Exploring the Functional Potential of the Broiler Gut Microbiome Using Shotgun Metagenomics
Abstract
1. Introduction
2. Materials and Methods
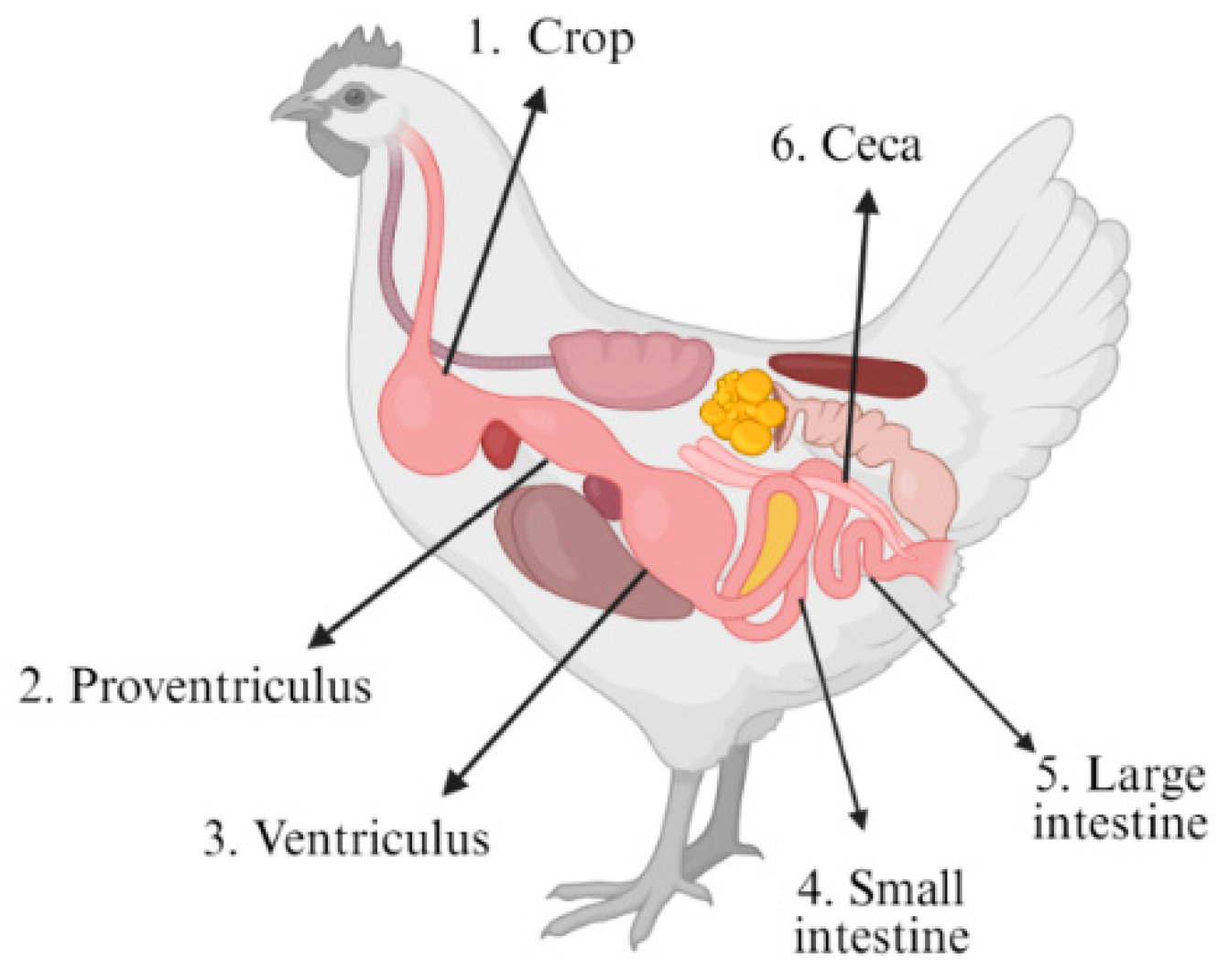
2.1. Sample Acquisition and Processing
2.2. DNA Extraction and Shotgun Metagenomic Sequencing
2.3. Bioinformatics Analysis
2.3.1. Metagenomic Read Processing, Assembly and Functional Annotation
2.3.2. Identification of Biosynthetic Gene Clusters and Virulence/Resistance Genes
2.3.3. KEGG-Based Functional Annotation
2.3.4. Taxonomy Profiling
3. Results
3.1. Assembly Quality Reports
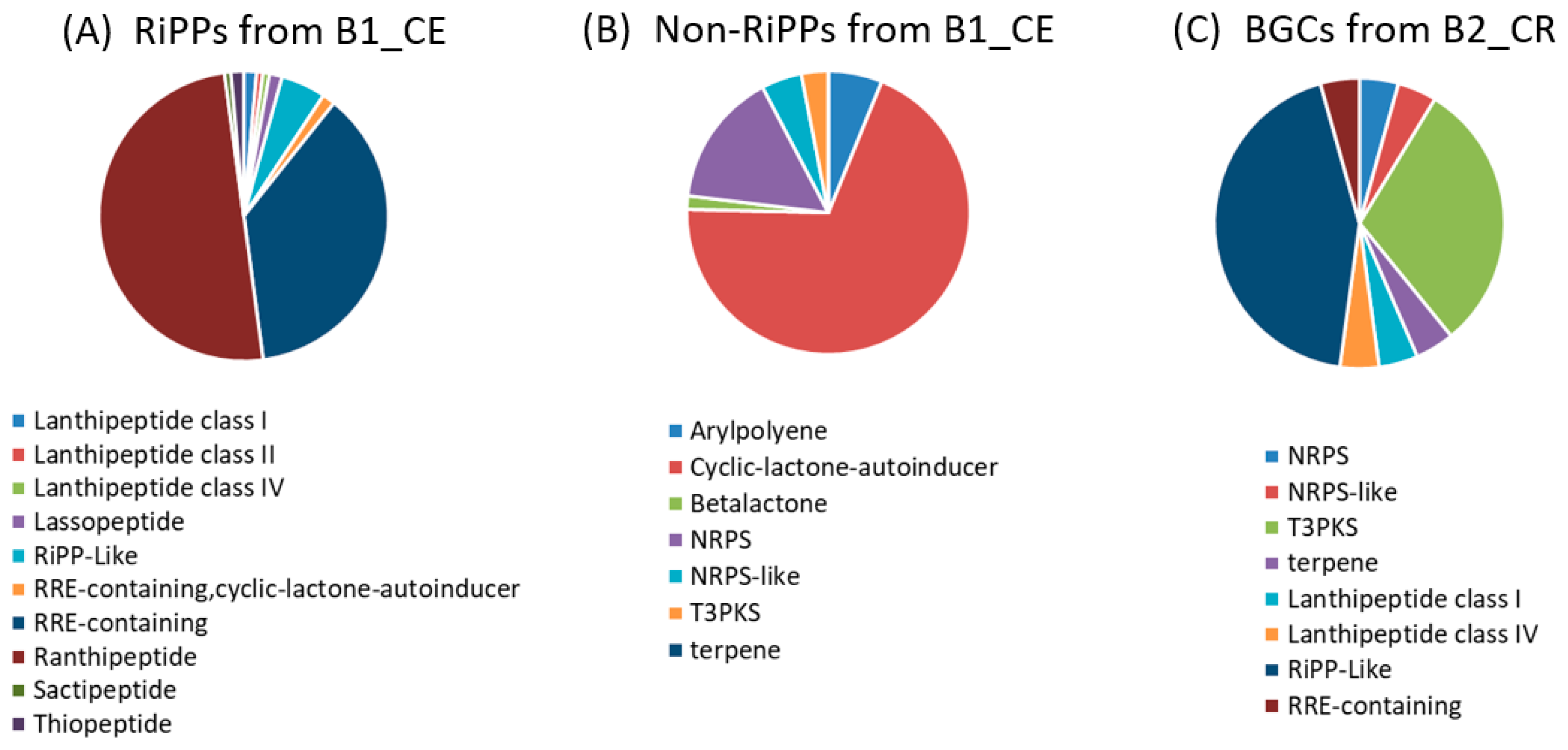
3.2. Identification of Biosynthetic Gene Clusters
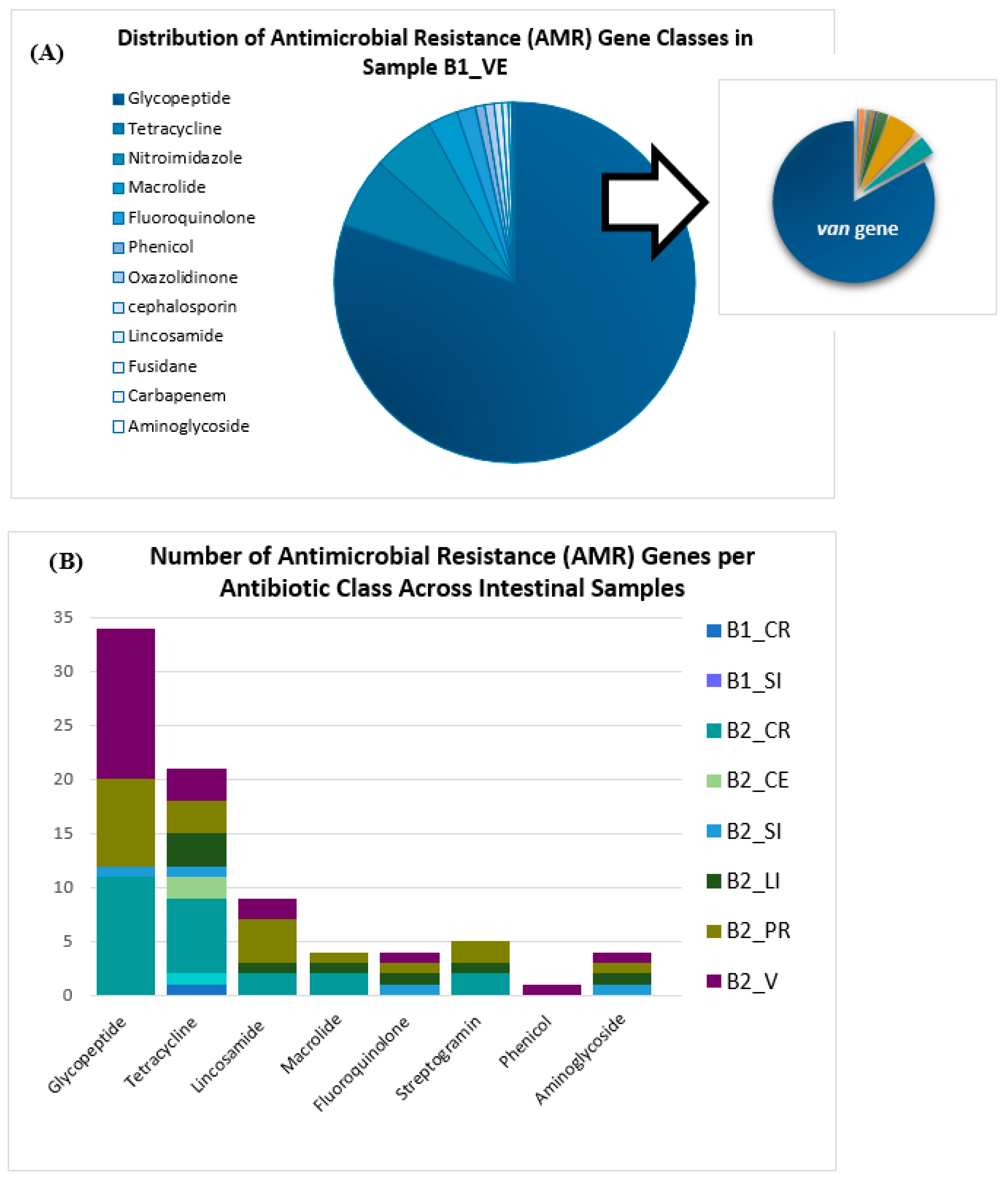
3.3. Antimicrobial Resistance Genes Profile
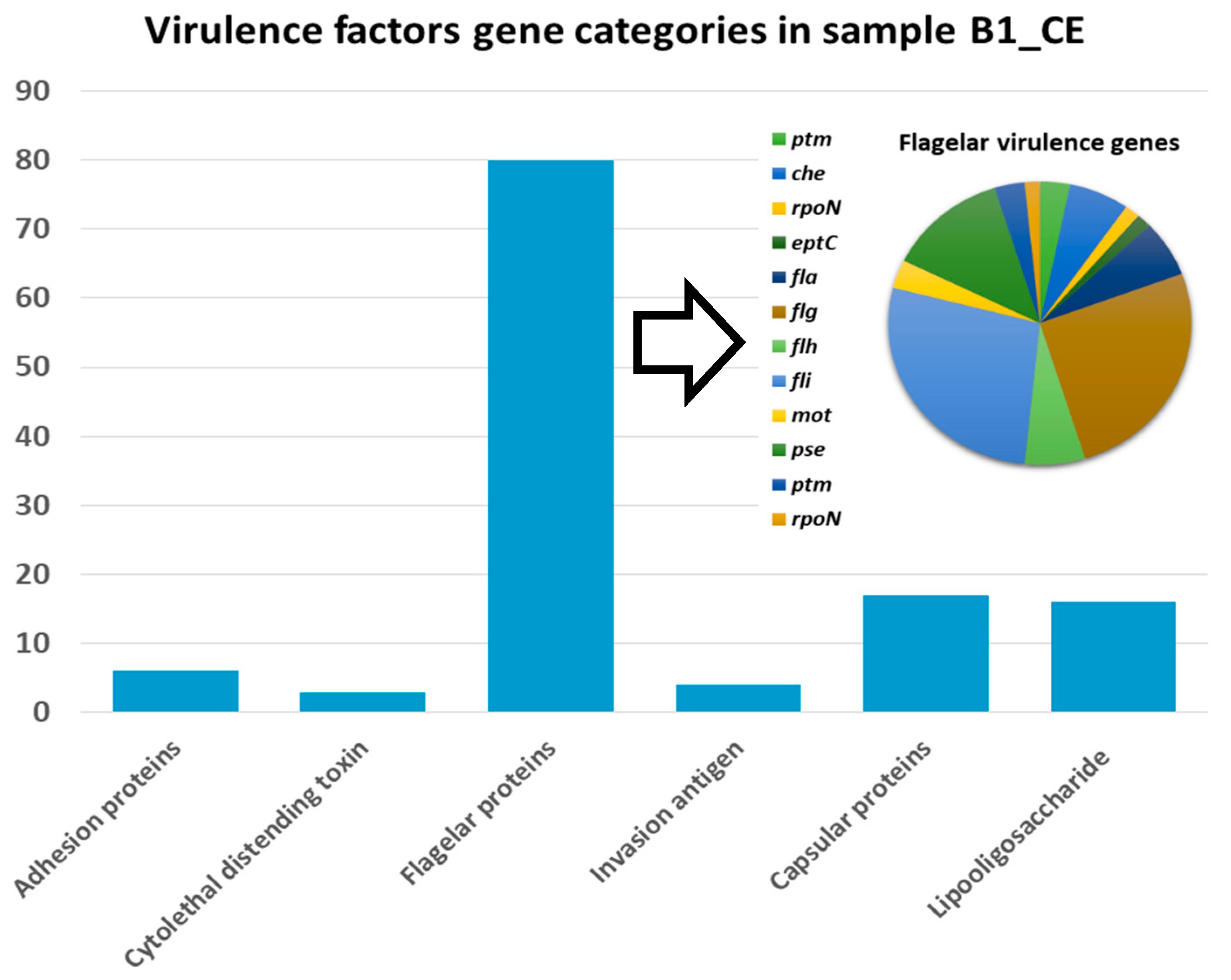
3.4. Identification of Virulence Factor Genes (VFGs)
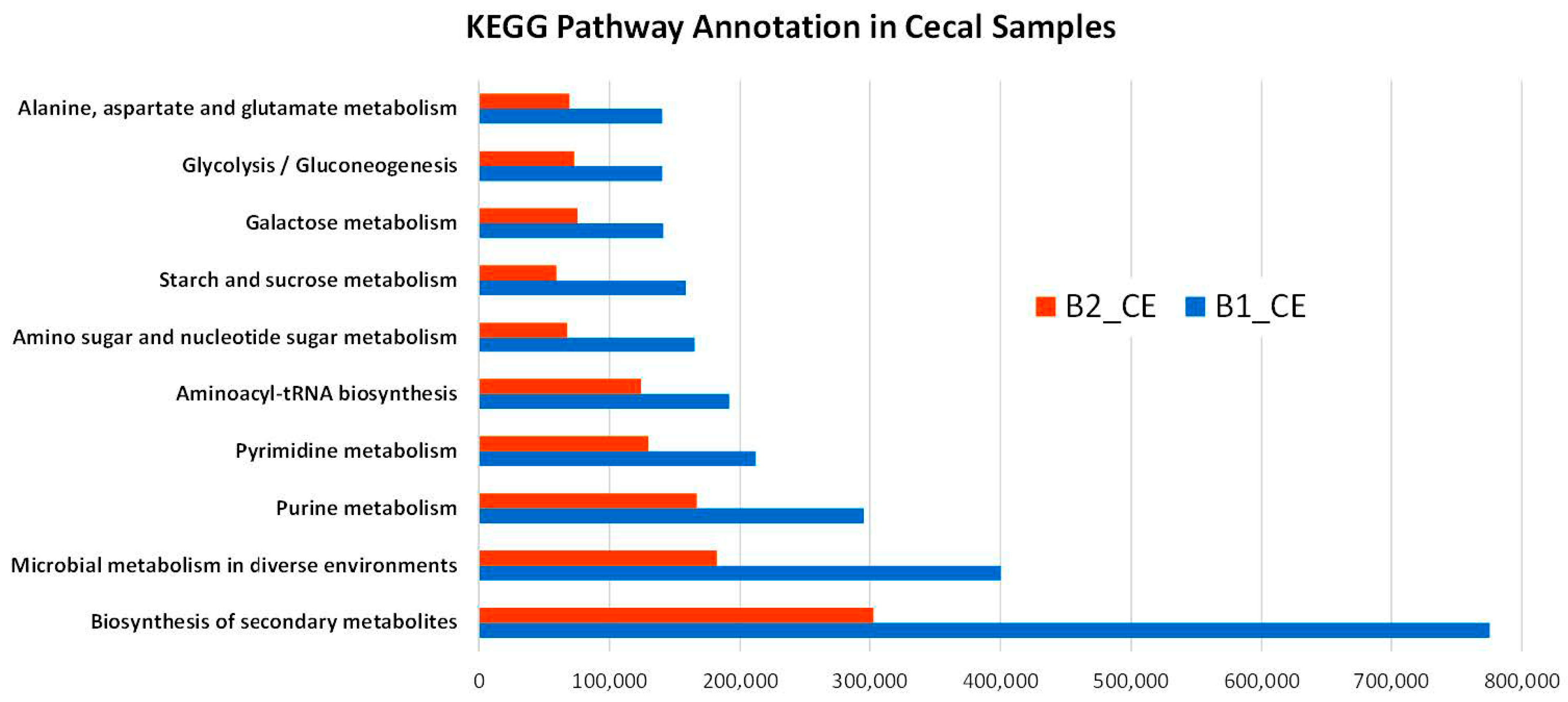
3.5. Functional Annotation of Metabolomic Compounds
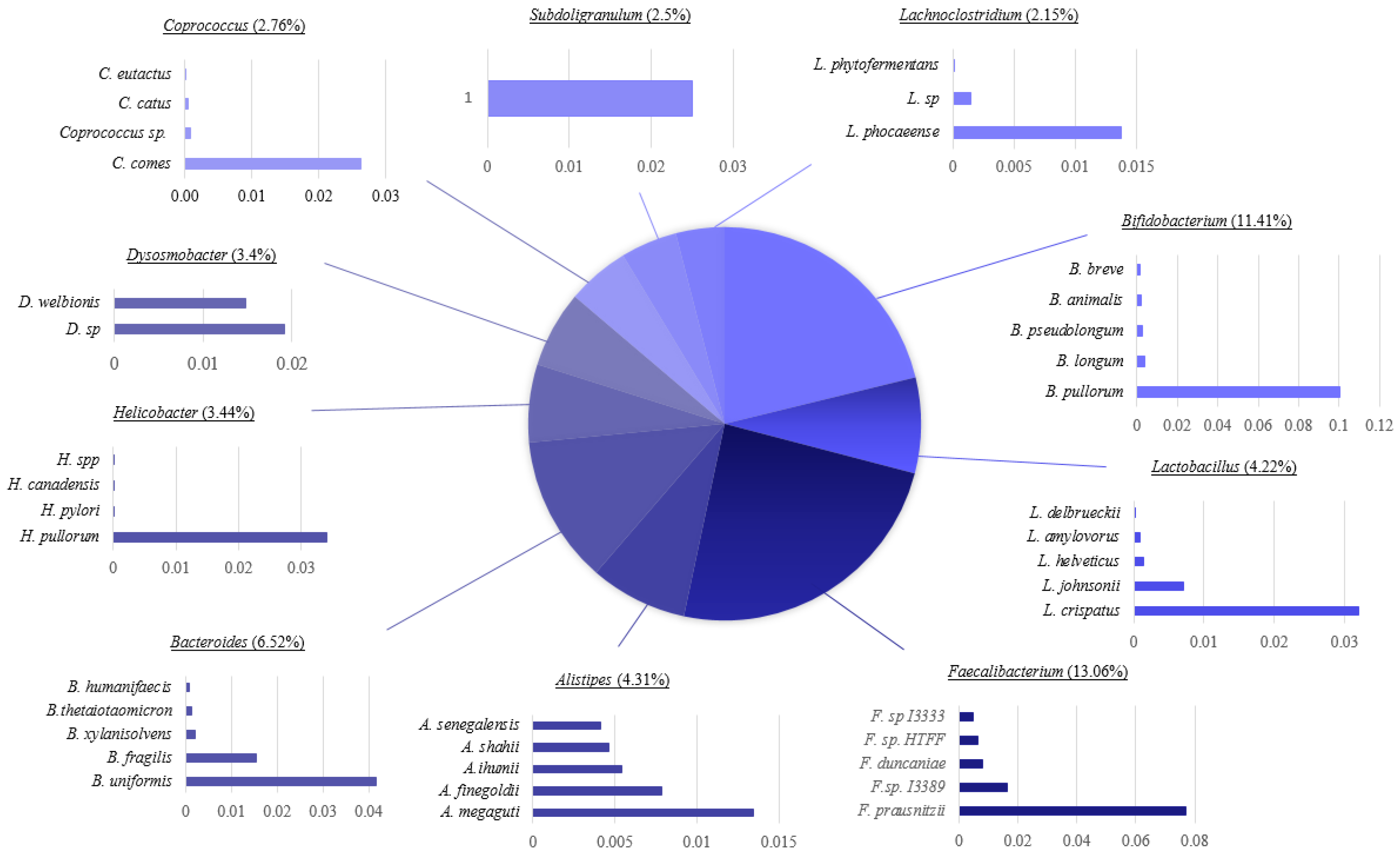
3.6. Microbial Taxonomic Profiling of Intestinal Microbiomes Across Sample Sites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- BD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- Mak, P.H.; Rehman, M.A.; Kiarie, E.G.; Topp, E.; Diarra, M.S. Production systems and important antimicrobial resistant-pathogenic bacteria in poultry: A review. J. Anim. Sci. Biotechnol. 2022, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Mughini-Gras, L.; Pasqualin, D.; Tarakdjian, J.; Santini, A.; Cunial, G.; Tonellato, F.; Schiavon, E.; Martino, G.D. Short-term and long-term effects of antimicrobial use on antimicrobial resistance in broiler and turkey farms. Avian Pathol. 2022, 51, 120–128. [Google Scholar] [CrossRef]
- Schmerold, I.; van Geijlswijk, I.; Gehring, R. European regulations on the use of antibiotics in veterinary medicine. Eur. J. Pharm. Sci. 2023, 189, 106473. [Google Scholar] [CrossRef]
- Cervantes, H.M. Antibiotic-free poultry production: Is it sustainable? J. Appl. Poult. Res. 2015, 24, 91–97. [Google Scholar] [CrossRef]
- Page, E.T.; Short, G.; Sneeringer, S.; Bowman, M. The Market for Chicken Raisedw Antibiotics, 2012–17; No. 264; U.S. Department of Agriculture, Economic Research Service: Washington, DC, USA, 2019.
- Rostagno, M.H. No antibiotic ever (NAE) versus conventional broiler production: It’s complicated. Proc. Ark. Nutr. Conf. 2023, 2023, 4. [Google Scholar]
- Markowiak, P.; Śliżewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Shehata, A.A.; Yalçın, S.; Latorre, J.D.; Basiouni, S.; Attia, Y.A.; Abd El-Wahab, A.; Visscher, C.; El-Seedi, H.R.; Huber, C.; Hafez, H.M.; et al. Probiotics, prebiotics, and phytogenic substances for optimizing gut health in poultry. Microorganisms 2022, 10, 395. [Google Scholar] [CrossRef]
- Ravikumar, R.; Chan, J.; Prabakaran, M. Vaccines against major poultry viral diseases: Strategies to improve the breadth and protective efficacy. Viruses 2022, 14, 1195. [Google Scholar] [CrossRef]
- Abd-El Wahab, A.; Basiouni, S.; El-Seedi, H.R.; Ahmed, M.F.E.; Bielke, L.R.; Hargis, B.; Tellez-Isaias, G.; Eisenreich, W.; Lehnherr, H.; Kittler, S.; et al. An overview of the use of bacteriophages in the poultry industry: Successes, challenges, and possibilities for overcoming breakdowns. Front. Microbiol. 2023, 14, 1136638. [Google Scholar] [CrossRef] [PubMed]
- Szott, V.; Reichelt, B.; Friese, A.; Roesler, U. A complex competitive exclusion culture reduces Campylobacter jejuni colonization in broiler chickens at slaughter age in vivo. Vet. Sci. 2022, 9, 181. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Haas, B.; Gottschalk, M.; Grenier, D. Antimicrobial potential of bacteriocins in poultry and swine production. Vet. Res. 2017, 48, 22. [Google Scholar] [CrossRef]
- Rodrigues, G.; Souza Santos, L.; Franco, O.L. Antimicrobial peptides controlling resistant bacteria in animal production. Front. Microbiol. 2022, 13, 874153. [Google Scholar] [CrossRef]
- Mamjoud, A.; Zirah, S.; Biron, E.; Fliss, O.; Fliss, I. In vitro insights into bacteriocin-mediated modulation of chicken cecal microbiota. Int. J. Mol. Sci. 2025, 26, 755. [Google Scholar] [CrossRef]
- Sugrue, I.; Ross, R.P.; Hill, C. Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. 2024, 22, 556–571. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef]
- Ongpipattanakul, C.; Desormeaux, E.K.; DiCaprio, A.; van der Donk, W.A.; Mitchell, D.A.; Nair, S.K. Mechanism of action of ribosomally synthesized and post-translationally modified peptides. Chem. Rev. 2022, 122, 14722–14814. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cantos, M.V.; Garcia-Morena, D.; Yi, Y.; Liang, L.; Gómez-Vázquez, E.; Kuipers, O.P. Bioinformatic mining for RiPP biosynthetic gene clusters in bacteroidales reveals possible new subfamily architectures and novel natural products. Front. Microbiol. 2023, 14, 1219272. [Google Scholar] [CrossRef]
- Savitskaya, A.; Masso-Silva, J.; Haddaoui, I.; Enany, S. Exploring the arsenal of antimicrobial peptides: Mechanisms, diversity, and applications. Biochimie 2023, 214, 216–227. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Ben Hamida, J.; Fliss, I. BACTIBASE: A new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualization. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Lv, L.J.; Li, S.H.; Wen, J.Y.; Wang, G.Y.; Li, H.; He, T.W.; Lv, Q.B.; Xiao, M.C.; Duan, H.L.; Chen, M.C.; et al. Deep metagenomic characterization of gut microbial community and function in preeclampsia. Front. Cell. Infect. Microbiol. 2022, 12, 933523. [Google Scholar] [CrossRef] [PubMed]
- Tenorio-Salgado, S.; Castelán-Sánchez, H.G.; Dávila-Ramos, S.; Huerta-Saquero, A.; Rodríguez-Morales, S.; Merino-Pérez, E.; Roa de la Fuente, L.F.; Solis-Pereira, S.E.; Pérez-Rueda, E.; Lizama-Uc, G. Metagenomic analysis and antimicrobial activity of two fermented milk kefir samples. Microbiologyopen 2021, 10, 1183. [Google Scholar] [CrossRef] [PubMed]
- Eltokhy, M.A.; Saad, B.T.; Eltayeb, W.N.; Alshahrani, M.Y.; Radwan, S.M.R.; Aboshanab, K.M.; Ashour, M.S.E. Metagenomic nanopore sequencing for exploring the nature of antimicrobial metabolites of Bacillus haynesii. AMB Express 2024, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Aruwa, C.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. Poultry Gut Health—Microbiome functions, environmental impacts, microbiome engineering and advancements in characterization technologies. J. Anim. Sci. Biotechnol. 2021, 12, 119. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Leonard, C. Streammd: Fast low-memory duplicate marking using a bloom filter. Bioinformatics 2023, 39, bta181. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A. Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 2010, 26, 449–457. [Google Scholar] [CrossRef]
- Avalos, M.; Garbeva, P.; Vader, L.; Van Wezel, G.P.; Dickschat, J.S.; Ulanova, D. Biosynthesis, evolution and ecology of microbial terpenoids. Nat. Prod. Rep. 2022, 39, 249–272. [Google Scholar] [CrossRef]
- Chen, N.; Liu, L.; Wang, J.; Mao, D.; Lu, H.; Shishido, T.K.; Zhi, S.; Chen, H.; He, S. Novel gene clusters for secondary metabolite synthesis in mesophotic sponge-associated bacteria. Microb. Biotechnol. 2025, 18, e70107. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Scanes, C.G. The global importance of poultry. Poult. Sci. 2007, 86, 1057–1058. [Google Scholar] [CrossRef]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef]
- Salama, Y.; Chennaoui, M. Understanding microbial contamination in meat and poultry production. J. Res. Agric. Food Sci. 2024, 1, 87–107. [Google Scholar] [CrossRef]
- Ríos-Castro, R.; Cabo, A.; Teira, E.; Cameselle, C.; Gouveia, S.; Payo, P.; Novoa, B.; Figueras, A. High-throughput sequencing as a tool for monitoring prokaryote communities in a wastewater treatment plant. Sci. Total Environ. 2023, 861, 160531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Li, D.; Zhang, W.; Jiang, M.; Chen, X.H.; Dong, M.S. Comparative analysis of the bacterial diversity of chinese fermented sausages using high-throughput sequencing. LWT 2021, 150, 111975. [Google Scholar] [CrossRef]
- Lahlali, R.; Ibrahim, D.S.S.; Belabess, Z.; Kadir Roni, M.Z.; Radouane, N.; Vicente, C.S.L.; Menéndez, E.; Mokrini, F.; Barka, E.A.; Galvão de Melo E Mota, M.; et al. High-throughput molecular technologies for unraveling the mystery of soil microbial community: Challenges and future prospects. Heliyon 2021, 7, e08142. [Google Scholar] [CrossRef] [PubMed]
- Stamilla, A.; Ruiz-Ruiz, S.; Artacho, A.; Pons, J.; Messina, A.; Lucia Randazzo, C.; Caggia, C.; Lanza, M.; Moya, A. Analysis of the microbial intestinal tract in broiler chickens during the rearing period. Biology 2021, 10, 942. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef]
- Gong, J.; Forster, R.J.; Yu, H.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M.; Chen, S. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 2002, 41, 171–179. [Google Scholar] [CrossRef]
- Mead, G.C. Microbes of the avian cecum: Types present and substrates utilized. J. Exp. Zool. 1989, 3 (Suppl. S3), 48–54. [Google Scholar] [CrossRef]
- Mohd Shaufi, M.A.; Sieo, C.C.; Chong, C.W.; Gan, H.M.; Ho, Y.W. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015, 7, 4. [Google Scholar] [CrossRef]
- Cao, L.; Do, T.; Link, A.J. Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs). J. Ind. Microbiol. Biotechnol. 2021, 48, kuab00. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Upton, M.; Cotter, P.D. Editorial: Bacteriocins and other ribosomally synthesized and post-translationally modified peptides (RiPPs) as alternatives to antibiotics. Front. Microbiol. 2021, 12, 3. [Google Scholar] [CrossRef]
- Vagstad, A.L. Engineering ribosomally synthesized and posttranslationally modified peptides as new antibiotics. Curr. Opin. Biotechnol. 2023, 80, 102891. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; Ji, X.; Zhao, R.; Li, G.; Gu, Y.; Shi, A.; Jiang, W.; Zhang, Q. The SCIFF-derived ranthipeptides participate in quorum sensing in solventogenic clostridia. Biotechnol. J. 2020, 15, e2000136. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kang, D.Y.; Choi, M.H.; Hong, J.S.; Kim, H.S.; Kim, Y.R.; Kim, Y.A.; Uh, Y.; Shin, K.S.; Shin, J.H.; et al. Fitness costs of Tn1546-type transposons harboring the vanA operon by plasmid type and structural diversity in Enterococcus faecium. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 62. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Chen, F.-J.; Huang, I.-W.; Wu, H.-C.; Kuo, S.-C.; Huang, T.-W.; Lauderdale, T.-L. Clonal expansion of Tn1546-like transposon-carrying vancomycin-resistant Enterococcus faecium, a nationwide study in Taiwan, 2004–2018. J. Glob. Antimicrob. Resist. 2024, 39, 100–108. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Occurrence, selection and spread of resistance to antimicrobial agents used for growth promotion for food animals in Denmark. APMIS Suppl. 2000, 108, 5–48. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- Teuber, M.; Meile, L.; Schwarz, F. Acquired antibiotic resistance in Lactic Acid Bacteria from food. Antonie Van Leeuwenhoek 1999, 76, 115–137. [Google Scholar] [CrossRef]
- Borgen, K.; Sørum, M.; Wasteson, Y.; Kruse, H. VanA-Type Vancomycin-Resistant Enterococci (VRE) remain prevalent in poultry carcasses 3 years after avoparcin was banned. Int. J. Food. Microbiol. 2001, 64, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Lopez, S.; Singh, A.; Harshey, R.M. Flagellar motility is mutagenic. Proc. Natl. Acad. Sci. USA 2024, 121, e2412541121. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. Targeting motility and chemotaxis as a strategy to combat bacterial pathogens. Microb. Biotechnol. 2023, 16, 2205–2211. [Google Scholar] [CrossRef]
- Haiko, J.; Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef]
- Minamino, T.; Kinoshita, M. Structure, assembly, and function of flagella responsible for bacterial locomotion. EcoSal Plus 2023, 11, eesp-0011-2023. [Google Scholar] [CrossRef]
- Tian, C.; Wang, L.; Liu, M.; Liu, J.; Qiu, M.; Chen, Y. Isolation and identification of chicken-derived Lactic Acid Bacteria: In vitro probiotic properties and antagonistic effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli. Microorganisms 2024, 12, 795. [Google Scholar] [CrossRef] [PubMed]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef]
- Abbas Hilmi, H.T.; Surakka, A.; Apajalahti, J.; Saris, P.E.J. Identification of the most abundant Lactobacillus species in the crop of 1- and 5-week-old broiler chickens. Appl. Environ. Microbiol. 2007, 73, 7867–7873. [Google Scholar] [CrossRef]
- Dec, M.; Nowaczek, A.; Stępień-Pyśniak, D.; Wawrzykowski, J.; Urban-Chmiel, R. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018, 18, 168. [Google Scholar] [CrossRef]
- Segura-Wang, M.; Grabner, N.; Koestelbauer, A.; Klose, V.; Ghanbari, M. Genome-resolved metagenomics of the chicken gut microbiome. Front. Microbiol. 2021, 12, 726923. [Google Scholar] [CrossRef]
- Zou, A.; Nadeau, K.; Xiong, X.; Wang, P.W.; Copeland, J.K.; Lee, J.Y.; Pierre, J.S.; Ty, M.; Taj, B.; Brumell, J.H.; et al. Systematic profiling of the chicken gut microbiome reveals dietary supplementation with antibiotics alters expression of multiple microbial pathways with minimal impact on community structure. Microbiome 2022, 10, 127. [Google Scholar] [CrossRef]
- Meng, J.X.; Li, M.H.; Wang, X.Y.; Li, S.; Zhang, Y.; Ni, H.B.; Ma, H.; Liu, R.; Yan, J.C.; Li, X.M.; et al. Temporal variability in the diversity, function and resistome landscapes in the gut microbiome of broilers. Ecotoxicol. Environ. Saf. 2025, 292, 117976. [Google Scholar] [CrossRef] [PubMed]
- Nene, M.; Kunene, N.W.; Pierneef, R.; Hadebe, K. Profiling the diversity of the village chicken faecal microbiota using 16S rRNA gene and metagenomic sequencing data to reveal patterns of gut microbiome signatures. Front. Microbiol. 2025, 15, 1487595. [Google Scholar] [CrossRef]
- Peña, N.; Lafuente, I.; Sevillano, E.; Feito, J.; Contente, D.; Muñoz-Atienza, E.; Cintas, L.M.; Hernández, P.E.; Borrero, J. Screening and genomic profiling of antimicrobial bacteria sourced from poultry slaughterhouse effluents: Bacteriocin production and safety evaluation. Genes 2024, 15, 1564. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, E.; Lafuente, I.; Peña, N.; Cintas, L.M.; Muñoz-Atienza, E.; Hernández, P.E.; Borrero, J. Isolation, genomics-based and biochemical characterization of bacteriocinogenic bacteria and their bacteriocins, sourced from the gastrointestinal tract of meat-producing pigs. Int. J. Mol. Sci. 2024, 25, 12210. [Google Scholar] [CrossRef] [PubMed]

| Paired_Non.Host_Reads | Contigs | Avg_Len (pb) | N50 | L50 | ||
|---|---|---|---|---|---|---|
| Broiler 1 | CR | 413,410 | 10,846 | 623 | 680 | 2758 |
| PR | 221,628 | 6881 | 576 | 611 | 2779 | |
| VE | 417,875 | 12,591 | 5996 | 631 | 3560 | |
| SI | 13,986 | 3717 | 515 | 528 | 1190 | |
| LI | 405,471 | 9913 | 6293 | 691 | 2371 | |
| CE | 24,098,100 | 500,066 | 9687 | 1516 | 50,213 | |
| Broiler 2 | CR | 5796,495 | 46,447 | 10,205 | 2170 | 3167 |
| PR | 374,194 | 11,295 | 6167 | 663 | 2950 | |
| VE | 1,014,566 | 18,603 | 705 | 734 | 4043 | |
| SI | 657,987 | 19,464 | 7914 | 937 | 3579 | |
| LI | 3,519,182 | 32,411 | 10,936 | 3013 | 1693 | |
| CE | 9,727,800 | 52,119 | 10,353 | 2788 | 2960 | |
| Avg | 511,689.75 | 10,253 | 2141.16 | 1246.833 | 6771.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña, N.; Lafuente, I.; Sevillano, E.; Feito, J.; Allendez, G.; Muñoz-Atienza, E.; Crispie, F.; Cintas, L.M.; Cotter, P.D.; Hernández, P.E.; et al. Exploring the Functional Potential of the Broiler Gut Microbiome Using Shotgun Metagenomics. Genes 2025, 16, 946. https://doi.org/10.3390/genes16080946
Peña N, Lafuente I, Sevillano E, Feito J, Allendez G, Muñoz-Atienza E, Crispie F, Cintas LM, Cotter PD, Hernández PE, et al. Exploring the Functional Potential of the Broiler Gut Microbiome Using Shotgun Metagenomics. Genes. 2025; 16(8):946. https://doi.org/10.3390/genes16080946
Chicago/Turabian StylePeña, Nuria, Irene Lafuente, Ester Sevillano, Javier Feito, Gastón Allendez, Estefanía Muñoz-Atienza, Fiona Crispie, Luis M. Cintas, Paul D. Cotter, Pablo E. Hernández, and et al. 2025. "Exploring the Functional Potential of the Broiler Gut Microbiome Using Shotgun Metagenomics" Genes 16, no. 8: 946. https://doi.org/10.3390/genes16080946
APA StylePeña, N., Lafuente, I., Sevillano, E., Feito, J., Allendez, G., Muñoz-Atienza, E., Crispie, F., Cintas, L. M., Cotter, P. D., Hernández, P. E., & Borrero, J. (2025). Exploring the Functional Potential of the Broiler Gut Microbiome Using Shotgun Metagenomics. Genes, 16(8), 946. https://doi.org/10.3390/genes16080946









