliferator-Activated Receptor (PPAR) Signaling in the of Hanwoo (Korean Cattle). Asian-Australas. J. Anim. Sci. 2015, 28, 1075. [Google Scholar] [CrossRef]
- Garcia-Bermudez, J.; Baudrier, L.; Bayraktar, E.C.; Shen, Y.; La, K.; Guarecuco, R.; Yucel, B.; Fiore, D.; Tavora, B.; Freinkman, E. Squalene Accumulation in Cholesterol Auxotrophic Lymphomas Prevents Oxidative Cell Death. Nature 2019, 567, 118–122. [Google Scholar] [CrossRef]
- Ha, J.; Kwon, S.; Hwang, J.H.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Park, H.C.; An, S.M.; Kim, C.W. Squalene Epoxidase Plays a Critical Role in Determining Pig Meat Quality by Regulating Adipogenesis, Myogenesis, and ROS Scavengers. Sci. Rep. 2017, 7, 16740. [Google Scholar] [CrossRef]
- Chang, C.; Yang, Y.; Zhou, L.; Baiyin, B.; Liu, Z.; Guo, L.; Ma, F.; Wang, J.; Chai, Y.; Shi, C. Candidate Genes and Gene Networks Change with Age in Japanese Black Cattle by Blood Transcriptome Analysis. Genes 2023, 14, 504. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhong, J.; Zhang, C.; Chai, Z.; Cao, H.; Wang, J.; Zhu, J.; Wang, J.; Ji, Q. The Whole-Transcriptome Landscape of Muscle and Adipose Tissues Reveals the CeRNA Regulation Network Related to Intramuscular Fat Deposition in Yak. BMC Genom. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S. Board-Invited Review: The Biology and Regulation of Preadipocytes and Adipocytes in Meat Animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [PubMed]
- Vilà-Brau, A.; De Sousa-Coelho, A.L.; Mayordomo, C.; Haro, D.; Marrero, P.F. Human HMGCS2 Regulates Mitochondrial Fatty Acid Oxidation and FGF21 Expression in HepG2 Cell Line. J. Biol. Chem. 2011, 286, 20423–20430. [Google Scholar] [CrossRef]
- Shaw, C.S.; Sherlock, M.; Stewart, P.M.; Wagenmakers, A.J.M. Adipophilin Distribution and Colocalisation with Lipid Droplets in Skeletal Muscle. Histochem. Cell Biol. 2009, 131, 575–581. [Google Scholar] [CrossRef]
- Davoli, R.; Gandolfi, G.; Braglia, S.; Comella, M.; Zambonelli, P.; Buttazzoni, L.; Russo, V. New SNP of the Porcine Perilipin 2 (PLIN2) Gene, Association with Carcass Traits and Expression Analysis in Skeletal Muscle. Mol. Biol. Rep. 2011, 38, 1575–1583. [Google Scholar] [CrossRef]
- Ryan, C.B.; Choi, J.S.; Al-Ali, H.; Lee, J.K. Myelin and Non-Myelin Debris Contribute to Foamy Macrophage Formation after Spinal Cord Injury. Neurobiol. Dis. 2022, 163, 105608. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, C.; Mirza, A.H.; Bamigbade, A.T.; Zhang, S.; Xu, S.; Liu, P. Rab18 Binds PLIN2 and ACSL3 to Mediate Lipid Droplet Dynamics. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2021, 1866, 158923. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The Perilipin Family of Lipid Droplet Proteins: Gatekeepers of Intracellular Lipolysis. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Mardani, I.; Tomas Dalen, K.; Drevinge, C.; Miljanovic, A.; Ståhlman, M.; Klevstig, M.; Scharin Täng, M.; Fogelstrand, P.; Levin, M.; Ekstrand, M. Plin2-Deficiency Reduces Lipophagy and Results in Increased Lipid Accumulation in the Heart. Sci. Rep. 2019, 9, 6909. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Z.; Lund, J.; Li, Y.; Knabenes, I.K.; Bakke, S.S.; Kase, E.T.; Lee, Y.K.; Kimmel, A.R.; Thoresen, G.H.; Rustan, A.C. Loss of Perilipin 2 in Cultured Myotubes Enhances Lipolysis and Redirects the Metabolic Energy Balance from Glucose Oxidation towards Fatty Acid Oxidation. J. Lipid Res. 2017, 58, 2147–2161. [Google Scholar] [CrossRef]
- Hussain, M.M.; Maxfield, F.R.; Mas-Oliva, J.; Tabas, I.; Ji, Z.-S.; Innerarity, T.L.; Mahley, R.W. Clearance of Chylomicron Remnants by the Low Density Lipoprotein Receptor-Related Protein/Alpha 2-Macroglobulin Receptor. J. Biol. Chem. 1991, 266, 13936–13940. [Google Scholar] [CrossRef]
- Huang, Z.H.; Reardon, C.A.; Mazzone, T. Endogenous ApoE Expression Modulates Adipocyte Triglyceride Content and Turnover. Diabetes 2006, 55, 3394–3402. [Google Scholar] [CrossRef]
- Matarese, V.; Bernlohr, D.A. Purification of Murine Adipocyte Lipid-Binding Protein. Characterization as a Fatty Acid-and Retinoic Acid-Binding Protein. J. Biol. Chem. 1988, 263, 14544–14551. [Google Scholar] [CrossRef]
- Albrecht, E.; Schering, L.; Liu, Y.; Komolka, K.; Kühn, C.; Wimmers, K.; Gotoh, T.; Maak, S. TRIENNIAL GROWTH AND DEVELOPMENT SYMPOSIUM: Factors Influencing Bovine Intramuscular Adipose Tissue Development and Cellularity. J. Anim. Sci. 2017, 95, 2244–2254. [Google Scholar] [CrossRef]
- Hoashi, S.; Hinenoya, T.; Tanaka, A.; Ohsaki, H.; Sasazaki, S.; Taniguchi, M.; Oyama, K.; Mukai, F.; Mannen, H. Association between Fatty Acid Compositions and Genotypes of FABP4 and LXR-Alpha in Japanese Black Cattle. BMC Genet. 2008, 9, 84. [Google Scholar] [CrossRef]
- Michal, J.J.; Zhang, Z.W.; Gaskins, C.T.; Jiang, Z. The Bovine Fatty Acid Binding Protein 4 Gene Is Significantly Associated with Marbling and Subcutaneous Fat Depth in Wagyu x Limousin F2 Crosses. Anim. Genet. 2006, 37, 400–402. [Google Scholar] [CrossRef]
- Blecha, I.M.Z.; Siqueira, F.; Ferreira, A.B.R.; Feijo, G.L.D.; Torres, R.A.d.A.; de Medeiros, S.R. Identification and Evaluation of Polymorphisms in FABP3 and FABP4 in Beef Cattle. Natl. Cent. Biotechnol. Inf. 2015, 14, 16353–16363. [Google Scholar] [CrossRef]
- Barendse, W.; Bunch, R.J.; Thomas, M.B.; Harrison, B.E. A Splice Site Single Nucleotide Polymorphism of the Fatty Acid Binding Protein 4 Gene Appears to Be Associated with Intramuscular Fat Deposition in Longissimus Muscle in Australian Cattle. Anim. Genet. 2009, 40, 770–773. [Google Scholar] [CrossRef]
- Han, X.; Jiang, T.; Yang, H.; Zhang, Q.; Wang, W.; Fan, B.; Liu, B. Investigation of Four Porcine Candidate Genes (H-FABP, MYOD1, UCP3 and MASTR) for Meat Quality Traits in Large White Pigs. Mol. Biol. Rep. 2012, 39, 6599–6605. [Google Scholar] [CrossRef]
- Shin, S.-C.; Heo, J.-P.; Chung, E.-R. Genetic Variants of the FABP4 Gene Are Associated with Marbling Scores and Meat Quality Grades in Hanwoo (Korean Cattle). Mol. Biol. Rep. 2012, 39, 5323–5330. [Google Scholar] [CrossRef]
- You, X.; Liu, Y.; Jiang, X.; Du, H.; Liu, Z.; Zhu, Q. Relationships between Single Nucleotide Polymorphisms of the H-FABP Gene and Slaughter and Meat Quality Traits in Chicken. Biochem. Genet. 2009, 47, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, Q.; Wang, Y.; Leng, L.; Zhang, Q.; Shang, Z.; Li, H. Adipocyte Fatty Acid-Binding Protein: An Important Gene Related to Lipid Metabolism in Chicken Adipocytes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 157, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Guillou, H.; Zadravec, D.; Martin, P.G.P.; Jacobsson, A. The Key Roles of Elongases and Desaturases in Mammalian Fatty Acid Metabolism: Insights from Transgenic Mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Westerberg, R.; Månsson, J.-E.; Golozoubova, V.; Shabalina, I.G.; Backlund, E.C.; Tvrdik, P.; Retterstøl, K.; Capecchi, M.R.; Jacobsson, A. ELOVL3 Is an Important Component for Early Onset of Lipid Recruitment in Brown Adipose Tissue. J. Biol. Chem. 2006, 281, 4958–4968. [Google Scholar] [CrossRef]
- Kitazawa, H.; Miyamoto, Y.; Shimamura, K.; Nagumo, A.; Tokita, S. Development of a High-Density Assay for Long-Chain Fatty Acyl-CoA Elongases. Lipids 2009, 44, 765–773. [Google Scholar] [CrossRef]
- Tong, H.L.; Yin, H.Y.; Zhang, W.W.; Hu, Q.; Li, S.F.; Yan, Y.Q.; Li, G.P. Transcriptional Profiling of Bovine Muscle-Derived Satellite Cells during Differentiation in Vitro by High Throughput RNA Sequencing. Cell. Mol. Biol. Lett. 2015, 20, 351–373. [Google Scholar] [CrossRef]
- Wang, L.; Valencak, T.G.; Shan, T. Fat Infiltration in Skeletal Muscle: Influential Triggers and Regulatory Mechanism. Iscience 2024, 27, 109221. [Google Scholar] [CrossRef]
- Kaur, J.; Reinhardt, D.P. Extracellular Matrix (ECM) Molecules. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Elsevier: Amsterdam, The Netherlands, 2015; pp. 25–45. [Google Scholar]
- Brachvogel, B.; Zaucke, F.; Dave, K.; Norris, E.L.; Stermann, J.; Dayakli, M.; Koch, M.; Gorman, J.J.; Bateman, J.F.; Wilson, R. Comparative Proteomic Analysis of Normal and Collagen IX Null Mouse Cartilage Reveals Altered Extracellular Matrix Composition and Novel Components of the Collagen IX Interactome. J. Biol. Chem. 2013, 288, 13481–13492. [Google Scholar] [CrossRef]
- Verma, P.; Dalal, K. ADAMTS-4 and ADAMTS-5: Key Enzymes in Osteoarthritis. J. Cell. Biochem. 2011, 112, 3507–3514. [Google Scholar] [CrossRef] [PubMed]
- Sheet, S.; Jang, S.S.; Lim, J.-A.; Park, W.; Kim, D. Molecular Signatures Diversity Unveiled through a Comparative Transcriptome Analysis of Longissimus Dorsi and Psoas Major Muscles in Hanwoo Cattle. Anim. Biotechnol. 2024, 35, 2379883. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yuan, S.; Zheng, K.; Liu, G.; Li, J.; Ye, B.; Yin, L.; Li, Y. IL-17 Signaling Pathway: A Potential Therapeutic Target for Reducing Skeletal Muscle Inflammation. Cytokine 2024, 181, 156691. [Google Scholar] [CrossRef] [PubMed]
- di Candia, A.M.; de Avila, D.X.; Moreira, G.R.; Villacorta, H.; Maisel, A.S. Growth Differentiation Factor-15, a Novel Systemic Biomarker of Oxidative Stress, Inflammation, and Cellular Aging: Potential Role in Cardiovascular Diseases. Am. Hear. J. Plus Cardiol. Res. Pract. 2021, 9, 100046. [Google Scholar] [CrossRef]
- Evangelakos, I. Relevance of Oxysterol 7-Alpha-Hydroxylase (CYP7B1) for Lipid Metabolism and Fatty Liver Diseas. Ph.D. Dissertation, University of Hamburg, Hamburg, Germany, 2022. [Google Scholar]
- Miyamori, H.; Yokokawa, T.; Miyakita, M.; Ozaki, K.; Goto, T.; Inoue, K.; Matsumura, S. CRTC1 in Mc4r-Expressing Cells Is Required for Peripheral Metabolism and Systemic Energy Homeostasis. Diabetes 2024, 73, db240014. [Google Scholar] [CrossRef]
- Kotikalapudi, N.; Ramachandran, D.; Vieira, D.; Rubio, W.B.; Gipson, G.R.; Troncone, L.; Vestal, K.; Maridas, D.E.; Rosen, V.; Yu, P.B.; et al. Acute regulation of murine adipose tissue lipolysis and insulin resistance by the TGFβ superfamily protein GDF3. Nat. Commun. 2025, 16, 4432. [Google Scholar] [CrossRef]
- Laing, N.G.; Dye, D.E.; Wallgren-Pettersson, C.; Richard, G.; Monnier, N.; Lillis, S.; Winder, T.L.; Lochmüller, H.; Graziano, C.; Mitrani-Rosenbaum, S. Mutations and Polymorphisms of the Skeletal Muscle A-actin Gene (ACTA1). Hum. Mutat. 2009, 30, 1267–1277. [Google Scholar] [CrossRef]
- Steinfeldt, J.; Becker, R.; Vergarajauregui, S.; Engel, F.B. Alternative Splicing of Pericentrin Contributes to Cell Cycle Control in Cardiomyocytes. J. Cardiovasc. Dev. Dis. 2021, 8, 87. [Google Scholar] [CrossRef]
- Cossu, G.; Borello, U. Wnt Signaling and the Activation of Myogenesis in Mammals. EMBO J. 1999, 18, 6867–6872. [Google Scholar] [CrossRef]
- Casey, T.M.; Walker, J.F.; Bhide, K.; Thimmapuram, J.; Schoonmaker, J.P. Global Transcriptional Differences in Myokine and Inflammatory Genes in Muscle of Mature Steer Progeny Are Related to Maternal Lactation Diet and Muscle Composition. Physiol. Genom. 2018, 50, 884–892. [Google Scholar] [CrossRef]
- Du, M.; Yin, J.; Zhu, M.J. Cellular Signaling Pathways Regulating the Initial Stage of Adipogenesis and Marbling of Skeletal Muscle. Meat Sci. 2010, 86, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Li, X.; Choi, Y.M.; Shin, S.; Oh, S.-A.; Suh, Y.; Nguyen, T.H.; Baik, M.; Hwang, S.; Lee, K. Differential Expressions of G0/G1 Switch Gene 2 and Comparative Gene Identification-58 Are Associated with Fat Content in Bovine Muscle. Lipids 2014, 49, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, R.; Lu, X.; Liu, Y.; He, W.; Li, Y.; Yu, H.; Qin, L.; Cao, Y.; Zhao, Z. RNA-Seq Analysis Identifies Differentially Expressed Genes in the Longissimus Dorsi of Wagyu and Chinese Red Steppe Cattle. Int. J. Mol. Sci. 2022, 24, 387. [Google Scholar] [CrossRef] [PubMed]
- Zuo, B.; Yang, H.; Lei, M.G.; Li, F.E.; Deng, C.Y.; Jiang, S.W.; Xiong, Y.Z. Association of the Polymorphism in GYS1 and ACOX1 Genes with Meat Quality Traits in Pigs. Animal 2007, 1, 1243–1248. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Zhou, Y.; Tang, C.; Zhang, W.; Zhong, Y.; Chen, Y.; Zhou, H.; Sheng, L. NIK Links Inflammation to Hepatic Steatosis by Suppressing PPARα in Alcoholic Liver Disease. Theranostics 2020, 10, 3579. [Google Scholar] [CrossRef]
- Goszczynski, D.E.; Mazzucco, J.P.; Ripoli, M.V.; Villarreal, E.L.; Rogberg-Muñoz, A.; Mezzadra, C.A.; Melucci, L.M.; Giovambattista, G. Genetic Characterisation of PPARG, CEBPA and RXRA, and Their Influence on Meat Quality Traits in Cattle. J. Anim. Sci. Technol. 2016, 58, 14. [Google Scholar] [CrossRef]
- Fernyhough, M.E.; Okine, E.; Hausman, G.; Vierck, J.L.; Dodson, M. V PPARγ and GLUT-4 Expression as Developmental Regulators/Markers for Preadipocyte Differentiation into an Adipocyte. Domest. Anim. Endocrinol. 2007, 33, 367–378. [Google Scholar] [CrossRef]
- Srivastava, R.A.K. Fenofibrate Ameliorates Diabetic and Dyslipidemic Profiles in KKAy Mice Partly via Down-Regulation of 11β-HSD1, PEPCK and DGAT2.: Comparison of PPARα, PPARγ, and Liver x Receptor Agonists. Eur. J. Pharmacol. 2009, 607, 258–263. [Google Scholar] [CrossRef]
- Michalik, L.; Auwerx, J.; Berger, J.P.; Chatterjee, V.K.; Glass, C.K.; Gonzalez, F.J.; Grimaldi, P.A.; Kadowaki, T.; Lazar, M.A.; O’Rahilly, S. International Union of Pharmacology. LXI. Peroxisome Proliferator-Activated Receptors. Pharmacol. Rev. 2006, 58, 726–741. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, Y.; Yuan, W.; Wang, M.; Zhou, Y.; Chen, K.; Huang, Q. Downregulation of Osteopontin Inhibits Browning of White Adipose Tissues through PI3K-AKT Pathway in C57BL/6 Mice. Eur. J. Pharmacol. 2020, 866, 172822. [Google Scholar] [CrossRef]
- Fayyad, A.M.; Khan, A.A.; Abdallah, S.H.; Alomran, S.S.; Bajou, K.; Khattak, M.N.K. Rosiglitazone Enhances Browning Adipocytes in Association with MAPK and PI3-K Pathways during the Differentiation of Telomerase-Transformed Mesenchymal Stromal Cells into Adipocytes. Int. J. Mol. Sci. 2019, 20, 1618. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Wu, J.; Qiao, M.; Xu, Z.; Peng, X.; Mei, S. Proteomic and Lipidomic Analyses Reveal Saturated Fatty Acids, Phosphatidylinositol, Phosphatidylserine, and Associated Proteins Contributing to Intramuscular Fat Deposition. J. Proteom. 2021, 241, 104235. [Google Scholar] [CrossRef]


| Breed | Total Reads | Mapped | Paired Seq | Read1 | Read2 | Properly Paired | Mate-Mapped | Singletons |
|---|---|---|---|---|---|---|---|---|
| Lim1 | 129.8 M | 123.5 M | 104.4 M | 52.2 M | 52.2 M | 94.5 M | 95.2 M | 2.91 M |
| Lim2 | 120.9 M | 113.2 M | 94.4 M | 47.2 M | 47.2 M | 82.5 M | 83.4 M | 3.34 M |
| Lim3 | 115.4 M | 108.1 M | 91.2 M | 45.6 M | 45.6 M | 80.3 M | 81.1 M | 2.80 M |
| Lim4 | 122.5 M | 114.5 M | 95.9 M | 47.9 M | 47.9 M | 84.0 M | 84.7 M | 3.23 M |
| Hf1 | 118.3 M | 94.6 M | 92.9 M | 46.5 M | 46.5 M | 46.1 M | 65.7 M | 3.49 M |
| Hf2 | 337.2 M | 312.6 M | 268.3 M | 134.1 M | 134.1 M | 232.9 M | 235.0 M | 8.61 M |
| Hf3 | 135.0 M | 122.6 M | 109.0 M | 54.5 M | 54.5 M | 91.4 M | 92.7 M | 3.97 M |
| Hf4 | 151.5 M | 145.6 M | 128.0 M | 64.0 M | 64.0 M | 88.6 M | 90.5 M | 11.0 M |
| Her1 | 143.3 M | 139.3 M | 112.6 M | 6.0 M | 59.8 M | 98.5 M | 103.5 M | 3.65 M |
| Her2 | 196.4 M | 187.2 M | 146.2 M | 73.1 M | 73.1 M | 131.9 M | 133.0 M | 3.89 M |
| Her3 | 174.2 M | 166.1 M | 135.1 M | 63.1 M | 45.1 M | 112.2 M | 113.2 M | 4.56 M |
| Her4 | 131.3 M | 126.0 M | 104.2 M | 52.1 M | 52.1 M | 85.2 M | 86.4 M | 3.64 M |
| Core Genes | Brief Description of Function | Reference |
| SMAD3 | Regulates adipogenesis and myogenesis via the TGFβ–SMAD pathway; negative regulator of adipogenesis | [21,22,23,24] |
| FABP4 | Transports long-chain fatty acids; IMF marker; QTL in many breeds | [57,58,59,60,61,62,63,64,65,66] |
| RXRA | Nuclear receptor; regulates lipid pathways (with PPARG); influences fat metabolism; known lipid regulator | [39,40,41,90] |
| SCD | Enzyme synthesizing MUFA; IMF and meat quality marker | [30,31,32,33,34,35] |
| PLIN2 | Protects lipid droplets from lipolysis; regulates fat storage | [48,49,50,51,52,53,54,89] |
| FGF10 | Regulates adipogenesis and preadipocyte proliferation via MAPK and pRb–C/EBP | [36,37,38] |
| ELOVL3 | Biosynthesis of long-chain fatty acids; activated in adipogenesis | [38,67,68,69,70] |
| WNT1 | Blocks adipogenesis; activates Wnt/β-catenin; influences MSC fate | [26,27,28,29] |
| CTNNB1 | Transcriptional activator in Wnt; muscle tissue remodeling | [25] |
| Contextual Genes | Brief Description of Function | Reference |
| SQLE, HMGCS2, APOE, NR1H4, ACOX1 | Involved in lipid biosynthesis, transport, oxidation, and adaptation | [42,43,47,55,56,78,87,88] |
| PRKCA, MC4R | Modulate insulin signaling and energy homeostasis | [44,45,46,79] |
| COL3A1, COL9A1, ADAMTS4, PCNT, ACTA1 | Extracellular matrix remodeling and cytoskeletal reorganization | [72,73,74,81,82] |
| IL10, IL17A, GDF15, GDF3 | Inflammatory response and immune-mediated tissue remodeling | [75,76,77,80] |
| NFATC3, HRAS, IBSP, LAMB3 | Participation in key signaling pathways regulating muscle and fat cell biology (Wnt, PI3K–Akt, PPAR) | [83,88,94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasab, S.E.; Dashab, G.R.; Rokouei, M.; Roudbari, Z.; Sadkowski, T. Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis. Genes 2025, 16, 984. https://doi.org/10.3390/genes16080984
Nasab SE, Dashab GR, Rokouei M, Roudbari Z, Sadkowski T. Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis. Genes. 2025; 16(8):984. https://doi.org/10.3390/genes16080984
Chicago/Turabian StyleNasab, Saideh Eskandri, Gholam Reza Dashab, Mohammad Rokouei, Zahra Roudbari, and Tomasz Sadkowski. 2025. "Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis" Genes 16, no. 8: 984. https://doi.org/10.3390/genes16080984
APA StyleNasab, S. E., Dashab, G. R., Rokouei, M., Roudbari, Z., & Sadkowski, T. (2025). Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis. Genes, 16(8), 984. https://doi.org/10.3390/genes16080984






