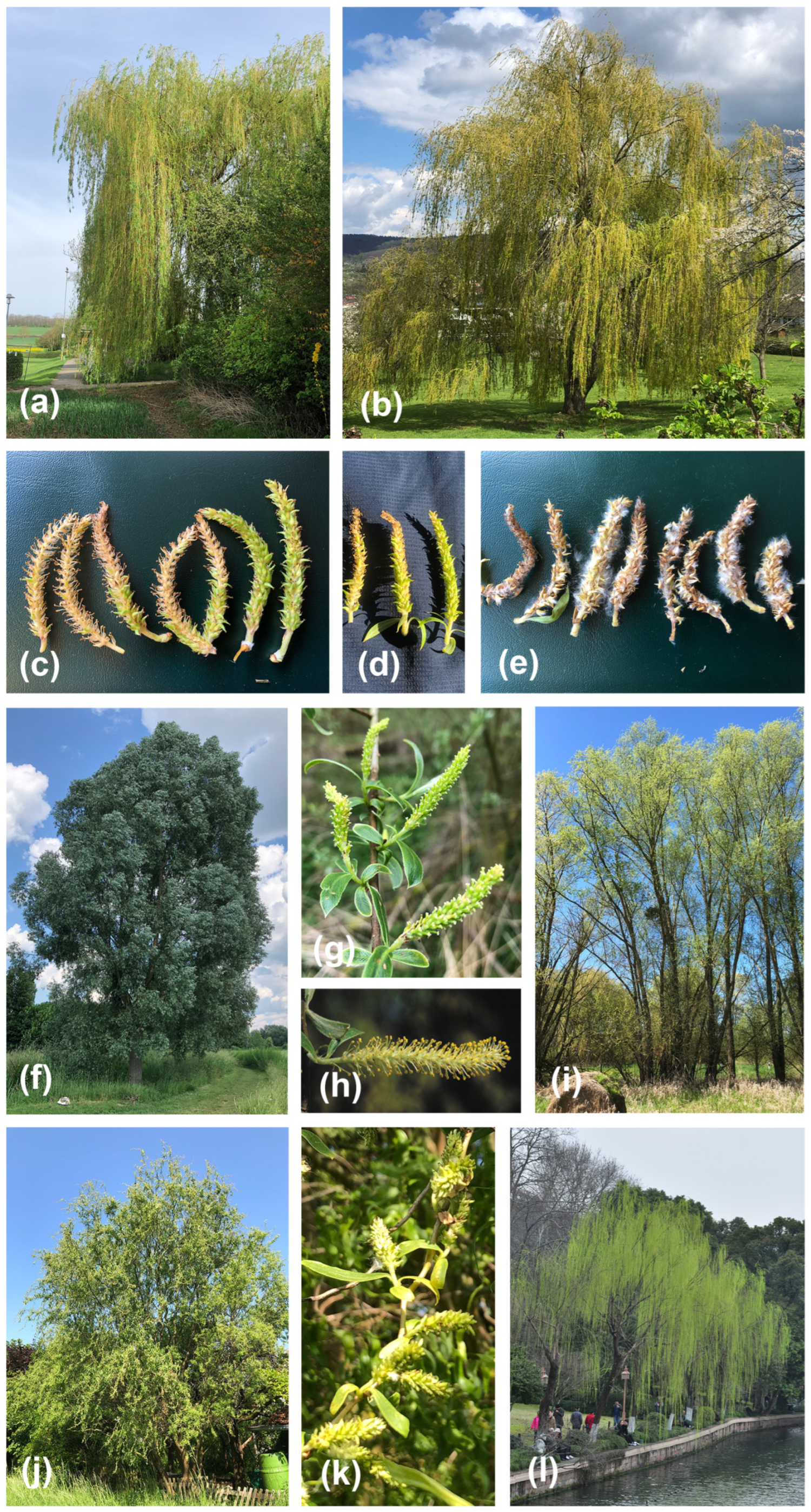
Hybridization Resulted in Shifts from Dioecy to Monoecy in Weeping Willows (Salix L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. DNA Extraction, RAD-Seq, and RAD-Loci Assembly
2.3. Analysis of Hybrid Structure
2.4. Identification of Sex-Specific Genomic Sequences
3. Results
3.1. RAD-Loci Assembly Results
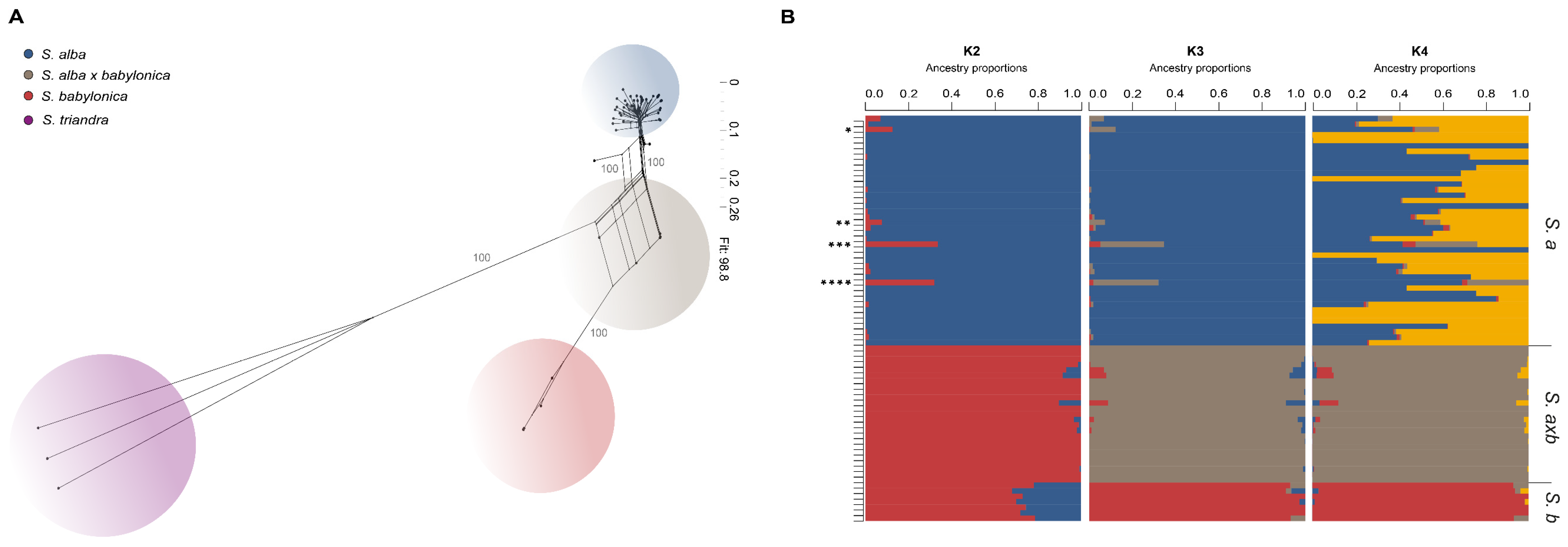
3.2. Analyses of Hybrid Origin
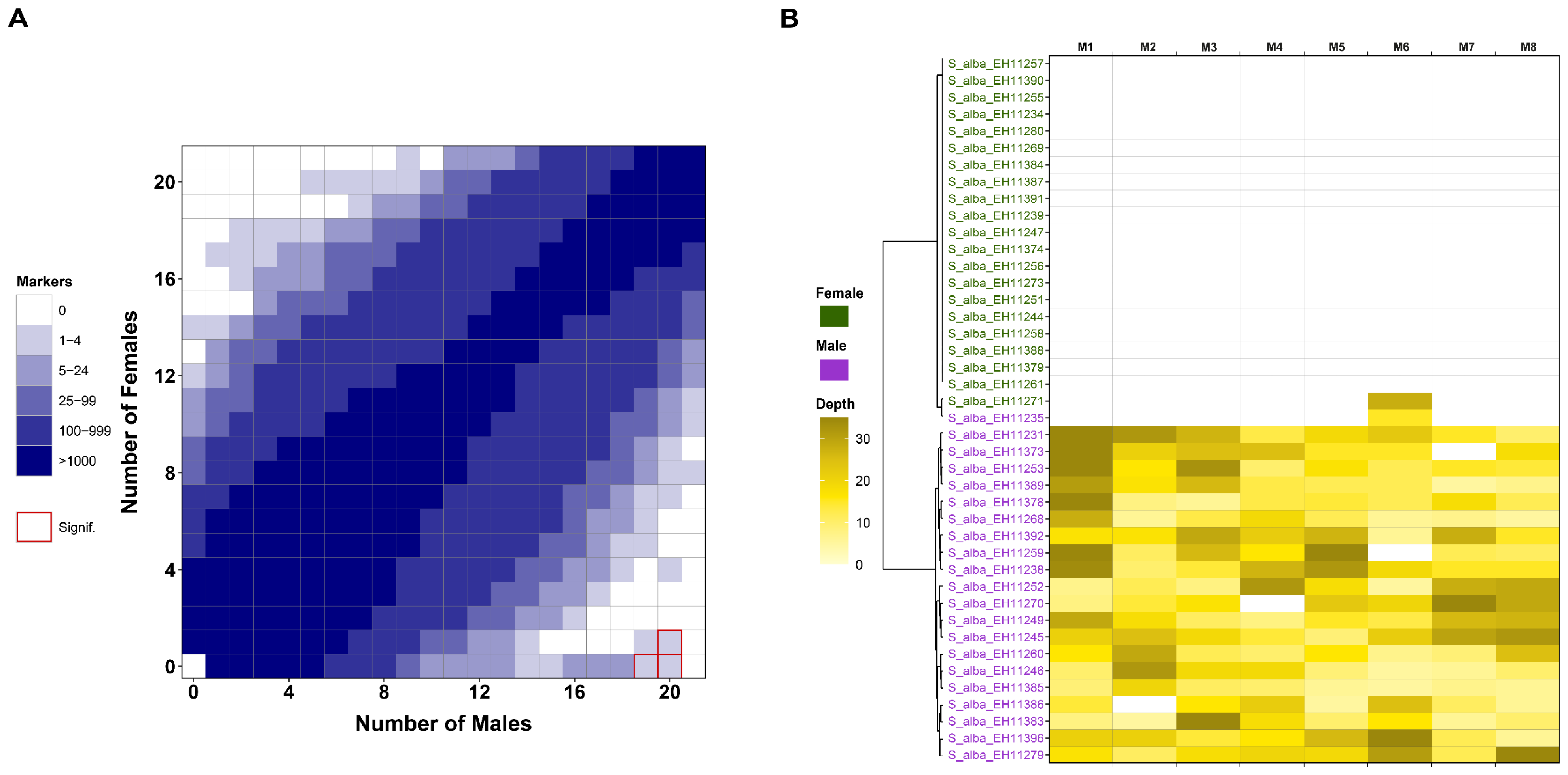
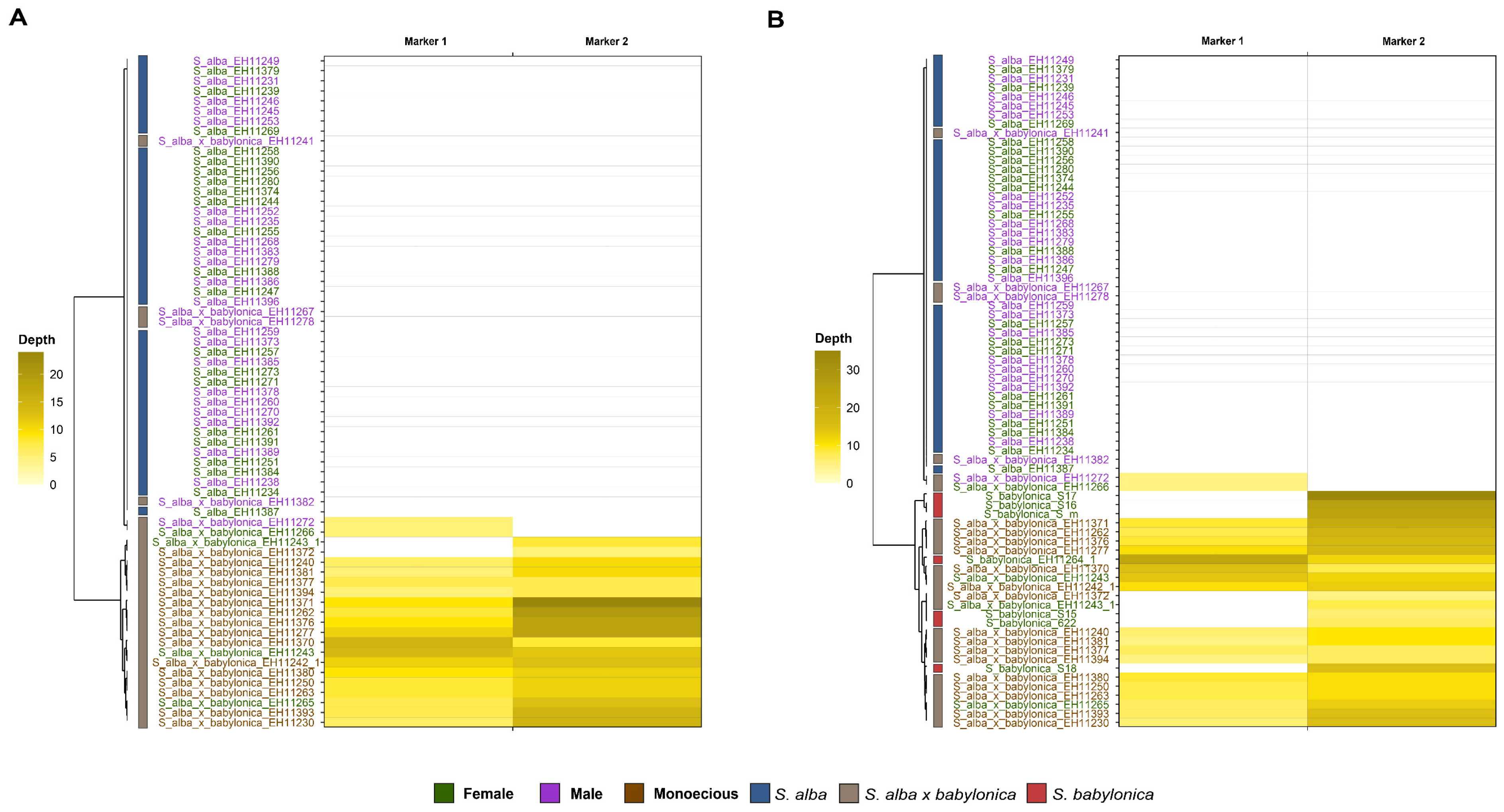
3.3. Sex Determination System (SDS) Analysis
4. Discussion
4.1. Genetic Composition of Hybrids and Comparison of Different Methods
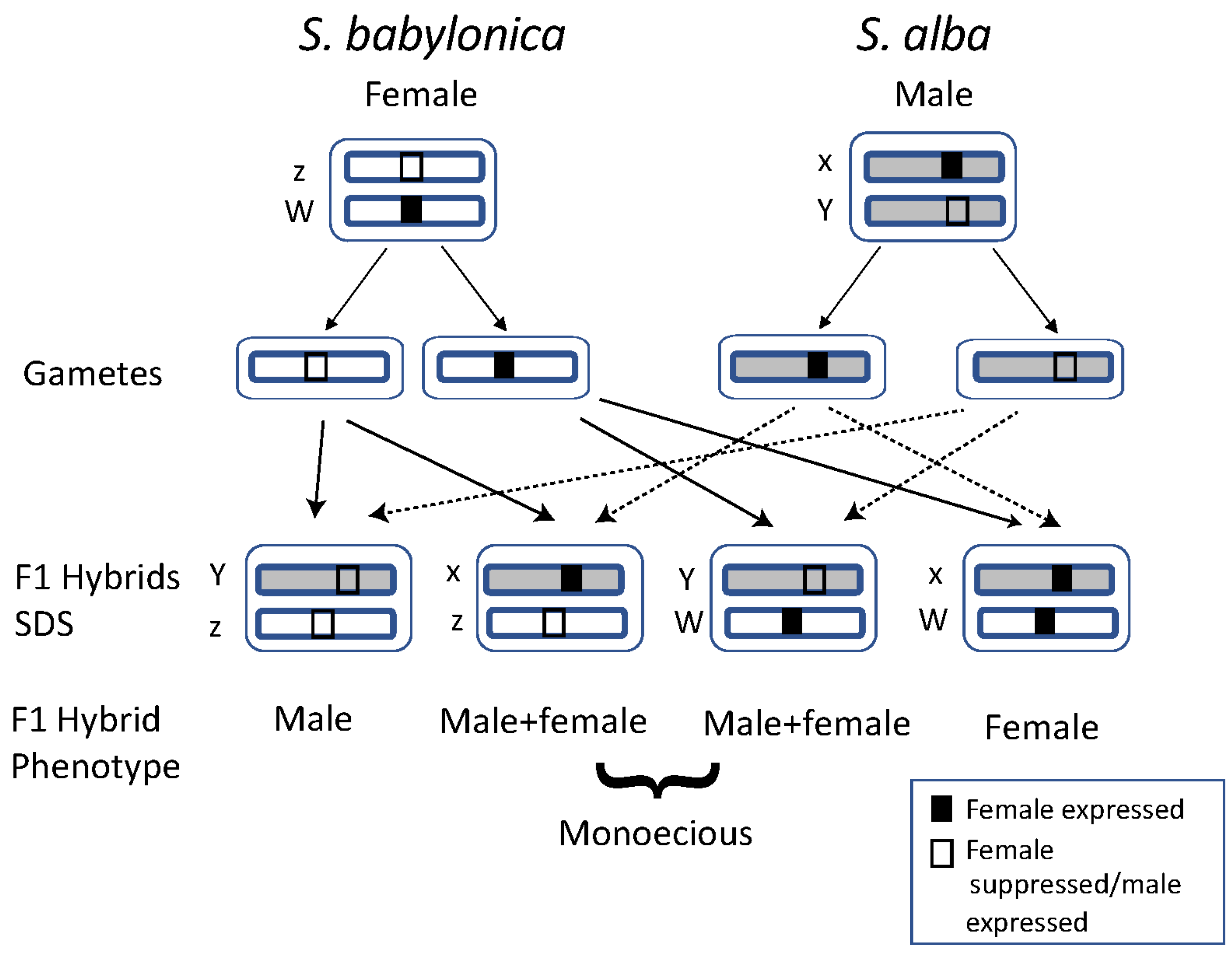
4.2. Putative Sex Determination System (SDS) in S. alba and the Combined SDS in S. alba × S. babylonica Hybrids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| S. | Salix |
| SDS | sex determination system |
| RADSeq | Restriction-site Associated DNA Sequencing |
| DArTseq | Diversity Array Technology |
| NCBI | National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/, accessed on 16 March 2025) |
Appendix A
| Character | S. alba | S. babylonica | S. alba × babylonica |
|---|---|---|---|
| Height | Up to 30 m | Up to 18 m | Up to 20 (30) m |
| Habit | Branchlets erect, sometimes apically slightly pendulous | Branchlets erect to pendulous 2 | Branchlets long pendulous from the basis |
| Branchlets | Brown, appressed hairy | Brownish, glabrous | Yellow, glabrous |
| Leave shape | Lanceolate | Narrow lanceolate to linear lanceolate | Narrow lanceolate to linear lanceolate |
| Stipules | Caduceous | Present during anthesis, ovate-lanceolate | Mostly caduceous; rarely present at anthesis, ovate-lanceolate |
| Leave indumentum (lower surface) | Densely silky hairy | Glabrous or sparsely pilose | Sparsely pilose, glabrescent |
| Sexes | Dioecious | Dioecious | Monoecious or dioecious |
| Female catkins (after anthesis) | 3.0–6.0 cm long | 2.0–3.0 cm long | 3.0–5.5 cm long |
| Ovary | Glabrous | Glabrous to hairy | Glabrous |
| Ovule no. per ovary | 8–24 3 | 2–6 | 2–8 |
References
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.W.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef]
- Arnold, M.L. Natural Hybridization and Evolution; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Gonzalez, A.; Lexer, C.; Cronk, Q.C.B. Adaptive introgression: A plant perspective. Biol. Lett. 2018, 14, 20170688. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Nyman, T.; Wang, D.C.; Argus, G.W.; Yang, Y.P.; Chen, J.H. Phylogeny of Salix subgenus Salix s.l. (Salicaceae): Delimitation, biogeography, and reticulate evolution. BMC Evol. Biol. 2015, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L.; Ballerini, E.S.; Brothers, A.N.; Brothers, A.N. Hybrid fitness, adaptation and evolutionary diversification: Lessons learned from Louisiana Irises. Heredity 2012, 108, 159–166. [Google Scholar] [CrossRef]
- Gramlich, S.; Wagner, N.D.; Hörandl, E. RAD-seq reveals genetic structure of the F-2-generation of natural willow hybrids (Salix L.) and a great potential for interspecific introgression. BMC Plant Biol. 2018, 18, 317. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.Y.; Zhou, R.Y.; Wang, Q.L.; Wang, L.S. Ratooning annual cotton (Gossypium spp.) for perennial utilization of heterosis. Front. Plant Sci. 2020, 11, 554970. [Google Scholar] [CrossRef]
- Natalini, A.; Acciarri, N.; Cardi, T. Breeding for Nutritional and Organoleptic Quality in Vegetable Crops: The Case of Tomato and Cauliflower. Agriculture 2021, 11, 606. [Google Scholar] [CrossRef]
- Ashraf, H.; Ghouri, F.; Baloch, F.S.; Nadeem, M.A.; Fu, X.; Shahid, M.Q. Hybrid Rice Production: A Worldwide Review of Floral Traits and Breeding Technology, with Special Emphasis on China. Plants 2024, 13, 578. [Google Scholar] [CrossRef]
- Meikle, R.D. Willows and Poplars of Great Britain and Ireland; Botanical Society of the British Islands: London, UK, 1984. [Google Scholar]
- Richards, J.A. Plant Breeding Systems, 2nd ed.; Chapman and Hall: London, UK, 1997; p. 529. [Google Scholar]
- Renner, S.S. The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 2014, 101, 1588–1596. [Google Scholar] [CrossRef]
- Baránková, S.; Pascual Díaz, J.; Sultana, N.; Alonso Lifante, M.; Balant, M.; Barros, K.; D’Ambrosio, U.; Malinská, H.; Peska, V.; Pérez Lorenzo, I.; et al. Sex-chrom, a database on plant sex chromosomes. New Phytol. 2020, 227, 1594–1604. [Google Scholar] [CrossRef]
- Charlesworth, D. Some thoughts about the words we use for thinking about sex chromosome evolution. Philos. Trans. R. Soc. B 2022, 377, 20210314. [Google Scholar] [CrossRef]
- Charlesworth, D.; Harkess, A. Why should we study plant sex chromosomes? Plant Cell 2024, 36, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.A.; Kersten, B.; Leite Montalvão, A.P.; Mähler, N.; Bernhardsson, C.; Bräutigam, K.; Carracedo Lorenzo, Z.; Hoenicka, H.; Kumar, V.; Mader, M.; et al. A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 2020, 6, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Gulyaev, S.; Cai, X.-J.; Guo, F.-Y.; Kikuchi, S.; Applequist, W.; Zhang, Z.-X.; Hörandl, E.; He, L. The phylogeny of Salix revealed by whole genome re-sequencing suggests different sex-determination systems in major groups of the genus. Ann. Bot. 2022, 129, 485–498. [Google Scholar] [CrossRef]
- He, L.; Jia, K.-H.; Zhang, R.-G.; Wang, Y.; Shi, T.-L.; Li, Z.-C.; Zeng, S.-W.; Cai, X.-J.; Wagner, N.D.; Hörandl, E.; et al. Chromosome-scale assembly of the genome of Salix dunnii reveals a male-heterogametic sex determination system on chromosome 7. Mol. Ecol. Resour. 2021, 21, 1966–1982. [Google Scholar] [CrossRef]
- He, L.; Wang, Y.; Wang, Y.; Zhang, R.-G.; Hörandl, E.; Ma, T.; Mao, Y.-F.; Mank, J.E.; Ming, R. Allopolyploidization from two dioecious ancestors leads to recurrent evolution of sex chromosomes. Nat. Commun. 2024, 15, 6893. [Google Scholar] [CrossRef]
- Xue, Z.-Q.; Applequist, W.L.; Hoerandl, E.; He, L. Sex chromosome turnover plays an important role in the maintenance of barriers to post-speciation introgression in willows. Evol. Lett. 2024, 8, 467–477. [Google Scholar] [CrossRef]
- Gerchen, J.F.; Veltsos, P.; Pannell, J.R. Recurrent allopolyploidization, Y-chromosome introgression and the evolution of sexual systems in the plant genus Mercurialis. Philos. Trans. Biol. Sci. 2022, 377, 20210224. [Google Scholar] [CrossRef]
- He, L.; Hörandl, E. Does polyploidy inhibit sex chromosome evolution in angiosperms? Front. Plant Sci. 2022, 13, 976765. [Google Scholar] [CrossRef]
- Argus, G.W. Infrageneric classification of Salix (Salicaceae) in the New World. Syst. Bot. Monogr. 1997, 52, 1–121. [Google Scholar] [CrossRef]
- Skvortsov, A. Willows of Russia and Adjacent Countries; University of Joensuu: Joensuu, Finland, 1999. (In English) [Google Scholar]
- Mirski, P. Exceptions from dioecy and sex lability in genus Salix. Dendrobiology 2014, 71, 167–171. [Google Scholar] [CrossRef][Green Version]
- Hardig, T.M.; Brunsfeld, S.J.; Fritz, R.S.; Morgan, M.; Orians, C.M. Morphological and molecular evidence for hybridization and introgression in a willow (Salix) hybrid zone. Mol. Ecol. 2000, 9, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Oberprieler, C.; Dietz, L.; Harlander, C.; Heilmann, J. Molecular and phytochemical evidence for the taxonomic integrity of Salix alba, S-fragilis, and their hybrid S. × rubens (Salicaceae) in mixed stands in SE Germany. Plant Syst. Evol. 2013, 299, 1107–1118. [Google Scholar] [CrossRef]
- Vasut, R.J.; Pospiskova, M.; Lukavsky, J.; Weger, J. Detection of hybrids in willows (Salix, Salicaceae) using genome-wide DArTseq markers. Plants 2024, 13, 639. [Google Scholar] [CrossRef]
- Marincek, P.; Pittet, L.; Wagner, N.D.; Hörandl, E. Evolution of a hybrid zone of two willow species (Salix L.) in the European Alps analyzed by RAD-seq and morphometrics. Ecol. Evol. 2023, 13, e9700. [Google Scholar] [CrossRef]
- Pittet, L.; Marincek, P.; Kosinski, P.; Wagner, N.D.; Hörandl, E. Hybrid zones in the European Alps impact the phylogeography of alpine vicariant willow species (Salix L.). Front. Plant Sci. 2025, 16, 1507275. [Google Scholar] [CrossRef]
- Newsholme, C. Willows: The Genus Salix; Timber Press: Portland, Oregon, 1992. [Google Scholar]
- Hörandl, E.; Florineth, F.; Hadacek, F. Weiden in Österreich und Angrenzenden Gebieten [Willows in Austria and Adjacent Regions], 2nd ed.; University of Agriculture: Vienna, Austria, 2012; p. 164. [Google Scholar]
- Barcaccia, G.; Meneghetti, S.; Albertini, E.; Triest, L.; Lucchin, M. Linkage mapping in tetraploid willows: Segregation of molecular markers and estimation of linkage phases support an allotetraploid structure for Salix alba × Salix fragilis interspecific hybrids. Heredity 2003, 90, 169–180. [Google Scholar] [CrossRef]
- Barcaccia, G.; Meneghetti, S.; Lucchin, M.; de Jong, H. Genetic segregation and genomic hybridization patterns support an allotetraploid structure and disomic inheritance for Salix species. Diversity 2014, 6, 633–651. [Google Scholar] [CrossRef]
- Fang, C.F.; Zhao, S.D.; Skvortsov, A. Salicaceae. In Flora of China; Flora of China Editorial Committee, Ed.; Missouri Botanical Garden: St. Louis, MO, USA, 1999; Volume 4, pp. 139–274. Available online: http://www.efloras.org (accessed on 16 March 2025).
- Wagner, N.D.; Marincek, P.; Pittet, L.; Hörandl, E. Insights into the Taxonomically Challenging Hexaploid Alpine Shrub Willows of Salix Sections Phylicifoliae and Nigricantes (Salicaceae). Plants 2023, 12, 1144. [Google Scholar] [CrossRef]
- Karbstein, K.; Tomasello, S.; Hodac, L.; Wagner, N.; Marincek, P.; Barke, B.H.; Paetzold, C.; Hörandl, E. Untying Gordian knots: Unraveling reticulate polyploid plant evolution by genomic data using the large Ranunculus auricomus species complex. New Phytol. 2022, 235, 2081–2098. [Google Scholar] [CrossRef]
- Moreau, E.L.P.; Medberry, A.N.; Honig, J.A.; Molnar, T.J. Genetic diversity analysis of big-bracted dogwood (Cornus florida and C. kousa) cultivars, interspecific hybrids, and wild-collected accessions using RADseq. PLoS ONE 2024, 19, e0307326. [Google Scholar] [CrossRef] [PubMed]
- Garner, A.G.; Goulet-Scott, B.E.; Hopkins, R. Phylogenomic analyses re-examine the evolution of reinforcement and hypothesized hybrid speciation in Phlox wildflowers. New Phytol. 2024, 243, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Florido, A.; González-Toral, C.; Maguilla, E.; Cires, E.; Díaz-Lifante, Z.; Andrés-Camacho, C.; Feliner, G.N.; Arroyo, J.; Escudero, M. Polyploidy and hybridization in the Mediterranean: Unravelling the evolutionary history of Centaurium (Gentianaceae). Ann. Bot. 2024, 134, 247–262. [Google Scholar] [CrossRef]
- Palombo, N.E.; Weiss-Schneeweiss, H.; García, C.C. Evolutionary relationships, hybridization and diversification under domestication of the locoto chile (Capsicum pubescens) and its wild relatives. Front. Plant Sci. 2024, 15, 1353991. [Google Scholar] [CrossRef]
- Feron, R.; Pan, Q.; Wen, M.; Imarazene, B.; Jouanno, E.; Anderson, J.; Herpin, A.; Journot, L.; Parrinello, H.; Klopp, C.; et al. RADSex: A computational workflow to study sex determination using restriction site-associated DNA sequencing data. Mol. Ecol. Resour. 2021, 21, 1715–1731. [Google Scholar] [CrossRef]
- Hobza, R.; Bacovsky, V.; Cegan, R.; Horakova, L.; Hubinsky, M.; Janicek, T.; Janousek, B.; Jedlicka, P.; Kruzlicova, J.; Kubat, Z.; et al. Sexy ways: Approaches to studying plant sex chromosomes. J. Exp. Bot. 2024, 75, 5204–5219. [Google Scholar] [CrossRef]
- Marchenko, A.M.; Kuzovkina, Y.A. Identification of hybrid formulae of a few willows (Salix) using ovule numbers. Silvae Genet. 2021, 70, 75–83. [Google Scholar] [CrossRef]
- Marchenko, A.M.; Kuzovkina, Y.A. The Ovule Number Variation Provides New Insights into Taxa Delimitation in Willows (Salix subgen. Salix; Salicaceae). Plants 2023, 12, 497. [Google Scholar] [CrossRef]
- Thibault, J. Nuclear DNA amount in pure species and hybrid willows (Salix): A flow cytometric investigation. Can. J. Bot. 1998, 76, 157–165. [Google Scholar] [CrossRef]
- Kosinski, P.; Sliwinska, E.; Hilpold, A.; Boratynski, A. DNA ploidy in Salix retusa agg. only partly in line with its morphology and taxonomy. Nord. J. Bot. 2019, 37, e02197. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 February 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.R.; Stevens, J.R.; Catchen, J.M. Lost in parameter space: A road map for stacks. Methods Ecol. Evol. 2017, 8, 1360–1373. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Bryant, D.; Moulton, V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef]
- Frichot, E.; Mathieu, F.; Trouillon, T.; Bouchard, G.; Francois, O. Fast and efficient estimation of individual ancestry coefficients. Genetics 2014, 196, 973–983. [Google Scholar] [CrossRef]
- Frichot, E.; Francois, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Knaus, B.J.; Grunwald, N.J. VCFR: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Mijangos, J.L.; Gruber, B.; Berry, O.; Pacioni, C.; Georges, A. dartR v2: An accessible genetic analysis platform for conservation, ecology and agriculture. Methods Ecol. Evol. 2022, 13, 2150–2158. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Wiens, B.J.; Decicco, L.H.; Colella, J.P. triangulaR: An R package for identifying AIMs and building triangle plots using SNP data from hybrid zones. Heredity 2025, 134, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.C.; Thompson, E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Wringe, B.F.; Stanley, R.R.E.; Jeffery, N.W.; Anderson, E.C.; Bradbury, I.R. hybriddetective: A workflow and package to facilitate the detection of hybridization using genomic data in r. Mol. Ecol. Resour. 2017, 17, e275–e284. [Google Scholar] [CrossRef]
- Feron, R. SexGenomicsToolkit/Radsex: 1.2.0. Version 1.2.0. [CrossRef]
- Wichura, M. Die Bastardbefruchtung im Pflanzenreich: Erläutert an den Bastarden der Weiden. E. Morgenstern: Breslau, Poland, 1865. [Google Scholar]
- Triest, L.; Trung, L.Q.; Talukder, A.; Van Puyvelde, K. Nuclear cyp73 intron fragment length polymorphism supports morphological analysis of Salix species and hybrids. Plant Biosyst. 2009, 143, 555–563. [Google Scholar] [CrossRef]
- Abbott, S.; Fairbanks, D.J. Experiments on Plant Hybrids by Gregor Mendel. Genetics 2016, 204, 407–422. [Google Scholar] [CrossRef]
- Fitzpatrick, B.M. Estimating ancestry and heterozygosity of hybrids using molecular markers. Bmc Evol. Biol. 2012, 12, 131. [Google Scholar] [CrossRef]
- Wagner, N.D.; He, L.; Hörandl, E. Phylogenomic relationships and evolution of polyploid Salix species revealed by RAD Sequencing data. Front. Plant Sci. 2020, 11, 1077. [Google Scholar] [CrossRef]
- Wang, L.; Yang, J.; Zhang, H.; Tao, Q.; Zhang, Y.; Dang, Z.; Zhang, F.; Luo, Z. Sequence coverage required for accurate genotyping by sequencing in polyploid species. Mol. Ecol. Resour. 2022, 22, 1417–1426. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef]
- Novembre, J. Pritchard, Stephens, and Donnelly on Population Structure. Genetics 2016, 204, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Wiens, B.J.; Colella, J.P. That’s not a hybrid: How to distinguish patterns of admixture and isolation by distance. Mol. Ecol. Resour. 2025, 25, e14039. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.J.; Van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 2018, 9, 3258. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Abbott, R.J.; Hegarty, M.J.; Hiscock, S.J.; Brennan, A.C. Homoploid hybrid speciation in action. Taxon 2010, 59, 1375–1386. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef]
- Cerca, J.; Maurstad, M.F.; Rochette, N.C.; Rivera-Colon, A.G.; Rayamajhi, N.; Catchen, J.M.; Struck, T.H. Removing the bad apples: A simple bioinformatic method to improve loci-recovery in de novo RADseq data for non-model organisms. Methods Ecol. Evol. 2021, 12, 805–817. [Google Scholar] [CrossRef]
- Renner, S.S.; Müller, N.A. Sex determination and sex chromosome evolution in land plants. Philos. Trans. R. Soc. B 2022, 377, 20210210. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alarcón-Bolaños, P.; Pittet, L.; He, L.; Hörandl, E. Hybridization Resulted in Shifts from Dioecy to Monoecy in Weeping Willows (Salix L.). Genes 2025, 16, 958. https://doi.org/10.3390/genes16080958
Alarcón-Bolaños P, Pittet L, He L, Hörandl E. Hybridization Resulted in Shifts from Dioecy to Monoecy in Weeping Willows (Salix L.). Genes. 2025; 16(8):958. https://doi.org/10.3390/genes16080958
Chicago/Turabian StyleAlarcón-Bolaños, Pablo, Loïc Pittet, Li He, and Elvira Hörandl. 2025. "Hybridization Resulted in Shifts from Dioecy to Monoecy in Weeping Willows (Salix L.)" Genes 16, no. 8: 958. https://doi.org/10.3390/genes16080958
APA StyleAlarcón-Bolaños, P., Pittet, L., He, L., & Hörandl, E. (2025). Hybridization Resulted in Shifts from Dioecy to Monoecy in Weeping Willows (Salix L.). Genes, 16(8), 958. https://doi.org/10.3390/genes16080958






