Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of DIR Gene Family Members in S. bicolor
2.2. Chromosomal Localization, Gene Structure, and Conserved Motif Analysis of SbDIR Genes
2.3. Construction of the Phylogenetic Tree of DIR Proteins
2.4. Analysis of Cis-Regulatory Elements in SbDIR Gene Promoters
2.5. Tissue-Specific Expression and Stress Response Analysis of S. bicolor DIR Genes
2.6. Total RNA Extraction and RT-qPCR
3. Results
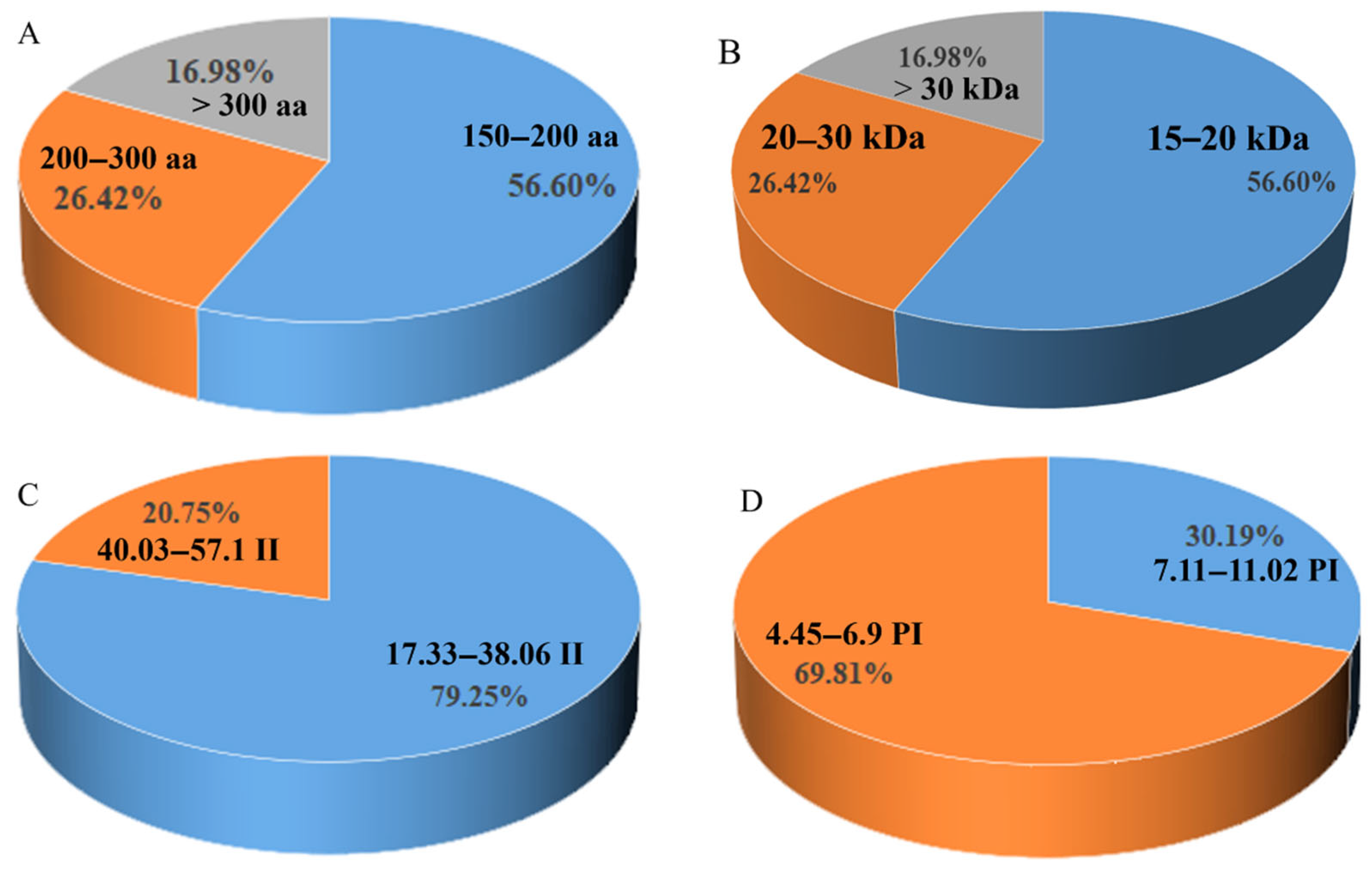
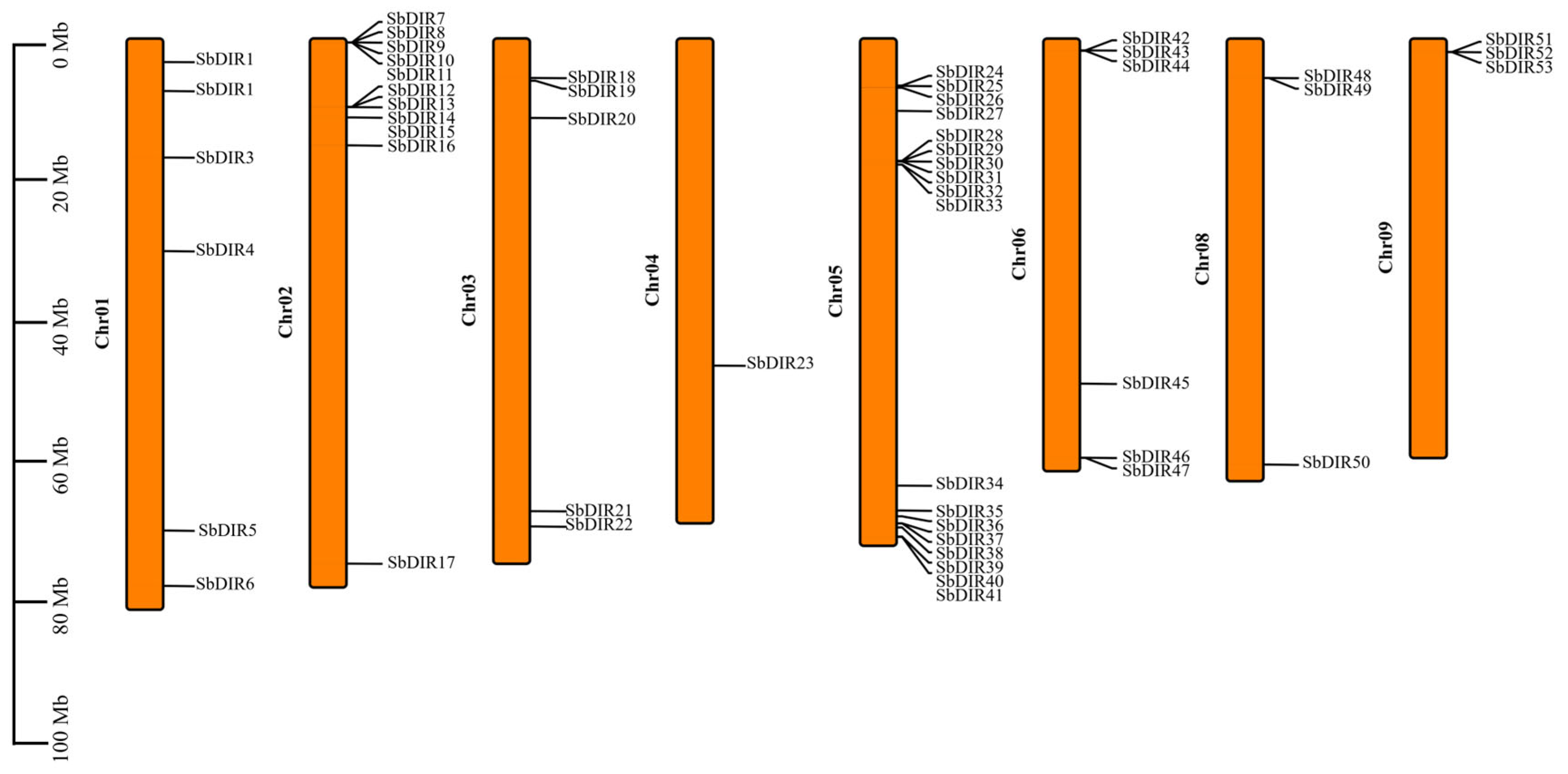
3.1. Identification and Chromosomal Localization of the DIR Gene Family in S. bicolor
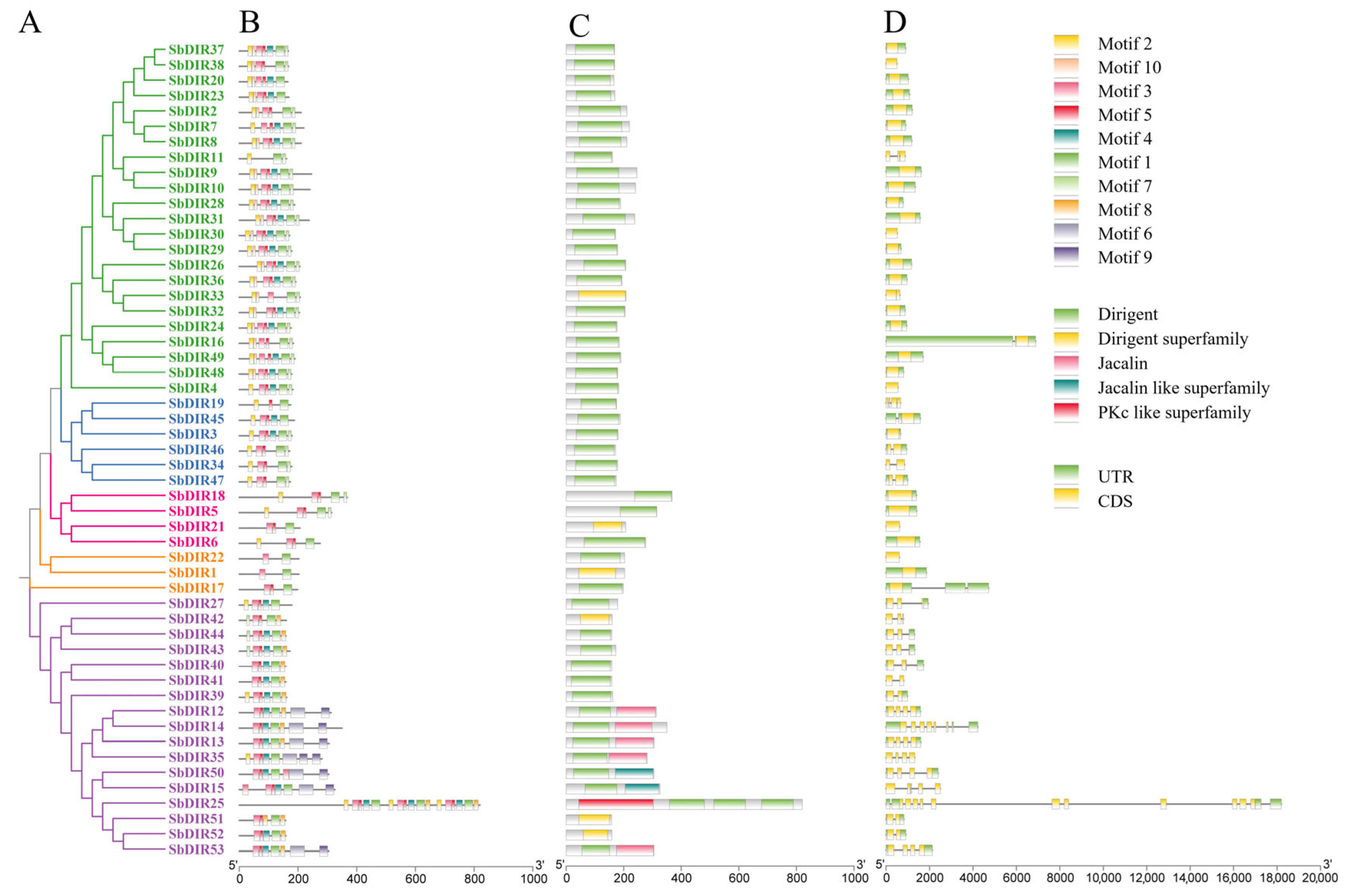
3.2. Structural and Conserved Motif Analysis of S. bicolor DIR Genes
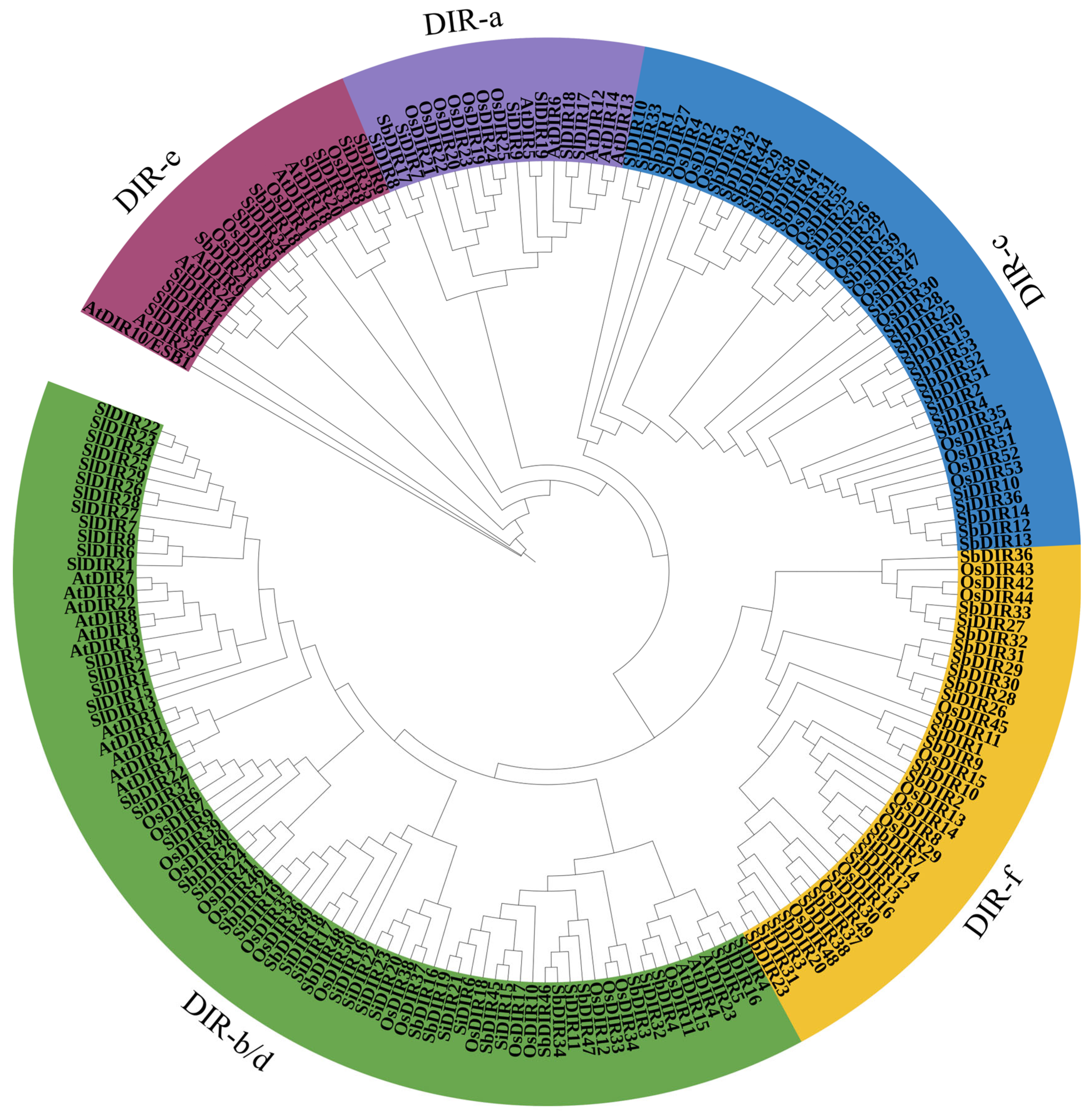
3.3. Phylogenetic Analysis of the S. bicolor DIR Gene Family
3.4. Tandem Duplication Acts as the Primary Driver for the Expansion of the SbDIR Gene Family
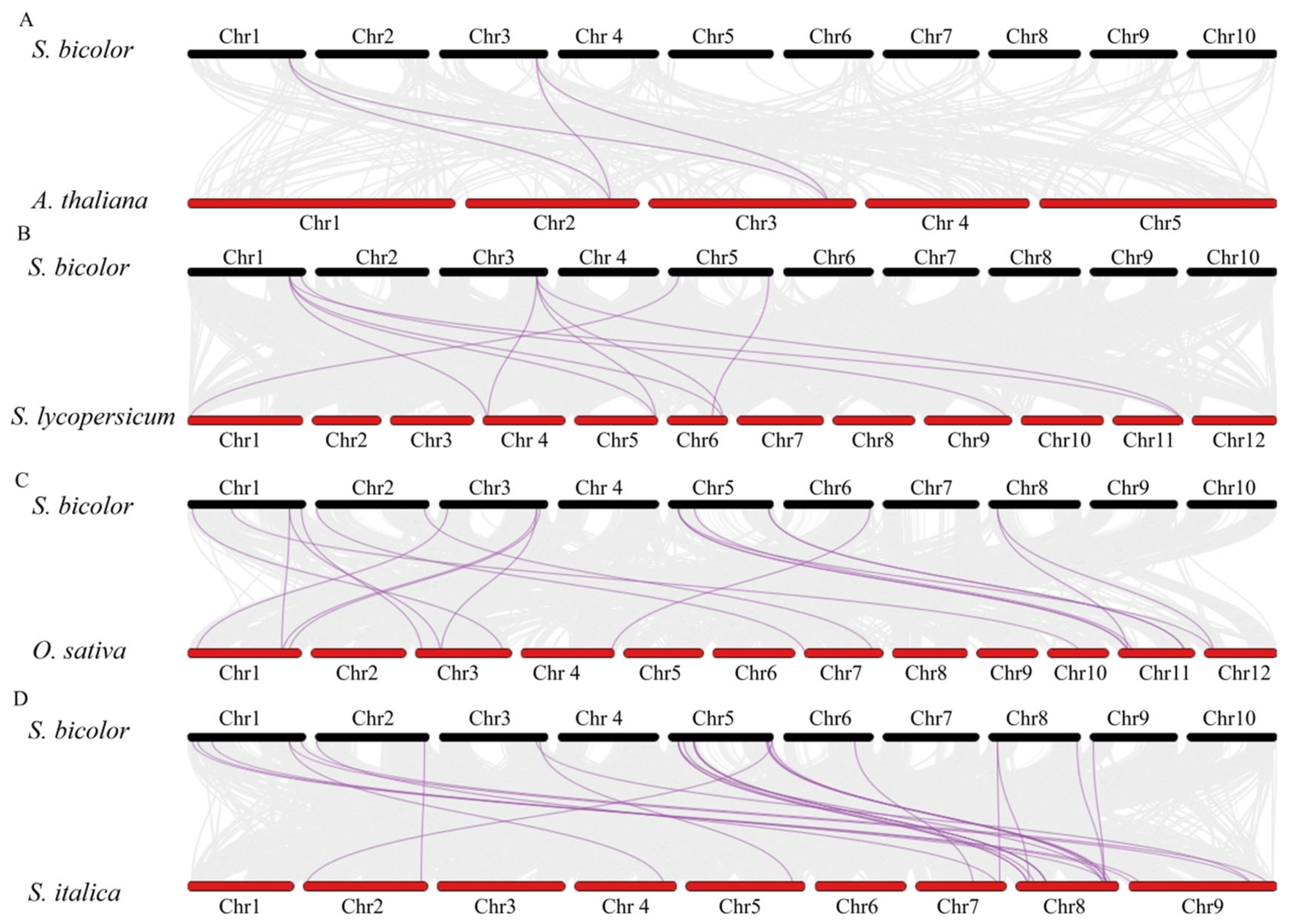
3.5. Synteny Analysis of S. bicolor and Other Species
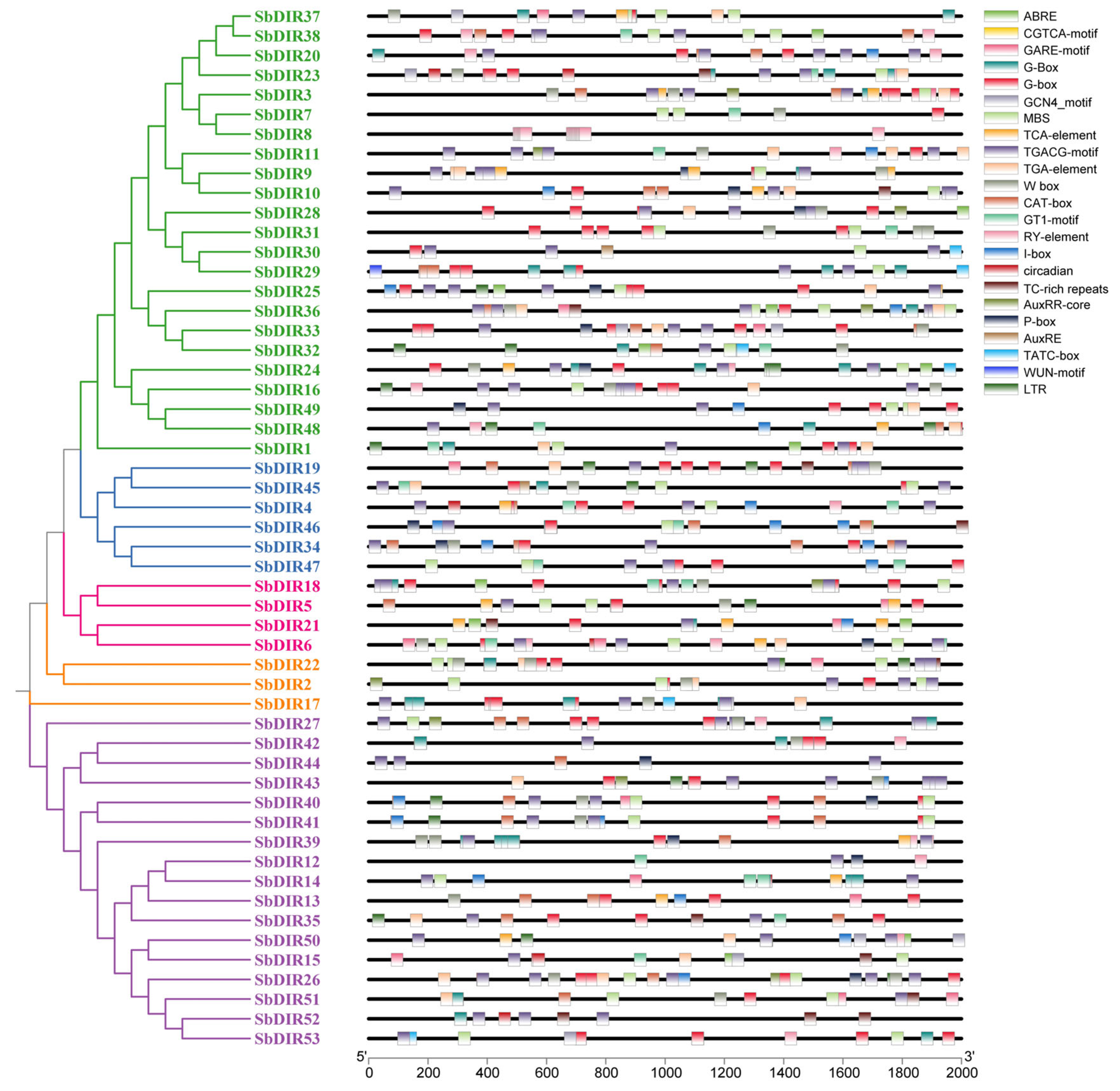
3.6. Cis-Acting Element Analysis in Promoter Regions of S. bicolor DIR Genes
3.7. Tissue-Specific Expression and Stress Response Patterns of S. bicolor DIR Genes
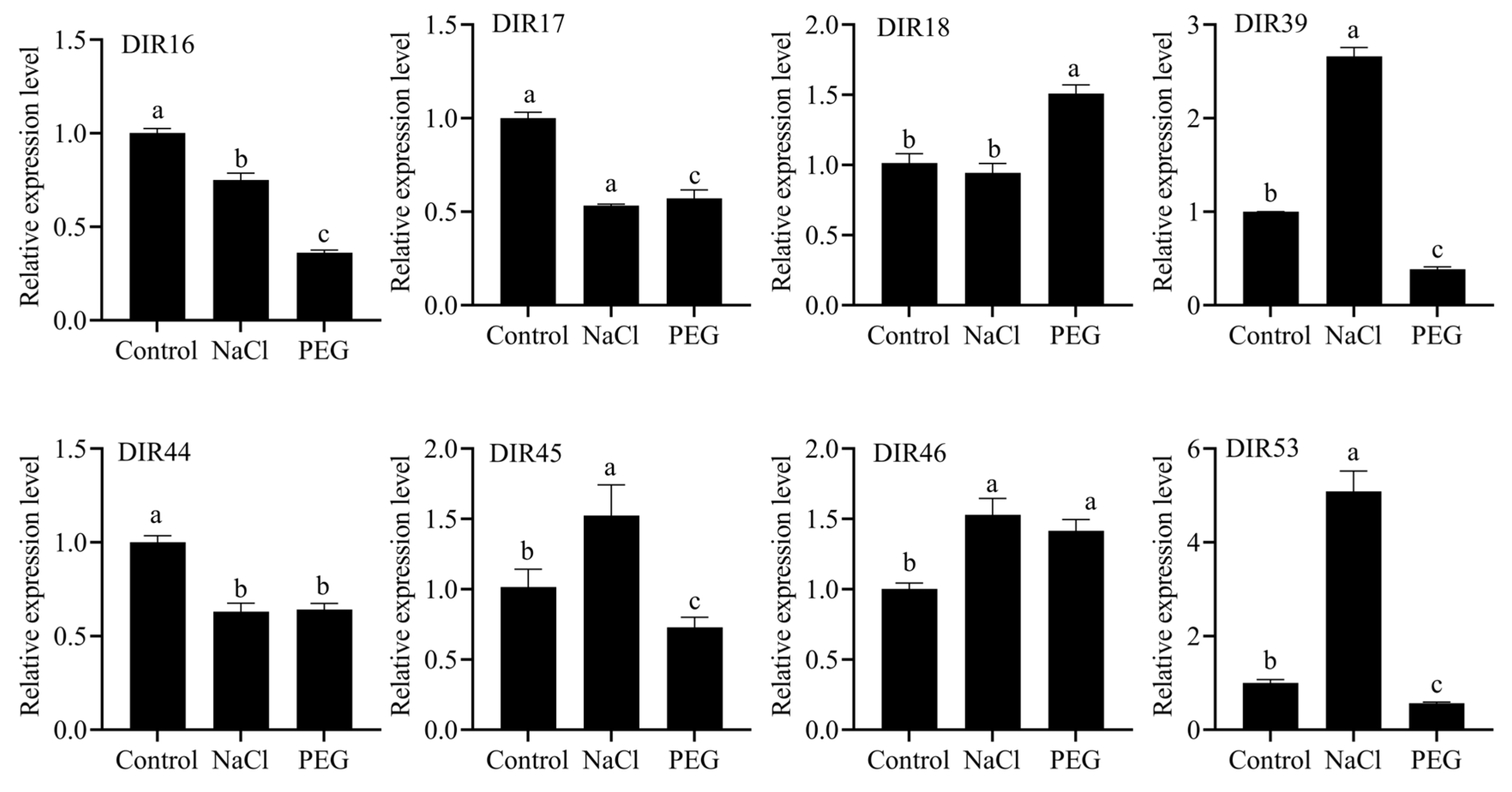
3.8. The Expression Level of the SbDIR Gene Under PEG and NaCl Stresses
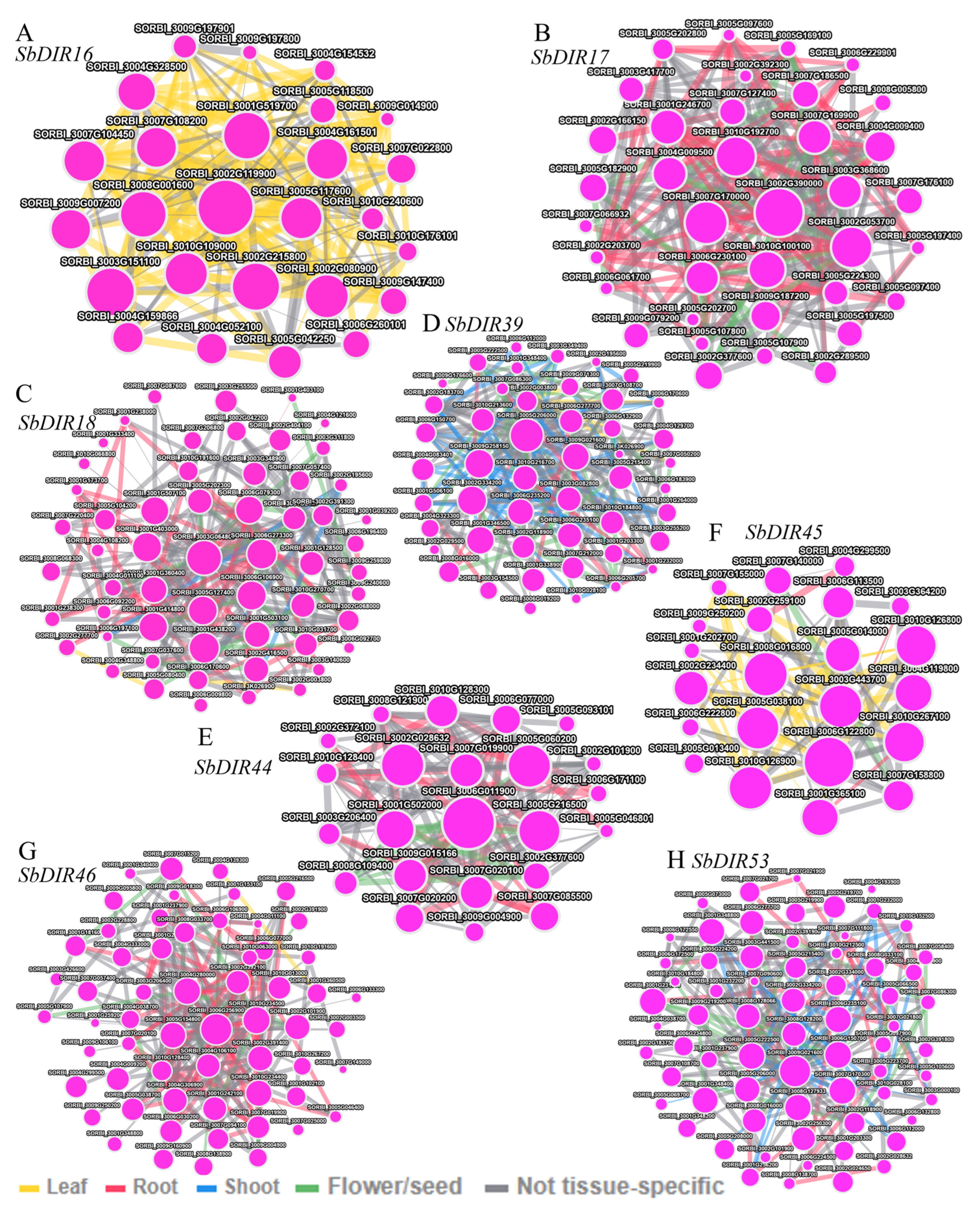
3.9. Gene Co-Expression Analysis
4. Discussion
4.1. Identification and Characterization of the DIR Gene Family in Sorghum
4.2. Phylogenetic Analysis of the SbDIR Gene Family
4.3. Analysis of Cis-Acting Elements in the Promoter Regions of SbDIR Genes
4.4. Tissue-Specific Expression Patterns and Stress Response Analysis of SbDIR Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein Without an Active Center. Science 1997, 275, 362–366. [Google Scholar] [CrossRef]
- Gang, D.R.; Costa, M.A.; Fujita, M.; Dinkova-Kostova, A.T.; Wang, H.B.; Burlat, V.; Martin, W.; Sarkanen, S.; Davin, L.B.; Lewis, N.G. Regiochemical control of monolignol radical coupling: A new paradigm for lignin and lignan biosynthesis. Chem. Biol. 1999, 6, 143–151. [Google Scholar] [CrossRef]
- Kim, M.K.; Jeon, J.-H.; Fujita, M.; Davin, L.B.; Lewis, N.G. The western red cedar (Thuja plicata) 8-8′ dirigent family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 2002, 49, 199–214. [Google Scholar] [CrossRef]
- Kim, K.-W.; Moinuddin, S.G.A.; Atwell, K.M.; Costa, M.A.; Davin, L.B.; Lewis, N.G. Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra Species. J. Biol. Chem. 2012, 287, 33957–33972. [Google Scholar] [CrossRef]
- Seneviratne, H.K.; Dalisay, D.S.; Kim, K.-W.; Moinuddin, S.G.; Yang, H.; Hartshorn, C.M.; Davin, L.B.; Lewis, N.G. Non-host disease resistance response in pea (Pisum sativum) pods: Biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 2015, 113, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: Integrated omics and maldi mass spectrometry imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xu, W.; Liu, T.; Chen, R.; Zhu, H.; Zhang, H.; Cai, C.; Li, S. Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence. Genomics 2021, 113, 979–990. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J.; et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genom. 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Duan, W.; Xue, B.; He, Y.; Liao, S.; Li, X.; Li, X.; Liang, Y.-K. Genome-wide identification and expression pattern analysis of dirigent members in the Genus Oryza. Int. J. Mol. Sci. 2023, 24, 7189. [Google Scholar] [CrossRef]
- Dokka, N.; Tyagi, S.; Ramkumar, M.; Rathinam, M.; Senthil, K.; Sreevathsa, R. Genome-wide identification and characterization of DIRIGENT gene family (CcDIR) in pigeonpea (Cajanus cajan L.) provide insights on their spatial expression pattern and relevance to stress response. Gene 2024, 914, 148417. [Google Scholar] [CrossRef] [PubMed]
- Xuan, X.; Su, S.; Chen, J.; Tan, J.; Yu, Z.; Jiao, Y.; Cai, S.; Zhang, Z.; Ramakrishnan, M. Evolutionary and functional analysis of the DIR gene family in Moso bamboo: Insights into rapid shoot growth and stress responses. Front. Plant Sci. 2025, 16, 1535733. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, M.; Yang, Y.; Tan, B.; Ren, D.; Tao, Y.; Li, R.; Zhao, Q. Comprehensive genome-wide analysis and functional characterization of the DIR gene family in Herpetospermum pedunculosum: Insights from HpDIR16 and HpDIR17. Plant Physiol. Biochem. 2025, 226, 110074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, Y.; Liang, X.; Zhuang, J.; Wang, X.; Qin, F.; Jiang, C. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 2022, 13, 2222. [Google Scholar] [CrossRef]
- Reboledo, G.; Del Campo, R.; Alvarez, A.; Montesano, M.; Mara, H.; Ponce de León, I. Physcomitrella patens Activates Defense Responses Against the Pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 2015, 16, 22280–22298. [Google Scholar] [CrossRef]
- Xue, B.; Duan, W.; Gong, L.; Zhu, D.; Li, X.; Li, X.; Liang, Y. The OsDIR55 gene increases salt tolerance by altering the root diffusion barrier. Plant J. 2024, 118, 1550–1568. [Google Scholar] [CrossRef]
- Zhao, Z.; Qin, T.; Zheng, H.; Guan, Y.; Gu, W.; Wang, H.; Yu, D.; Qu, J.; Wei, J.; Xu, W. Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought. Plants 2025, 14, 785. [Google Scholar] [CrossRef]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W.; et al. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef]
- Ralph, S.; Park, J.-Y.; Bohlmann, J.; Mansfield, S.D. Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in Spruce (Picea spp.). Plant Mol. Biol. 2006, 60, 21–40. [Google Scholar] [CrossRef]
- Ralph, S.G.; Jancsik, S.; Bohlmann, J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 2007, 68, 1975–1991. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Möller, C.; Hensel, G.; Kumlehn, J.; Höfle, C.; Hückelhoven, R.; Schaffrath, U. Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular poaceae-specific proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef]
- Esch, L.; Schaffrath, U. An update on jacalin-like lectins and their role in plant defense. Int. J. Mol. Sci. 2017, 18, 1592. [Google Scholar] [CrossRef] [PubMed]
- Fontanet-Manzaneque, J.B.; Laibach, N.; Herrero-García, I.; Coleto-Alcudia, V.; Blasco-Escámez, D.; Zhang, C.; Orduña, L.; Alseekh, S.; Miller, S.; Bjarnholt, N.; et al. Untargeted mutagenesis of brassinosteroid receptor SbBRI1 confers drought tolerance by altering phenylpropanoid metabolism in Sorghum bicolor. Plant Biotechnol. J. 2024, 12, 3406–3423. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Ge, F.; Du, H.; Sun, Y.; Sui, Y.; Tang, S.; Shen, Z.; Li, X.; Zhang, H.; Mei, C.; et al. A comprehensive omics resource and genetic tools for functional genomics research and genetic improvement of sorghum. Mol. Plant 2025, 18, 703–719. [Google Scholar] [CrossRef]
- Xue, B.; Liang, Z.; Li, D.; Liu, Y.; Liu, C. Genome-wide identification and expression analysis of CASPL gene family in Zea mays (L.). Front. Plant Sci. 2024, 15, 1477383. [Google Scholar] [CrossRef]
- Xue, B.; Liang, Z.; Liu, Y.; Li, D.; Cao, P.; Liu, C. Comparative Analysis of Casparian Strip Membrane Domain Protein Family in Oryza sativa (L.) and Arabidopsis thaliana (L.). Int. J. Mol. Sci. 2024, 25, 9858. [Google Scholar] [CrossRef]
- Zhou, Y.; Sukul, A.; Mishler-Elmore, J.W.; Faik, A.; Held, M.A. PlantNexus: A Gene Co-expression Network Database and Visualization Tool for Barley and Sorghum. Plant Cell Physiol. 2022, 63, 565–572. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Sun, Z.; Zhang, Y.; Meng, C.; Chen, B.; Wang, G.; Ke, H.; Wu, J.; Yan, Y.; et al. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes. BMC Plant Biol. 2023, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.U.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Feng, T.; Liu, C.-B.; Huang, H.; Wang, Y.-L.; Fu, X.; Han, M.-L.; Zhang, X.; Huang, X.; Wu, J.-C.; et al. The evolutionary innovation of root suberin lamellae contributed to the rise of seed plants. Nat. Plants 2023, 12, 1968–1977. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-Q.; Huang, J.-Q.; Reyt, G.; Song, T.; Love, A.; Tiemessen, D.; Xue, P.-Y.; Wu, W.-K.; George, M.W.; Chen, X.-Y.; et al. A dirigent protein complex directs lignin polymerization and assembly of the root diffusion barrier. Science 2023, 382, 464–471. [Google Scholar] [CrossRef]
- Gong, L.; Li, B.; Zhu, T.; Xue, B. Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 2023, 14, 1243806. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Lin, J.L.; Fang, X.; Li, J.X.; Chen, Z.W.; Wu, W.K.; Guo, X.X.; Liu, N.J.; Huang, J.F.; Chen, F.Y.; Wang, L.J.; et al. Dirigent gene editing of gossypol enantiomers for toxicity-depleted cotton seeds. Nat. Plants 2023, 9, 605–615. [Google Scholar] [CrossRef]
- Manna, M.; Rengasamy, B.; Sinha, A.K. Nutrient and Water Availability Influence Rice Physiology, Root Architecture and Ionomic Balance via Auxin Signalling. Plant Cell Environ. 2025, 48, 2691–2705. [Google Scholar] [CrossRef]
- Bárzana, G.; Carvajal, M. Genetic regulation of water and nutrient transport in water stress tolerance in roots. J. Biotechnol. 2020, 324, 134–142. [Google Scholar] [CrossRef]
- Reyt, G.; Ramakrishna, P.; Salas-González, I.; Fujita, S.; Love, A.; Tiemessen, D.; Lapierre, C.; Morreel, K.; Calvo-Polanco, M.; Flis, P.; et al. Two chemically distinct root lignin barriers control solute and water balance. Nat. Commun. 2021, 12, 2320. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, L.; Wang, Z.; Shang, H.; Liu, X.; Zhu, Y.; Qi, D.; Deng, X. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 2009, 19, 347–352. [Google Scholar] [CrossRef]
- Jin-Long, G.; Li-Ping, X.; Jing-Ping, F.; Ya-Chun, S.; Hua-Ying, F.; You-Xiong, Q.; Jing-Sheng, X. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 2012, 31, 1801–1812. [Google Scholar] [CrossRef]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Jing, T.; Liang, S.; Wang, H.; Guo, X.; Ma, Q.; Wang, J.; Wang, K.; He, X.; Zhao, H.; et al. Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.). Genes 2025, 16, 973. https://doi.org/10.3390/genes16080973
Liu S, Jing T, Liang S, Wang H, Guo X, Ma Q, Wang J, Wang K, He X, Zhao H, et al. Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.). Genes. 2025; 16(8):973. https://doi.org/10.3390/genes16080973
Chicago/Turabian StyleLiu, Shipeng, Tingrui Jing, Shuang Liang, Hairuo Wang, Xinyi Guo, Quan Ma, Junshen Wang, Kai Wang, Xiaolong He, Haibin Zhao, and et al. 2025. "Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.)" Genes 16, no. 8: 973. https://doi.org/10.3390/genes16080973
APA StyleLiu, S., Jing, T., Liang, S., Wang, H., Guo, X., Ma, Q., Wang, J., Wang, K., He, X., Zhao, H., Jiang, W., & Zhang, X. (2025). Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.). Genes, 16(8), 973. https://doi.org/10.3390/genes16080973




