A Study on the Diagnostic and Prognostic Value of Extrachromosomal Circular DNA in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Samples
2.2. EccDNA Enrichment and Sequencing
2.3. EccDNA Identification and Annotation
2.4. Genomic Features of eccDNA
2.5. Transcriptome Analysis
2.6. Prognostic Analysis
2.7. Construction of eccDNA-Based Diagnostic and Prognostic Prediction Model
3. Results
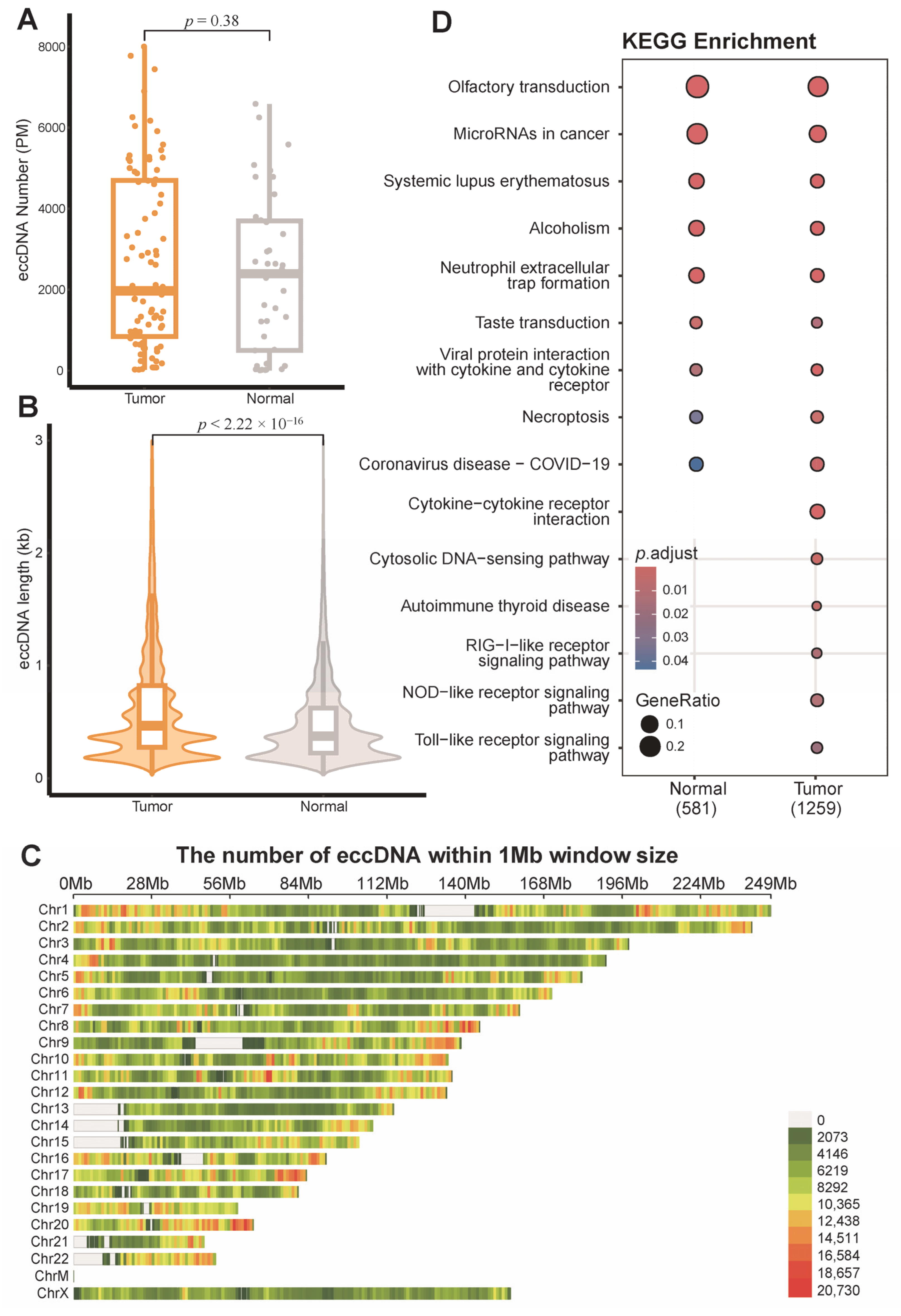
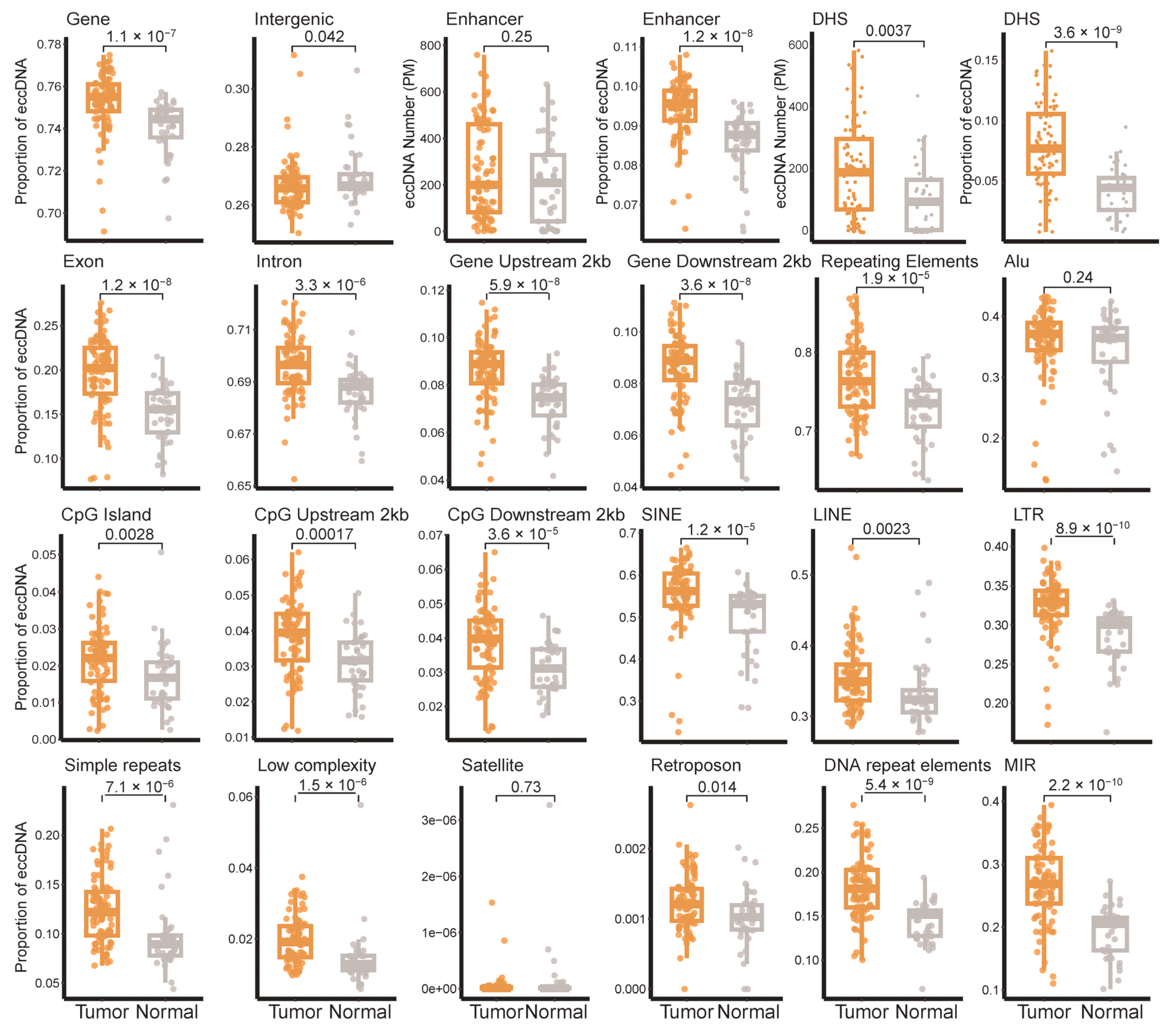
3.1. EccDNA Landscape in Breast Tumor and Adjacent Non-Tumor Tissues
3.2. Distribution of eccDNA Across Breast Cancer Subtypes
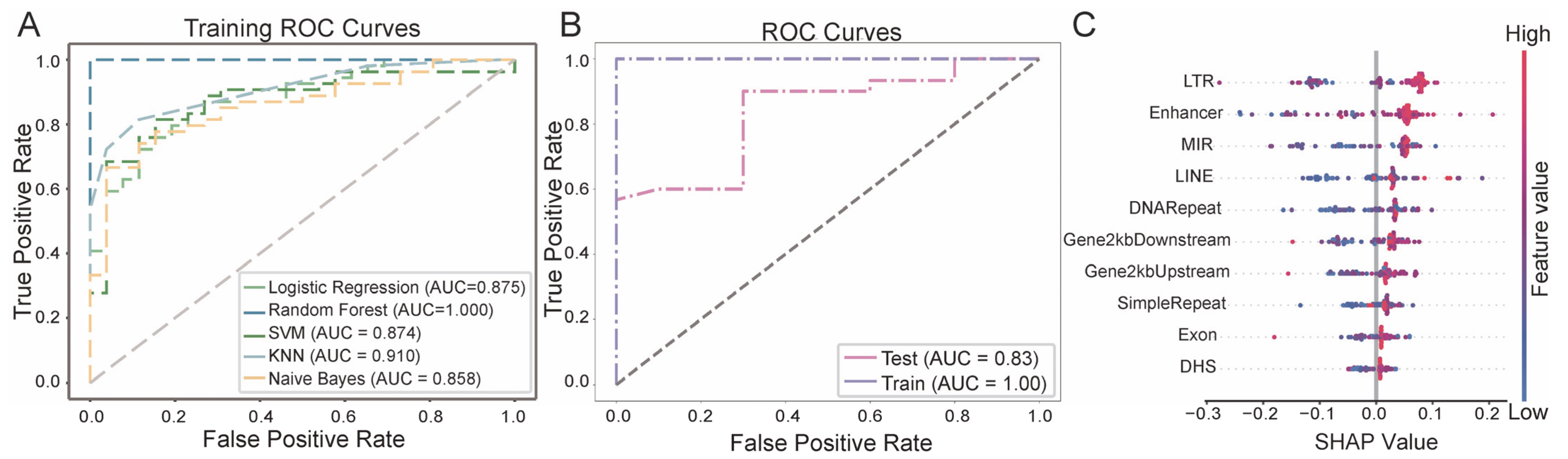
3.3. Breast Cancer Diagnostic Model Based on eccDNA Features
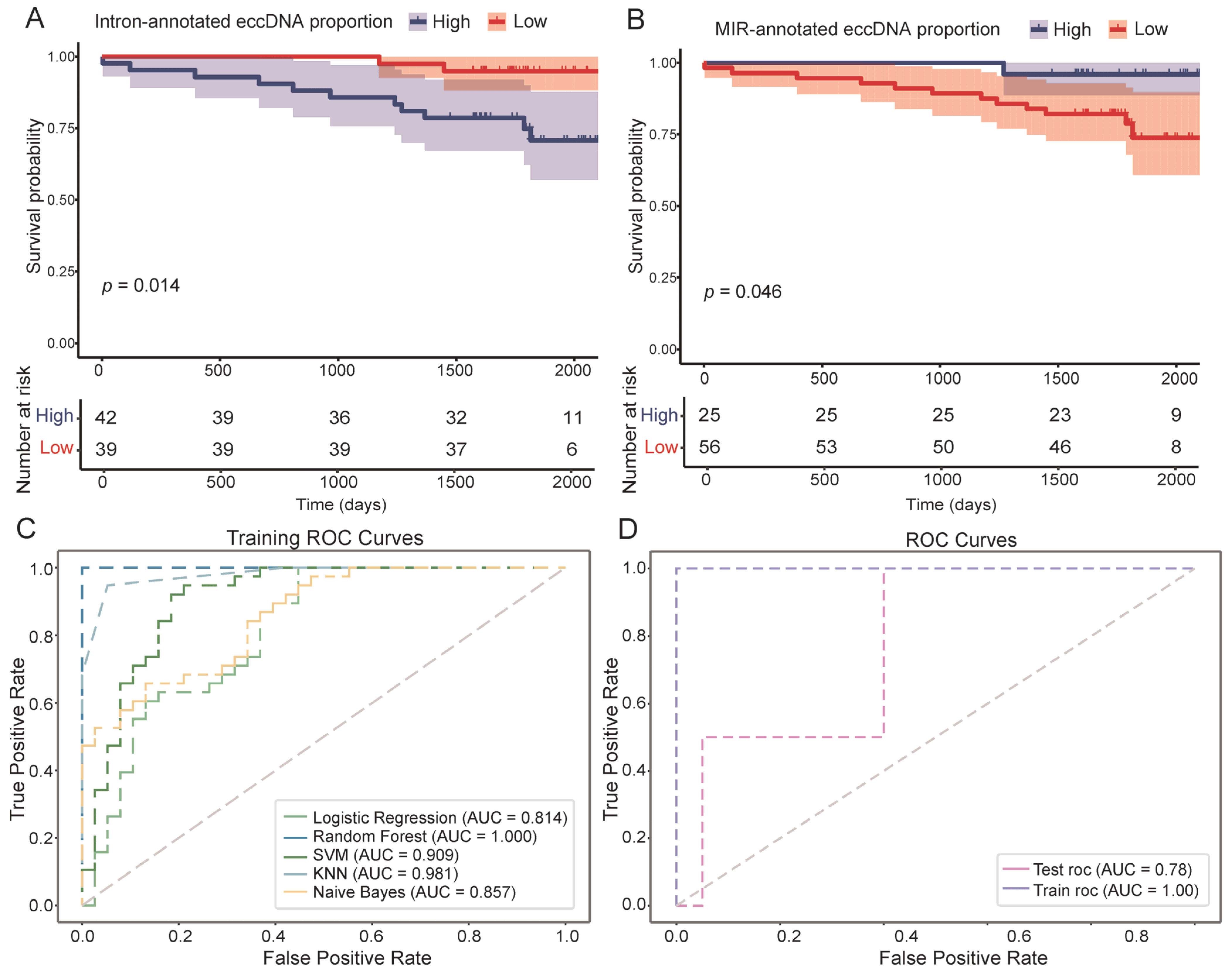
3.4. EccDNA Features as Indicators of DFS in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| eccDNA | Extrachromosomal circular DNA |
| ERBB2/HER2 | Human epidermal growth factor receptor 2 |
| DFS | Disease-free survival |
| LINE | Long interspersed nuclear element |
| SINE | Short interspersed nuclear element |
| MIR | Mammalian-wide interspersed repeat |
| LTR | Long terminal repeat element |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| UTR | Untranslated region |
| DHS | DNase I hypersensitive site |
| ROC | Receiver operating characteristic |
| AUC | Area under the receiver operating characteristic curve |
| SVM | Support vector machine |
| KNN | K-nearest neighbors |
| DEG | Differentially expressed gene |
| ANOVA | Analysis of variance |
| SD | Standard deviation |
| FDR | False discovery rate |
| GO | Gene Ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
References
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Gaubatz, J.W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 1990, 237, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, T.; Kumar, P.; Koseoglu, M.M.; Dutta, A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018, 34, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Needham-VanDevanter, D.R.; Yucel, J.; Windle, B.E.; Wahl, G.M. Amplified human MYC oncogenes localized to replicating submicroscopic circular DNA molecules. Proc. Natl. Acad. Sci. USA 1988, 85, 4804–4808. [Google Scholar] [CrossRef] [PubMed]
- Helmsauer, K.; Valieva, M.E.; Ali, S.; Chamorro González, R.; Schöpflin, R.; Röefzaad, C.; Bei, Y.; Dorado Garcia, H.; Rodriguez-Fos, E.; Puiggròs, M.; et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat. Commun. 2020, 11, 5823. [Google Scholar] [CrossRef]
- Yi, E.; Chamorro González, R.; Henssen, A.G.; Verhaak, R.G.W. Extrachromosomal DNA amplifications in cancer. Nat. Rev. Genet. 2022, 23, 760–771. [Google Scholar] [CrossRef]
- Turner, K.M.; Deshpande, V.; Beyter, D.; Koga, T.; Rusert, J.; Lee, C.; Li, B.; Arden, K.; Ren, B.; Nathanson, D.A.; et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017, 543, 122–125. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Bafna, V.; Mischel, P.S. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat. Rev. Cancer 2019, 19, 283–288. [Google Scholar] [CrossRef]
- Morton, A.R.; Dogan-Artun, N.; Faber, Z.J.; MacLeod, G.; Bartels, C.F.; Piazza, M.S.; Allan, K.C.; Mack, S.C.; Wang, X.; Gimple, R.C.; et al. Functional Enhancers Shape Extrachromosomal Oncogene Amplifications. Cell 2019, 179, 1330–1341.e13. [Google Scholar] [CrossRef]
- deCarvalho, A.C.; Kim, H.; Poisson, L.M.; Winn, M.E.; Mueller, C.; Cherba, D.; Koeman, J.; Seth, S.; Protopopov, A.; Felicella, M.; et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat. Genet. 2018, 50, 708–717. [Google Scholar] [CrossRef]
- Kaufman, R.J.; Brown, P.C.; Schimke, R.T. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc. Natl. Acad. Sci. USA 1979, 76, 5669–5673. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Song, T.; Liu, S.; Liu, Z.; Wang, X.; Li, Y.; Liu, D. Circle-map profiling of extrachromosomal circular DNA as diagnostic biomarkers for lung cancer. Precis. Clin. Med. 2024, 7, pbae006. [Google Scholar] [CrossRef]
- Xu, Z.; He, J.; Han, P.; Dai, P.; Lv, W.; Liu, N.; Liu, L.; Liu, L.; Pan, X.; Xiang, X.; et al. Plasma extrachromosomal circular DNA is a pathophysiological hallmark of short-term intensive insulin therapy for type 2 diabetes. Clin. Transl. Med. 2023, 13, e1437. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Bang, Y.J. HER2-targeted therapies—A role beyond breast cancer. Nat. Rev. Clin. Oncol. 2020, 17, 33–48. [Google Scholar] [CrossRef]
- Moasser, M.M. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007, 26, 6469–6487. [Google Scholar] [CrossRef] [PubMed]
- Vicario, R.; Peg, V.; Morancho, B.; Zacarias-Fluck, M.; Zhang, J.; Martínez-Barriocanal, Á.; Navarro Jiménez, A.; Aura, C.; Burgues, O.; Lluch, A.; et al. Patterns of HER2 Gene Amplification and Response to Anti-HER2 Therapies. PLoS ONE 2015, 10, e0129876. [Google Scholar] [CrossRef]
- Yang, J.; Liu, X.; Huang, Y.; He, L.; Zhang, W.; Ren, J.; Wang, Y.; Wu, J.; Wu, X.; Shan, L.; et al. TRPS1 drives heterochromatic origin refiring and cancer genome evolution. Cell Rep. 2021, 34, 108814. [Google Scholar] [CrossRef]
- Bao, Y.; Sui, X.; Wang, X.; Qu, N.; Xie, Y.; Cong, Y.; Cao, X. Extrachromosomal circular DNA landscape of breast cancer with lymph node metastasis. Int. J. Cancer 2024, 155, 756–765. [Google Scholar] [CrossRef]
- Sheng, Z.; Wang, X.; Zheng, Y.; Duan, W.; Cui, J.; Gu, L.; Gao, X.; Ma, J.; Cui, M.; Luo, H.; et al. Genome-wide characterization of extrachromosomal circular DNA in breast cancer and its potential role in carcinogenesis and cancer progression. Cell Rep. 2024, 43, 114845. [Google Scholar] [CrossRef]
- Ouyang, Y.; Lu, W.; Wang, Y.; Wang, B.; Li, F.; Li, X.; Bai, Y.; Wang, Y. Integrated analysis of mRNA and extrachromosomal circular DNA profiles to identify the potential mRNA biomarkers in breast cancer. Gene 2023, 857, 147174. [Google Scholar] [CrossRef]
- Yang, F.; Su, W.; Chung, O.W.; Tracy, L.; Wang, L.; Ramsden, D.A.; Zhang, Z.Z. Retrotransposons hijack alt-EJ for DNA replication and eccDNA biogenesis. Nature 2023, 620, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yu, J.; Zhu, W.; Hong, X.; Xu, Z.; Mao, S.; Huang, L.; Han, P.; He, C.; Song, C.; et al. Unveiling the mysteries of extrachromosomal circular DNA: From generation to clinical relevance in human cancers and health. Mol. Cancer 2024, 23, 276. [Google Scholar] [CrossRef]
- Møller, H.D.; Mohiyuddin, M.; Prada-Luengo, I.; Sailani, M.R.; Halling, J.F.; Plomgaard, P.; Maretty, L.; Hansen, A.J.; Snyder, M.P.; Pilegaard, H.; et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 2018, 9, 1069. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Taubert, H.; Lange, H.; Kriese, K.; Schmitt, W.D.; Hoffmann, S.; Bartel, F.; Hauptmann, S. Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol. Rep. 2009, 22, 393–400. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jjingo, D.; Conley, A.B.; Wang, J.; Mariño-Ramírez, L.; Lunyak, V.V.; Jordan, I.K. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mob. DNA 2014, 5, 14. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, K.; Jia, X.; Du, S.; Wang, D.; Wang, L.; Qu, H.; Zhu, S.; Wang, Y.; Wang, Z.; et al. A novel extrachromosomal circular DNA related genes signature for overall survival prediction in patients with ovarian cancer. BMC Med. Genom. 2023, 16, 140. [Google Scholar] [CrossRef]
- Li, Z.; Wei, C.; Zhang, Z.; Han, L. ecGBMsub: An integrative stacking ensemble model framework based on eccDNA molecular profiling for improving IDH wild-type glioblastoma molecular subtype classification. Front. Pharmacol. 2024, 15, 1375112. [Google Scholar] [CrossRef]
- Li, Z.; Wang, B.; Liang, H.; Li, Y.; Zhang, Z.; Han, L. A three-stage eccDNA based molecular profiling significantly improves the identification, prognosis assessment and recurrence prediction accuracy in patients with glioma. Cancer Lett. 2023, 574, 216369. [Google Scholar] [CrossRef]
- Lu, W.; Yao, L.; Wang, Y.; Li, F.; Zhou, B.A.-O.; Ming, W.; Jiang, Y.; Liu, X.; Liu, Y.; Sun, X.A.-O.; et al. Characterization of extrachromosomal circular DNA associated with genomic repeat sequences in breast cancer. Int. J. Cancer 2025, 157, 384–397. [Google Scholar] [CrossRef]
- Nelson, J.R. Random-primed, Phi29 DNA polymerase-based whole genome amplification. Curr. Protoc. Mol. Biol. 2014, 105, 15.13.1–15.13.16. [Google Scholar] [CrossRef]
- Johne, R.; Müller, H.; Rector, A.; van Ranst, M.; Stevens, H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Prada-Luengo, I.; Krogh, A.; Maretty, L.; Regenberg, B. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinform. 2019, 20, 663. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Frankish, A.; Carbonell-Sala, S.; Diekhans, M.; Jungreis, I.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Arnan, C.; Barnes, I.; et al. GENCODE: Reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 2023, 51, D942–D949. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zhang, Y.; Huang, Q.; Meng, J.; Ding, R.; Chang, Y.; Xiong, L.; Guo, Z. EnhancerDB: A resource of transcriptional regulation in the context of enhancers. Database 2019, 2019, bay141. [Google Scholar] [CrossRef]
- Tempel, S. Using and understanding RepeatMasker. Methods Mol. Biol. 2012, 859, 29–51. [Google Scholar]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Ming, W.; Zhu, Y.; Bai, Y.; Gu, W.; Li, F.; Hu, Z.; Xia, T.; Dai, Z.; Yu, X.; Li, H.; et al. Radiogenomics analysis reveals the associations of dynamic contrast-enhanced-MRI features with gene expression characteristics, PAM50 subtypes, and prognosis of breast cancer. Front. Oncol. 2022, 12, 943326. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- McHugh, M.L. Multiple comparison analysis testing in ANOVA. Biochem. Med. 2011, 21, 203–209. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Martínez Pérez, J.A.; Pérez Martínez, P.S. [Survival analysis]. SEMERGEN 2023, 49, 101986. [Google Scholar] [CrossRef]
- Fachal, L.; Aschard, H.; Beesley, J.; Barnes, D.R.; Allen, J.; Kar, S.; Pooley, K.A.; Dennis, J.; Michailidou, K.; Turman, C.; et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet. 2020, 52, 56–73. [Google Scholar] [CrossRef]
- Li, M.; Schweiger, M.W.; Ryan, D.J.; Nakano, I.; Carvalho, L.A.; Tannous, B.A. Olfactory receptor 5B21 drives breast cancer metastasis. iScience 2021, 24, 103519. [Google Scholar] [CrossRef]
- Tang, J.; Ahmad, A.; Sarkar, F.H. The role of microRNAs in breast cancer migration, invasion and metastasis. Int. J. Mol. Sci. 2012, 13, 13414–13437. [Google Scholar] [CrossRef]
- Tan, H.; Wang, S.; Huang, F.; Tong, Z. Association between breast cancer and thyroid cancer risk: A two-sample Mendelian randomization study. Front. Endocrinol. 2023, 14, 1138149. [Google Scholar] [CrossRef]
- Nielsen, S.M.; White, M.G.; Hong, S.; Aschebrook-Kilfoy, B.; Kaplan, E.L.; Angelos, P.; Kulkarni, S.A.; Olopade, O.I.; Grogan, R.H. The Breast-Thyroid Cancer Link: A Systematic Review and Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2016, 25, 231–238. [Google Scholar] [CrossRef]
- Bernatsky, S.; Ramsey-Goldman, R.; Urowitz, M.B.; Hanly, J.G.; Gordon, C.; Petri, M.A.; Ginzler, E.M.; Wallace, D.J.; Bae, S.C.; Romero-Diaz, J.; et al. Cancer Risk in a Large Inception Systemic Lupus Erythematosus Cohort: Effects of Demographic Characteristics, Smoking, and Medications. Arthritis Care Res. 2021, 73, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.T.; Minopoli, M.; Del Vecchio, S.; Carriero, M.V. The Emerging Role of Neutrophil Extracellular Traps (NETs) in Tumor Progression and Metastasis. Front. Immunol. 2020, 11, 1749. [Google Scholar] [CrossRef] [PubMed]
- Salman, N.A.; Davies, G.; Majidy, F.; Shakir, F.; Akinrinade, H.; Perumal, D.; Ashrafi, G.H. Association of High Risk Human Papillomavirus and Breast cancer: A UK based Study. Sci. Rep. 2017, 7, 43591. [Google Scholar] [CrossRef]
- Yu, L.; Liu, P. Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduct. Target. Ther. 2021, 6, 170. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, H.; Wang, J.; Chen, J.; Guo, Z.; Liu, Y.; Hua, H. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J. Hematol. Oncol. 2023, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Khademalhosseini, M.; Arababadi, M.K. Toll-like receptor 4 and breast cancer: An updated systematic review. Breast Cancer 2019, 26, 265–271. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, L.; Chen, W.; Chang, Z.; Tu, J.; Qin, Y.; Yao, Y.; Dong, M.; Ding, J.; Li, S.; et al. Microbiota enterotoxigenic Bacteroides fragilis-secreted BFT-1 promotes breast cancer cell stemness and chemoresistance through its functional receptor NOD1. Protein Cell 2024, 15, 419–440. [Google Scholar] [CrossRef]
| Information | Sample Number (81 in Total) | |
|---|---|---|
| Age | 25~68 (Mean: 48.7) | |
| ER status | Positive | 56 |
| Negative | 25 | |
| PR status | Positive | 50 |
| Negative | 31 | |
| HER2 status | Positive | 14 |
| Negative | 67 | |
| Ki-67 status | High | 63 |
| Low | 18 | |
| Immunohistochemical subtype | Luminal-A | 14 |
| Luminal-B | 44 | |
| HER2-enriched | 7 | |
| Triple-negative | 16 | |
| Pathological stage | I | 26 |
| II | 45 | |
| III | 10 | |
| eccDNA Feature | Hazard Ratios (95% Confidence Interval) | p-Value |
|---|---|---|
| Intron-annotated eccDNA proportion | 0.185 (0.0410–0.838) | 0.0285 |
| Repeat-annotated eccDNA proportion | 0.268 (0.0737–0.977) | 0.0459 |
| MIR-annotated eccDNA proportion | 6.17 (0.801–47.6) | 0.0806 |
| DNA repeat-annotated eccDNA proportion | 6.10 (0.790–47.2) | 0.0829 |
| Exon-annotated eccDNA proportion | 6.29 (0.817–48.5) | 0.0775 |
| Gene Upstream-annotated eccDNA proportion | 6.20 (0.805–47.8) | 0.0797 |
| LINE-annotated eccDNA proportion | 7.17 (0.930–55.4) | 0.0587 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Lu, W.; Yao, L.; Bai, Y. A Study on the Diagnostic and Prognostic Value of Extrachromosomal Circular DNA in Breast Cancer. Genes 2025, 16, 802. https://doi.org/10.3390/genes16070802
Li F, Lu W, Yao L, Bai Y. A Study on the Diagnostic and Prognostic Value of Extrachromosomal Circular DNA in Breast Cancer. Genes. 2025; 16(7):802. https://doi.org/10.3390/genes16070802
Chicago/Turabian StyleLi, Fuyu, Wenxiang Lu, Lingsong Yao, and Yunfei Bai. 2025. "A Study on the Diagnostic and Prognostic Value of Extrachromosomal Circular DNA in Breast Cancer" Genes 16, no. 7: 802. https://doi.org/10.3390/genes16070802
APA StyleLi, F., Lu, W., Yao, L., & Bai, Y. (2025). A Study on the Diagnostic and Prognostic Value of Extrachromosomal Circular DNA in Breast Cancer. Genes, 16(7), 802. https://doi.org/10.3390/genes16070802






