Abstract
Background/Objectives: The influence of single-nucleotide polymorphisms (SNPs) on hepatocellular carcinoma (HCC) in terms of etiological factors remains to be explored. This study evaluated the distribution of PNPLA3 rs738409, TM6SF2 rs58542926, and HSD17B13 rs6834314 and overall survival of HCC patients with metabolic dysfunction-associated steatotic liver disease (MASLD-HCC) and viral-related HCC (VIRAL-HCC). Methods: This study included 564 patients with HCC: 254 with MASLD-HCC and 310 with VIRAL-HCC. The SNPs were determined by real-time PCR using TaqMan assays. Results: The mean ages of patients with MASLD-HCC and VIRAL-HCC were 68.4 vs. 60.9 years (p < 0.001), with a significant difference between groups. The prevalence of PNPLA3 GG genotype in MASLD-HCC was significantly higher in MASLD-HCC than in VIRAL-HCC (24.0% vs. 15.5%, OR = 1.86, 95% CI = 1.14–3.05, p = 0.009). Similarly, the prevalence of TM6SF2 TT genotype in MASLD-HCC and VIRAL-HCC was 7.1% vs. 2.6% (OR = 3.39, 95% CI = 1.36–9.21, p = 0.003), while HSD17B13 GG genotype in the corresponding groups was 7.1% vs. 12.6% (OR = 0.53, 95% CI = 0.27–1.01, p = 0.039). The overall median survival of MASLD-HCC was significantly shorter than that of the VIRAL-HCC group (42 vs. 66 months, p = 0.035). In Cox regression hazard analysis, HSD17B13 GG genotype was significantly associated with a lower mortality rate in MASLD-HCC (HR = 0.38, 95% CI = 0.18–0.81, p = 0.011). In contrast, PNPLA3 and TM6SF2 were not associated with overall survival in patients with MASLD-HCC or VIRAL-HCC. Conclusions: Our data demonstrated that the prevalence of the SNPs significantly differed between MASLD-HCC and VIRAL-HCC. The HSD176B13 GG genotype was also associated with a survival benefit in Thai patients with MASLD-HCC. Thus, assessing the HSD176B13 genotype might be beneficial in risk stratification and potential therapeutic inhibition of HSD17B13 among patients with MASLD-HCC.
Keywords:
hepatocellular carcinoma; steatotic liver disease; polymorphisms; PNPLA3; TM6SF2; HSD17B13 1. Introduction
Hepatocellular carcinoma (HCC) represents the most aggressive and common liver cancer, representing approximately 90% of all cases on a global scale [1]. Notably, the HCC incidence ratio of men to women is approximately 3:1, with a 5-year survival rate of 20%, as patients have been diagnosed at advanced stages when curative treatment is not possible [2]. HCC commonly develops under the backdrop of chronic liver disease, cirrhosis with diverse etiologies: Hepatitis B Virus (HBV), Hepatitis C Virus (HCV), excessive alcohol consumption, genetic risks, metabolic dysfunction-associated steatotic liver disease (MASLD) [3]. Accordingly, the distribution of underlying etiological factors alters depending on the epidemiological patterns and underlying cirrhosis [2]. Although chronic HBV infection is the key risk factor, accounting for over 50% of HCC cases [2], HBV vaccination has remarkably reduced the prevalence of infection and the incidence of HCC globally [4]. Additionally, the accessibility of direct-acting antiviral agents (DAAs) decreases the worldwide burden of HCC-related incidence and mortality [5]. Apart from the viral infection, non-infectious risk factors leading to HCC occurrence have alarmingly increased, characterized by abnormal metabolic functions [6].
MASLD, previously known as non-alcoholic fatty liver disease (NAFLD), is diagnosed by the existence of hepatic steatosis (≥5%), complemented by at least one cardiometabolic disorder [7]. MASLD is a multi-system disorder connected to metabolic syndrome, including type 2 diabetes mellitus, insulin resistance, obesity, dyslipidemia, and hypertension [7]. The progression of HCC in MASLD is complicated, originating from fat deposition in hepatocytes and associated lipotoxicity, fibrogenesis, hepatocyte regeneration, and genetic mutations, which lead to an increased tendency of HCC development [7]. The overall global prevalence of MASLD-HCC is 30% [8], with a regional rating of 33% in southeast-Asian Countries, including Thailand [9]. Interestingly, the corresponding rates for MASLD-HCC in females become almost parallel to the male approximations [10]. Even though cirrhosis leads to a 10-fold increase in HCC development, it is worth noting that approximately 20% of MASLD-HCC cases arise at an early stage of HCC, regardless of cirrhosis [11].
Several recent studies have indicated the role of single-nucleotide polymorphisms (SNPs) in the development and progression of MASLD [12,13]. Among them, the patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene displays an essential role in lipid metabolism by regulating the rate-limiting step in triglyceride hydrolysis [14]. Indeed, the PNPLA3 rs738409 polymorphism represents one of the strongest and most consistent SNPs related to increased risk of MASLD progression, which potentially could lead to HCC development [15]. The transmembrane 6 superfamily member 2 (TM6SF2) gene, functioning in lipid metabolism, is also related to an increased risk of MASLD progression [16]. The minor allele of the TM6SF2 rs58542926 variant could lead to severe hepatic steatosis progression in terms of impaired lipid metabolism, and its association is independent of PNPLA3 rs738409’s effect [17]. An additional SNP associated with MASLD is the variant in the Hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13) gene, which plays a key role in retinoid metabolism through its enzymatic activity and catalyzing retinol oxidation to retinaldehyde [18]. Interestingly, the loss-of-function HSD17B13 variant has been recognized as a hepatoprotective factor of MASLD, and this HSD17B13 rs6834314 G allele was independently correlated with decreasing incidence of liver-related consequences [19]. Although these SNPs are involved in the pathogenesis of MASLD, it remains uncertain whether they are related to the development and prognosis of HCC, considering different underlying liver diseases. Therefore, it would be necessary to explore the impact of the SNPs on cancer development in patients with related HCC (MASLD-HCC) and virus-related HCC (VIRAL-HCC).
In this study, we aimed to investigate the potential impact of these genetic variants on cancer development among patients, specifically concerning the etiological factors of HCC. In addition, we determined whether these SNPs influenced the overall survival of individuals with MASLD-HCC compared to those with VIRAL-HCC.
2. Materials and Methods
2.1. Study Population
This study comprised 564 HCC participants, recruited after a radiologic- and/or pathologic-confirmed diagnosis of HCC from King Chulalongkorn Memorial Hospital (Bangkok, Thailand) between 2014 and 2024. Among them, 254 participants were MASLD-HCC, and 310 patients were VIRAL-HCC [228 with chronic HBV infection (HBV-HCC), and 82 with chronic HCV infection (HCC-HCV)]. Regarding the MASLD-HCC group, the diagnosis was based on evidence of the presence of liver steatosis alongside a clinical phenotype of metabolic dysfunction without seropositivity for HBsAg and anti-HCV. Additionally, 180 healthy individuals without any liver disease were included as the control group (“Healthy controls”). Blood samples that had been collected were separated, and then DNA was extracted and stored at −80 °C. This research had received the approval of the Institutional Review Board (IRB No. 0585-67) and was conducted following the Declaration of Helsinki. All the participants had provided written informed consent, and all the methods were carried out according to the standard guidelines.
2.2. Diagnosis and Follow-Up of HCC
According to the recommendation of the American Association for the Study of Liver Disease (AASLD), the diagnosis of HCC was conducted using standard imaging investigations and/or histology. Diagnosing criteria of HCC were based on specific lesions with hyperattenuation at the arterial phase and hypoattenuation at the portal phase in dynamic Computed Tomography (CT) scans or Magnetic Resonance Imaging (MRI) [20]. If typical imaging features were inconclusive, liver biopsy or fine needle aspiration was further investigated. Patients’ demographic data, such as age, gender, BMI, and clinical information, including tumor size, extrahepatic metastasis, ascites, and laboratory information, were recorded. Cirrhosis was defined using the Child–Pugh score, while HCC staging was categorized based on the Barcelona Clinic Liver Cancer staging system (BCLC) [21]. After being diagnosed with HCC, the patients were treated with appropriate modalities, including surgical resection, liver transplantation, local ablative treatment, systemic therapies, and supportive care. These treatments depended on various factors, including HCC stages and the patient’s overall health. The follow-up period was performed at the diagnostic date of HCC, until either the date of death or the complete registration date, between September 2014 and January 2025.
2.3. DNA Preparation and SNP Genotyping
Genomic DNA was isolated from 100 μL of peripheral blood mononuclear cells (PBMCs) utilizing the phenol-chloroform-isoamyl alcohol technique. DNA quality was assessed by a spectrophotometer (NanoDrop 2000c, Thermo Scientific, Wilmington, DE, USA). The three polymorphisms, PNPLA3, TM6SF2, and HSD17B13, were analyzed using TaqMan real-time PCR protocols. The reaction mixture contained 4 µL of 2.5× master mix (5 PRIME, QIAGEN Beverly Inc., Beverly, MA, USA), 0.25 µL of 40× primer and probe mixture from the TaqMan SNP Genotyping Assay (assay ID: C_7241_10, Applied Biosystems, Waltham, MA, USA), 50–100 ng of genomic DNA, and nuclease-free water, reaching a total volume of 10 µL. Following this, real-time PCR was carried out on a QuantStudio3 Real-time PCR system (Applied Biosystems, USA). The process involved a denaturation step at 95 °C for 10 min, followed by 50 amplification cycles comprising denaturation at 92 °C for 10 s and annealing/extension at 60 °C for 1 min. Fluorescent signals (FAM and VIC) were recorded at each cycle’s end. Each experiment included positive and negative controls to interpret data accuracy. The allelic discrimination plot was evaluated using the QuantStudioTM 3 Real-Time PCR System (Thermo Fisher Scientific, USA).
2.4. Statistical Analysis
Statistical analysis was performed using the STATA software (version 18.0) for Mac (College Station, TX, USA, Stata Corp. 2023). A comparison between groups was analyzed using the Chi-square or Fisher’s exact test for the categorical variables, which were presented as frequencies and percentages. In addition, the Student’s t-test or Mann–Whitney U test was applied to analyze continuous variables, with mean ± standard deviation (SD). The Hardy–Weinberg equilibrium (HWE) was determined using Pearson’s Chi-Square to evaluate the distribution of allele and genotype frequencies. The odds ratio (OR) with a 95% confidence interval (CI) between the two study groups was documented. A logistic regression analysis was conducted to assess factors associated with MASLD-HCC. Moreover, the overall survival of patients with HCC was assessed using the Kaplan–Meier and the log-rank test. Subsequently, Cox Regression was applied to evaluate independent associations between genetic variables, clinical risk factors, and mortality following the HCC diagnosis. The hazard ratio was presented with a forest plot using GraphPad Prism (GraphPad Software, Version 9.3.1, Boston, MA, USA). The p-value < 0.05 was regarded as statistically significant.
3. Results
3.1. Patient Characteristics
Baseline characteristics, including the study population’s demographic, clinical, and laboratory features, categorized into healthy controls, the MASLD-HCC, and VIRAL-HCC groups, are summarized in Table 1. Patients with MASLD-HCC were significantly older and had higher body mass index (BMI) than the VIRAL-HCC group. Additionally, the prevalence of diabetes mellitus and hypertension was significantly higher in the MASLD-HCC group than in the VIRAL-HCC group. Compared to the VIRAL-HCC group, MASLD-HCC patients exhibited lower mean hemoglobin, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels, but higher platelet counts. Regarding tumor characteristics, patients with MASLD-HCC had more advanced HCC at initial presentation than the other group, in terms of tumor size, BCLC stages, and extrahepatic metastasis. Nevertheless, the proportion of cirrhosis was higher in the VIRAL-HCC group compared to the MASLD-HCC group. Of note, alpha-fetoprotein (AFP) levels were higher in the VIRAL-HCC group than in the MASLD-HCC group.

Table 1.
Baseline characteristics of healthy controls and patients with MASLD-HCC and VIRAL-HCC.
3.2. Distribution of SNPs in Each Studied Group
The genotyping assays for the three SNPs—PNPLA3 rs738409, TM6SF2 rs58542926, and HSD17B13 rs6834314—were conducted on samples from healthy controls and all HCC patients. The genotype frequencies of each SNP across the entire cohort showed no deviations from the Hardy–Weinberg equilibrium (p > 0.05), as described in Table S1. The genotype and allele frequencies of SNPs in healthy controls and the two study groups are demonstrated in Table 2. The frequencies of the PNPLA3 GG and CG + GG genotypes were significantly lower in healthy controls compared to the MASLD-HCC group (OR 0.36; 95% CI 0.19–0.66; p < 0.001, and OR 0.59; 95% CI 0.39–0.91; p = 0.009, respectively), but their frequencies did not significantly differ from the VIRAL-HCC group. Similarly, the frequencies of TM6SF2 CT, TT and CT + TT genotypes were lower in healthy controls vs. the MASLD-HCC (OR 0.49; 95% CI 0.30–0.80; p = 0.003, OR 0.06; 95% CI 0.001–0.4; p < 0.001, and OR 0.41; 95% CI 0.25–0.66; p < 0.001, respectively). Additionally, the HSD17B13 AG, GG, and AG + GG genotypes were higher in healthy controls vs. the MASLD-HCC group (OR 1.70; 95% CI 1.11–2.62; p = 0.01, OR 2.52; 95% CI 1.20–5.31; p = 0.006, and OR 1.82; 95% CI 1.21–2.75; p = 0.002, respectively). The frequencies of HSD17B13 AG and AG+GG genotypes were also higher in healthy controls vs. the VIRAL-HCC group (OR 1.71; 95% CI 1.13–2.60; p = 0.008, and OR 1.62; 95% CI 1.09–2.41; p = 0.01, respectively).

Table 2.
Distribution of genotype and allele frequencies of the SNPs in healthy controls, patients with MASLD-HCC, and VIRAL-HCC.
The frequencies of the PNPLA3 GG genotype were significantly higher in the MASLD-HCC group, compared to VIRAL-HCC (OR 1.86; 95% CI 1.14–3.05; p = 0.009). Correspondingly, the frequencies of TM6SF2 CT, TT and CT+ TT genotypes were higher in the MASLD-HCC, compared with the VIRAL-HCC group (OR 1.88; 95% CI 1.24–2.83; p = 0.002, OR 3.39; 95% CI 1.36–9.21; p = 0.003, and OR 2.05; 95% CI 1.39–3.02; p <0.001 respectively). Furthermore, the minor allele frequency of the TM6SF2 T allele in the MASLD-HCC group was significantly higher than in the VIRAL-HCC group (OR 2.32; 95% CI 1.03–5.42; p = 0.027). In contrast, the HSD17B13 GG genotype among MASLD-HCC patients was significantly lower than that of the VIRAL-HCC group (OR 0.53; 95% CI 0.27–0.97; p = 0.039), while the minor allele frequency of the HSD17B13 G allele was comparable between the two study groups.
3.3. Association of Factors with the Overall Survival of Patients with HCC
The mean and median follow-up times per participant were 1.8 and 1 year, respectively. The overall median survival time (MST) of HCC patients was 51 months, with MASLD-HCC significantly shorter than the VIRAL-HCC group (42 vs. 66 months, p = 0.035), as shown in Table 3. The overall survival of patients with HCC was significantly associated with HSD17B13 rs6834314, underlying etiologies, tumor size, and the presence of extrahepatic metastasis. However, there was no association with PNPLA3 rs738409, TM6SF2 rs58542926, age, sex, diabetes, hypertension, cirrhosis, Child–Pugh score, or BCLC stage.

Table 3.
Factors associated with the overall survival of patients with HCC.
The prognostic association of the studied SNPs was also investigated using the Kaplan–Meier method (Figure 1). For PNPLA3 rs738409, the overall survival of HCC patients carrying the CC + CG and GG genotypes was not significantly different (p = 0.489 by log-rank test) (Figure 1A). Similarly, for the prognostic role of TM6SF2 rs58542926, the overall survival of patients carrying the CC + CT and TT genotypes was similar in all HCC cases (p = 0.744) (Figure 1B). In contrast, the overall survival of patients with HCC concerning HSD17B13 rs6834314 differed significantly between patients carrying the AA + AG and GG genotypes (p = 0.008) (Figure 1C). Among the MASLD-HCC group, the HSD17B13 GG genotype was associated with the overall survival, while the PNPLA3 rs738409 and TM6SF2 rs58542926 did not show any association. Regarding the VIRAL-HCC group, none of the studied SNPs showed a significant correlation with overall survival (Figure 2, Figures S1 and S2).
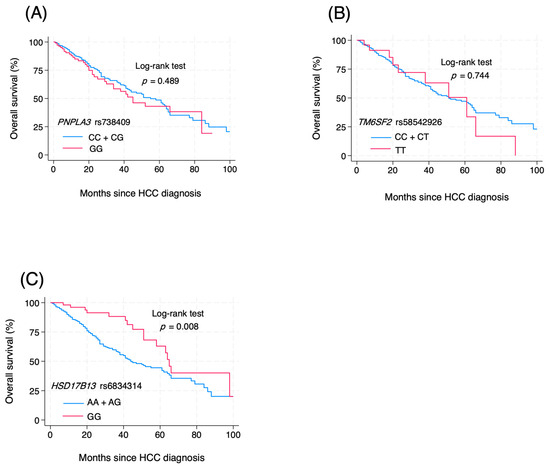
Figure 1.
Effect of genetic polymorphisms on the prognosis of HCC (A) PNPLA3 rs738409 CC + CG vs. GG; (B) TM6SF2 rs58542926 CC+CT vs. TT; (C) HSD17B13 rs6834314 AA + AG vs. GG.
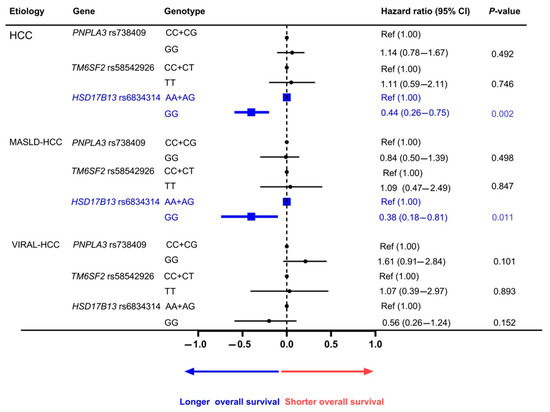
Figure 2.
Adjusted correlation between genetic polymorphisms and overall survival of HCC.
4. Discussion
HCC represents the most prevalent form of primary liver cancer, comprising around 90%. The development of HCC is associated with various risk factors, which include chronic viral infections, environmental influences, genetic predispositions, and metabolic syndrome. Recent studies have indicated that MASLD is becoming increasingly common and is a growing concern. MASLD is a multi-system condition associated with several risk factors, including the influence of genetic variations that play a crucial role in the progression of advanced disease. This study aimed to evaluate the impact of the genetic polymorphisms PNPLA3 rs738409, TM6SF2 rs58542926, and HSD7B13 rs6834314 on susceptibility to hepatocellular carcinoma (HCC) in a cohort of HCC patients with two different etiologies: MASLD-HCC and VIRAL-HCC, as well as to determine whether these genetic variations correlated with the prognosis of HCC. We selected these polymorphisms for analysis in this study due to their strong association with the development and progression of MASLD in Asian populations [13].
Our data demonstrated that the prevalence of the SNPs differed between MASLD-HCC and VIRAL-HCC. Specifically, PNPLA3 GG and TM6SF2 TT genotypes had greater distribution in patients with MASLD-HCC compared to the VIRAL-HCC group. In contrast, the HSD17B13 GG genotype was less prevalent in the MASLD-HCC than the VIRAL-HCC group. Our results also indicated that overall survival following the diagnosis of HCC was associated with HSD17B13 rs6834314 genotype, particularly among patients with MASLD-HCC, as individuals with the HSD7B13 GG genotype seemed to exhibit a survival advantage compared to those with the other genotypes. Conversely, no significant evidence linked the genetic susceptibility variants in PNPLA3 and TM6SF2 to the overall survival of patients with MASLD-HCC and VIRAL-HCC.
Advancements in genomic research have revolutionized the approaches for identifying the genetic factors related to MASLD development. Recent studies have recognized that genome-wide and exome-wide association studies have identified associations between specific genetic variants, particularly SNPs, and the multi-factorial process of liver carcinogenesis [22]. HSD17B13, a member of the 17-beta hydroxysteroid dehydrogenase family, is highly expressed in the lipid droplets of hepatocytes and plays a key role in retinoid metabolism, converting retinol to retinaldehyde through its retinol dehydrogenase activity [23]. An intergenic variant, rs6834314 HSD17B13, triggers the substitution of adenine (A) with guanine (G), resulting in both major and minor alleles, and leading to a loss of function. Although the pathogenic role of HSD17B13 genetic variants in MASLD has not yet been completely understood, several reports have demonstrated their relation to the clinical spectrum of MASLD.
For instance, a recent Japanese study revealed that the HSD17B13 rs6834314 AG + GG genotypes were related to enhanced steatosis but displayed a reduced effect of PNPLA3 on advanced fibrosis [24]. A study of the Caucasian population with biopsy-proven MASLD indicated that the G minor allele of HSD17B13 rs6834314 was potentially related to steatosis development, and it might decrease liver inflammation and ballooning [25]. Additionally, a long-term follow-up of multi-ethnic Asian people with MASLD demonstrated that the HSD17B13 rs6834314 SNP was linked to decreased steatotic liver disease and lower hepatic complications [19]. A recent report demonstrated that HCC patients harboring the HSD17B13 rs72613567 TA variant displayed a survival benefit after diagnosing HCC [26], which aligned well with our report. Together, these data indicate that HSD17B13 polymorphism could be a protective factor for MASLD progression by mitigating liver-related complications. The molecular mechanism by which HSD17B13 variants are responsible for the hepatoprotective effects of HCC remains to be elucidated. Previous data, including histological evidence, have reported that loss-of-function variants may reduce retinol dehydrogenase activity, leading to a decrease in inflammatory and fibrogenic processes [23,25]. In this context, the potential application of inhibiting HSD17B13 as a therapeutic approach for HCC is currently being investigated, which might be valuable in treating this aggressive cancer [27].
Notably, we did not demonstrate a correlation between the PNPLA3 rs738409 and TM6SF2 rs58542926 polymorphisms and overall survival in our HCC cohort. PNPLA3 is a transmembrane protein expressed principally in the liver and displays hydrolase activity against triglycerides [28]. Thus, the loss-of-function PNPLA3 rs738409 variant is associated with increased intrahepatic triglyceride deposition, leading to an inflammatory process, progressive liver disease, and the development of HCC [29]. A meta-analysis of Western populations has shown that this polymorphism is linked to an augmented risk of HCC in patients with MASLD and ALD [30,31]. Additionally, data in Asian patients have indicated that this variant is associated with advanced fibrosis and HCC in patients with NAFLD [32,33]. In a recent meta-analysis, a global pattern of PNPLA3 GG has been revealed, indicating its association with an increased risk of liver-related complications and worse clinical outcomes among patients with MASLD [34]. For instance, some studies demonstrated that the PNPLA3 rs738409 variant might determine susceptibility to HCC development and poor prognosis after diagnosis [35,36]. In contrast, our study did not support these findings, as the SNP exhibited only the susceptibility of MASLD-HCC occurrence but had a neutral effect on overall survival. The explanation for this discrepancy was unclear, but it might suggest that the differences in ethnicities or clinical characteristics in each cohort could impact the diverse associations among reports.
TM6SF2, predominantly expressed in the liver and kidney, has been shown to play a key role in modulating lipid metabolism, particularly in the liver. TM6SF2 is mainly involved in the secretion of intrahepatic triglycerides, thereby modulating the intracellular lipid content [37]. The genetic variant of TM6SF2 rs58542926, which leads to reduced protein expression, is linked to higher intrahepatic triglyceride accumulation [38]. Of note, persons carrying the variant have exhibited an increased risk for MASLD development but a decreasing cardiovascular disease risk [39]. Like the PNPLA3 variant, TM6SF2 rs58542926 contributes to the progressive liver disease in patients with MASLD, initiating from simple steatosis to progressive fibrosis and cirrhosis [40]. According to a meta-analysis, the minor allele T of TM6SF2 rs58542926 is associated with a higher risk of severe steatosis and fibrosis in children and adults [41]. Furthermore, a European cohort also indicated that the TM6SF2 variant was significantly associated with liver-related complications in advanced liver disease [42]. The meta-analysis findings also suggest a significant association of TM6SF2 polymorphism with HCC risk, particularly in patients with steatotic liver disease [43]. However, the available data concerning the role of this polymorphism in HCC prognosis are lacking and need further investigations.
This study had some limitations, primarily due to its case–control design, which comprised only Thai patients and may not be applicable to other ethnic populations. Moreover, this study did not analyze other genetic variants associated with MASLD, such as MBOAT7 rs641738 and GCKR rs1260326. The explanations were based on a relatively low MBOAT7 rs641738 T allele frequency in Asian people, and the role of the variant GCKR rs1260326 regarding MASLD development might diverge across different Asian populations [44,45]. Despite these limitations, our data suggested that the prevalence of the polymorphisms PNPLA3 rs738409, TM6SF2 rs58542926, and HSD17B13 rs6834314 significantly differed between MASLD-HCC and VIRAL-HCC. Additionally, the HSD176B13 GG genotype was associated with a survival benefit in Thai patients with MASLD-HCC after being diagnosed with the cancer. Our findings aligned with previous data indicating the consistency of the hepatoprotective effect of the HSD17B13 polymorphism across ethnic groups. Indeed, several oligonucleotide-based agents targeting HSD17B13 are currently being developed in clinical trials, with promising outcomes [46]. Thus, genetic determination of the variant could lead to targeting HSD17B13 or modulating its activity, which may offer personalized therapeutic options for managing patients with MASLD-HCC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16070808/s1, Figure S1: Effect of genetic polymorphisms on the prognosis of MASLD-HCC; Figure S2: Effect of genetic polymorphisms on the prognosis of VIRAL-HCC; Table S1: Hardy–Weinberg Equilibrium (HWE); Table S2: Factors associated with the survival of patients with MASLD-HCC; Table S3. Factors associated with the survival of patients with VIRAL-HCC.
Author Contributions
Conceptualization, T.C.Z.A., N.C. and P.T.; methodology, T.C.Z.A.; formal analysis, P.T.; investigation, T.C.Z.A., N.C. and P.T.; resources, M.C., K.P., N.C. and P.T.; data curation, P.T.; writing—original draft preparation, T.C.Z.A.; writing—review and editing B.B. and P.T.; visualization, T.C.Z.A., B.B., N.C. and P.T.; project administration, T.C.Z.A.; supervision, N.C. and P.T.; funding acquisition, P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation (PMU-B, grant number B36G660010) and the Center of Excellence in Hepatitis and Liver Cancer, Chulalongkorn University.
Institutional Review Board Statement
The study was reviewed and approved by the Ethics Committee and the Institutional Review Board (IRB) at the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB 0585-67), and the study was conducted following the Declaration of Helsinki.
Informed Consent Statement
All individuals involved in this study have already provided written informed consent.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank all members of the Center of Excellence in Hepatitis and Liver Cancer, Chulalongkorn University, for their technical assistance.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Abbreviations
| HCC | Hepatocellular carcinoma |
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| NAFLD | Non-alcoholic fatty liver disease |
| BCLC | Barcelona Clinic Liver Cancer |
| PNPLA3 | Patatin-like phospholipase domain-containing protein 3 |
| TM6SF2 | Transmembrane 6 superfamily member 2 |
| HSD17B13 | Hydroxysteroid 17-β dehydrogenase 13 |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| GWAS | Genome-wide association study |
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
| OR | Odds ratio |
| CI | Confidence interval |
| OS | Overall survival |
| MST | Median survival time |
| MAF | Minor allele frequency |
References
- Rumgay, H.; Ferlay, J.; de Martel, C.; Georges, D.; Ibrahim, A.S.; Zheng, R.; Wei, W.; Lemmens, V.; Soerjomataram, I. Global, regional and national burden of primary liver cancer by subtype. Eur. J. Cancer 2022, 161, 108–118. [Google Scholar] [CrossRef]
- Ferrante, N.D.; Pillai, A.; Singal, A.G. Update on the Diagnosis and Treatment of Hepatocellular Carcinoma. Gastroenterol. Hepatol. 2020, 16, 506–516. [Google Scholar]
- Singal, A.G.; Kanwal, F.; Llovet, J.M. Global trends in hepatocellular carcinoma epidemiology: Implications for screening, prevention and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 864–884. [Google Scholar] [CrossRef] [PubMed]
- Al-Busafi, S.A.; Alwassief, A. Global Perspectives on the Hepatitis B Vaccination: Challenges, Achievements, and the Road to Elimination by 2030. Vaccines 2024, 12, 288. [Google Scholar] [CrossRef]
- Zhang, H.; Quadeer, A.A.; McKay, M.R. Direct-acting antiviral resistance of Hepatitis C virus is promoted by epistasis. Nat. Commun. 2023, 14, 7457. [Google Scholar] [CrossRef] [PubMed]
- Phoolchund, A.G.S.; Khakoo, S.I. MASLD and the Development of HCC: Pathogenesis and Therapeutic Challenges. Cancers 2024, 16, 259. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023, 79, 1542–1556. [Google Scholar] [CrossRef]
- Taru, M.-G.; Lupsor-Platon, M. Exploring Opportunities to Enhance the Screening and Surveillance of Hepatocellular Carcinoma in Non-Alcoholic Fatty Liver Disease (NAFLD) through Risk Stratification Algorithms Incorporating Ultrasound Elastography. Cancers 2023, 15, 4097. [Google Scholar] [CrossRef]
- Miao, L.; Targher, G.; Byrne, C.D.; Cao, Y.Y.; Zheng, M.H. Current status and future trends of the global burden of MASLD. Trends Endocrinol. Metab. 2024, 35, 697–707. [Google Scholar] [CrossRef]
- Tan, D.J.H.; Setiawan, V.W.; Ng, C.H.; Lim, W.H.; Muthiah, M.D.; Tan, E.X.; Dan, Y.Y.; Roberts, L.R.; Loomba, R.; Huang, D.Q. Global burden of liver cancer in males and females: Changing etiological basis and the growing contribution of NASH. Hepatology 2023, 77, 1150–1163. [Google Scholar] [CrossRef]
- Kanwal, F.; Kramer, J.R.; Mapakshi, S.; Natarajan, Y.; Chayanupatkul, M.; Richardson, P.A.; Li, L.; Desiderio, R.; Thrift, A.P.; Asch, S.M.; et al. Risk of Hepatocellular Cancer in Patients With Non-Alcoholic Fatty Liver Disease. Gastroenterology 2018, 155, 1828–1837.e1822. [Google Scholar] [CrossRef] [PubMed]
- Seko, Y.; Yamaguchi, K.; Itoh, Y. The genetic backgrounds in nonalcoholic fatty liver disease. Clin. J. Gastroenterol. 2018, 11, 97–102. [Google Scholar] [CrossRef]
- Sookoian, S.; Rotman, Y.; Valenti, L. Genetics of Metabolic Dysfunction-associated Steatotic Liver Disease: The State of the Art Update. Clin. Gastroenterol. Hepatol. 2024, 22, 2177–2187.e2173. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef]
- Liu, Y.L.; Patman, G.L.; Leathart, J.B.S.; Piguet, A.C.; Burt, A.D.; Dufour, J.F.; Day, C.P.; Daly, A.K.; Reeves, H.L.; Anstee, Q.M. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J. Hepatol. 2014, 61, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Mahdessian, H.; Taxiarchis, A.; Popov, S.; Silveira, A.; Franco-Cereceda, A.; Hamsten, A.; Eriksson, P.; van’t Hooft, F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. USA 2014, 111, 8913–8918. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Reeves, H.L.; Burt, A.D.; Tiniakos, D.; McPherson, S.; Leathart, J.B.S.; Allison, M.E.D.; Alexander, G.J.; Piguet, A.-C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309. [Google Scholar] [CrossRef]
- Ma, Y.; Karki, S.; Brown, P.M.; Lin, D.D.; Podszun, M.C.; Zhou, W.; Belyaeva, O.V.; Kedishvili, N.Y.; Rotman, Y. Characterization of essential domains in HSD17B13 for cellular localization and enzymatic activity. J. Lipid Res. 2020, 61, 1400–1409. [Google Scholar] [CrossRef]
- Ting, Y.W.; Kong, A.S.; Zain, S.M.; Chan, W.K.; Tan, H.L.; Mohamed, Z.; Pung, Y.F.; Mohamed, R. Loss-of-function HSD17B13 variants, non-alcoholic steatohepatitis and adverse liver outcomes: Results from a multi-ethnic Asian cohort. Clin. Mol. Hepatol. 2021, 27, 486–498. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M.; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Trepo, E.; Valenti, L. Update on NAFLD genetics: From new variants to the clinic. J. Hepatol. 2020, 72, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Amangurbanova, M.; Huang, D.Q.; Loomba, R. Review article: The role of HSD17B13 on global epidemiology, natural history, pathogenesis and treatment of NAFLD. Aliment. Pharmacol. Ther. 2023, 57, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Seko, Y.; Yamaguchi, K.; Tochiki, N.; Yano, K.; Takahashi, A.; Okishio, S.; Kataoka, S.; Okuda, K.; Umemura, A.; Moriguchi, M.; et al. Attenuated effect of PNPLA3 on hepatic fibrosis by HSD17B13 in Japanese patients with non-alcoholic fatty liver disease. Liver Int. 2020, 40, 1686–1692. [Google Scholar] [CrossRef]
- Ma, Y.; Belyaeva, O.V.; Brown, P.M.; Fujita, K.; Valles, K.; Karki, S.; de Boer, Y.S.; Koh, C.; Chen, Y.; Du, X.; et al. 17-Beta Hydroxysteroid Dehydrogenase 13 Is a Hepatic Retinol Dehydrogenase Associated With Histological Features of Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 1504–1519. [Google Scholar] [CrossRef]
- Innes, H.; Morgan, M.Y.; Hampe, J.; Stickel, F.; Buch, S. The rs72613567:TA polymorphism in HSD17B13 is associated with survival benefit after development of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2023, 58, 623–631. [Google Scholar] [CrossRef]
- Valenti, L.; Casirati, E. Editorial: Unveiling new horizons for liver steatosis genetic variants beyond hepatocellular carcinoma diagnosis—Exploring the potential of HSD17B13 inhibition. Aliment. Pharmacol. Ther. 2023, 58, 727–728. [Google Scholar] [CrossRef]
- Jenkins, C.M.; Mancuso, D.J.; Yan, W.; Sims, H.F.; Gibson, B.; Gross, R.W. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 2004, 279, 48968–48975. [Google Scholar] [CrossRef]
- Tavaglione, F.; Pennisi, G.; Pelusi, S. PNPLA3 I148M and Hepatocellular Carcinoma. Liver Int. 2025, 45, e70051. [Google Scholar] [CrossRef]
- Singal, A.G.; Manjunath, H.; Yopp, A.C.; Beg, M.S.; Marrero, J.A.; Gopal, P.; Waljee, A.K. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: A meta-analysis. Am. J. Gastroenterol. 2014, 109, 325–334. [Google Scholar] [CrossRef]
- Trepo, E.; Nahon, P.; Bontempi, G.; Valenti, L.; Falleti, E.; Nischalke, H.D.; Hamza, S.; Corradini, S.G.; Burza, M.A.; Guyot, E.; et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014, 59, 2170–2177. [Google Scholar] [CrossRef] [PubMed]
- Seko, Y.; Sumida, Y.; Tanaka, S.; Mori, K.; Taketani, H.; Ishiba, H.; Hara, T.; Okajima, A.; Umemura, A.; Nishikawa, T.; et al. Development of hepatocellular carcinoma in Japanese patients with biopsy-proven non-alcoholic fatty liver disease: Association between PNPLA3 genotype and hepatocarcinogenesis/fibrosis progression. Hepatol. Res. 2017, 47, 1083–1092. [Google Scholar] [CrossRef]
- Ueyama, M.; Nishida, N.; Korenaga, M.; Korenaga, K.; Kumagai, E.; Yanai, H.; Adachi, H.; Katsuyama, H.; Moriyama, S.; Hamasaki, H.; et al. The impact of PNPLA3 and JAZF1 on hepatocellular carcinoma in non-viral hepatitis patients with type 2 diabetes mellitus. J. Gastroenterol. 2016, 51, 370–379. [Google Scholar] [CrossRef]
- Souza, M.; Al-Sharif, L.; Diaz, I.; Mantovani, A.; Villela-Nogueira, C.A. Global Epidemiology and Implications of PNPLA3 I148M Variant in Metabolic Dysfunction–Associated Steatotic Liver Disease: A Systematic Review and Meta-analysis. J. Clin. Exp. Hepatol. 2025, 15, 102495. [Google Scholar] [CrossRef]
- Hassan, M.M.; Kaseb, A.; Etzel, C.J.; El-Serag, H.; Spitz, M.R.; Chang, P.; Hale, K.S.; Liu, M.; Rashid, A.; Shama, M.; et al. Genetic variation in the PNPLA3 gene and hepatocellular carcinoma in USA: Risk and prognosis prediction. Mol. Carcinog. 2013, 52 (Suppl. 1), E139–E147. [Google Scholar] [CrossRef]
- Valenti, L.; Motta, B.M.; Soardo, G.; Iavarone, M.; Donati, B.; Sangiovanni, A.; Carnelutti, A.; Dongiovanni, P.; Rametta, R.; Bertelli, C.; et al. PNPLA3 I148M polymorphism, clinical presentation, and survival in patients with hepatocellular carcinoma. PLoS ONE 2013, 8, e75982. [Google Scholar] [CrossRef]
- Luo, F.; Oldoni, F.; Das, A. TM6SF2: A Novel Genetic Player in Nonalcoholic Fatty Liver and Cardiovascular Disease. Hepatol. Commun. 2022, 6, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjaerg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Sookoian, S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology 2015, 62, 1742–1756. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Seth, D.; Day, C.P. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology 2016, 150, 1728–1744.e1727. [Google Scholar] [CrossRef]
- Li, X.-Y.; Liu, Z.; Li, L.; Wang, H.-J.; Wang, H. TM6SF2 rs58542926 is related to hepatic steatosis, fibrosis and serum lipids both in adults and children: A meta-analysis. Front. Endocrinol. 2022, 13, 1026901. [Google Scholar] [CrossRef] [PubMed]
- Balcar, L.; Scheiner, B.; Urheu, M.; Weinberger, P.; Paternostro, R.; Simbrunner, B.; Semmler, G.; Willheim, C.; Pinter, M.; Ferenci, P.; et al. The impact of transmembrane 6 superfamily 2 (TM6SF2) rs58542926 on liver-related events in patients with advanced chronic liver disease. Dig. Liver Dis. 2023, 55, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zhang, J.; Mei, T.T.; Guo, H.Q.; Wei, X.H.; Zhang, W.Y.; Liu, Y.L.; Liang, S.; Fan, Z.P.; Ma, L.X.; et al. Association of TM6SF2 rs58542926 T/C gene polymorphism with hepatocellular carcinoma: A meta-analysis. BMC Cancer 2019, 19, 1128. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.K.; An, J.N.; Joo, S.K.; Kim, D.; Lee, S.; Bae, J.M.; Park, J.H.; Kim, J.H.; Chang, M.S.; Kim, W. Association Between a Polymorphism in MBOAT7 and Chronic Kidney Disease in Patients With Biopsy-Confirmed Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2020, 18, 2837–2839.e2832. [Google Scholar] [CrossRef]
- Sulaiman, S.A.; Dorairaj, V.; Adrus, M.N.H. Genetic Polymorphisms and Diversity in Nonalcoholic Fatty Liver Disease (NAFLD): A Mini Review. Biomedicines 2022, 11, 106. [Google Scholar] [CrossRef]
- Caddeo, A.; Romeo, S. Precision medicine and nucleotide-based therapeutics to treat steatotic liver disease. Clin. Mol. Hepatol. 2025, 31, S76–S93. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).