Genetic Characterization of Wild Soybean Collected from Zhejiang Province in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. SSR Analysis
2.3. Trait Analysis
2.4. Seed Quality Analysis
3. Results
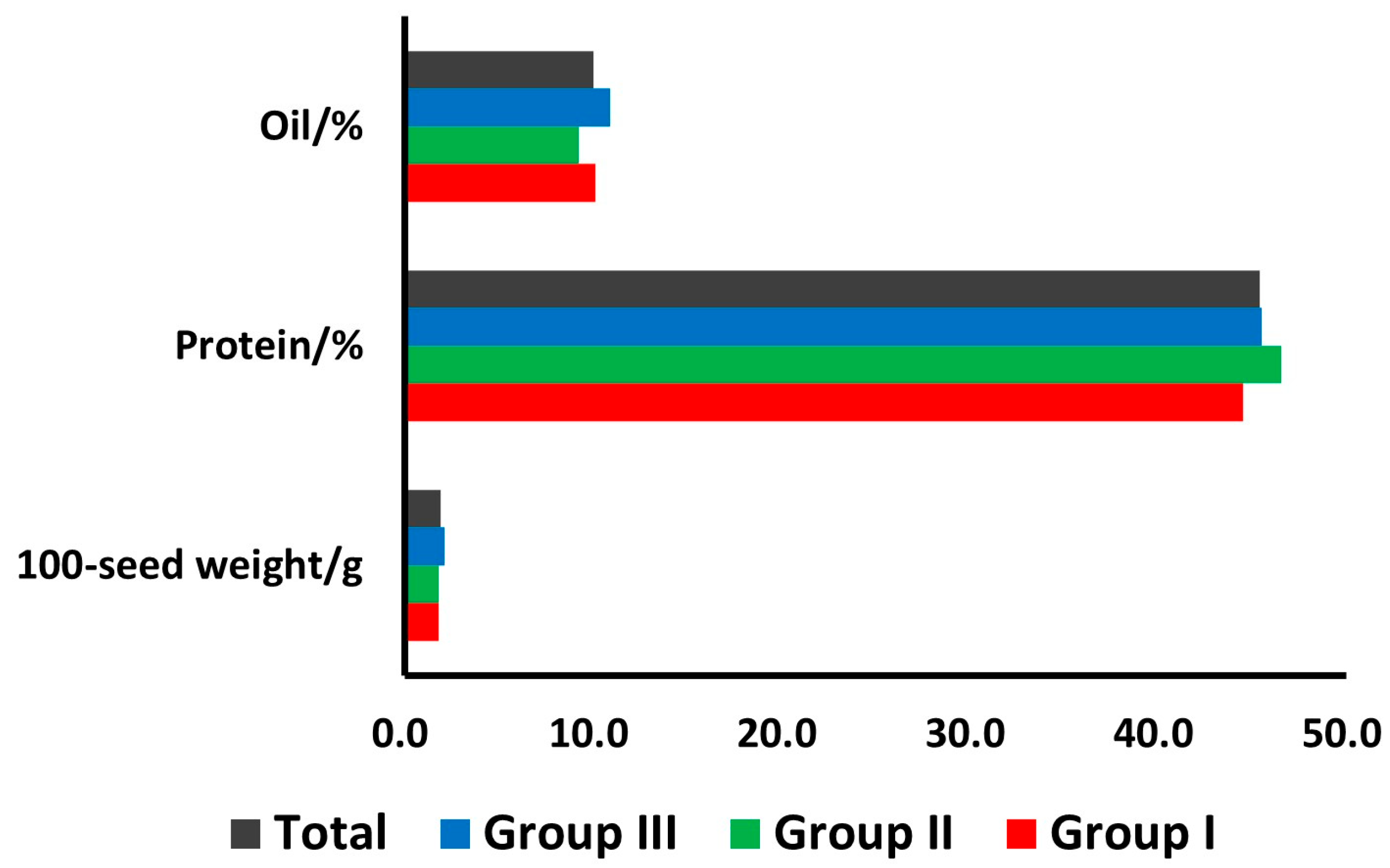
3.1. Seed Characteristics and Quality Traits
3.2. Genetic Variation
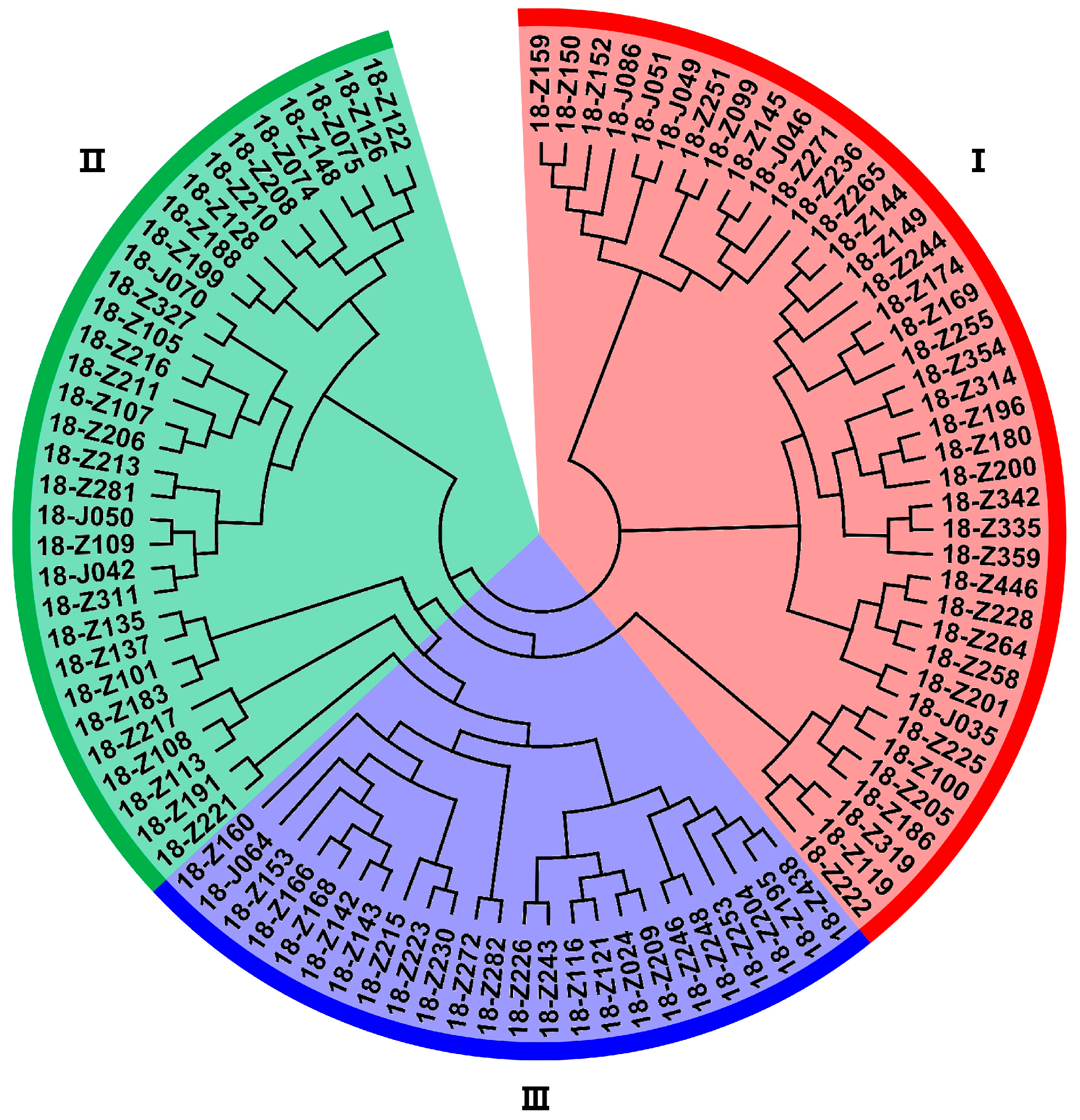
3.3. Clustering Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, Y.; Zhuang, B.; Zhao, H.; He, M. The genetic diversity of annual wild soybeans grown in China. Theor. Appl. Genet. 2001, 103, 98–103. [Google Scholar] [CrossRef]
- Wen, Z.; Ding, Y.; Zhao, T.; Gai, J. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor. Appl. Genet. 2009, 119, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Li, J.; Guo, W.; Liu, X.; Zhang, L.; Dong, Y. Phenotypic traits and diversity of different 100-seed weight accessions of wild soybean (Glycine soja Sieb. & Zucc.) in China. Can. J. Plant Sci. 2017, 97, 449–456. [Google Scholar]
- Zhao, H.; Wang, Y.; Xing, F.; Liu, X.; Yuan, C.; Qi, G.; Guo, J.; Dong, Y. The genetic diversity and geographic differentiation of the wild soybean in Northeast China based on nuclear microsatellite variation. Int. J. Genom. 2018, 2018, 8561458. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, P.; Ding, X.; Wang, Y.; Qi, G.; Yu, J.; Zeng, Y.; Cai, D.; Yang, X.; Yang, J.; et al. The population divergence and genetic basis of local adaptation of wild soybean (Glycine soja) in China. Plants 2023, 12, 4128. [Google Scholar] [CrossRef]
- Yu, X.; Fu, X.; Yang, Q.; Jin, H.; Zhu, L.; Yuan, F. Genetic and phenotypic characterization of soybean landraces collected from the Zhejiang Province in China. Plants 2024, 13, 353. [Google Scholar] [CrossRef]
- Wang, L.; Guan, Y.; Guan, R.; Li, Y.; Ma, Y.; Dong, Z.; Liu, X.; Zhang, H.; Zhang, Y.; Liu, Z.; et al. Establishment of Chinese soybean Glycine max core collections with agronomic traits and SSR markers. Euphytica 2006, 151, 215–223. [Google Scholar] [CrossRef]
- An, W.; Zhao, H.; Dong, Y.; Wang, Y.; Li, Q.; Zhuang, B.; Gong, L.; Liu, B. Genetic diversity in annual wild soybean (Glycine Soja Sieb. Et Zucc.) and cultivated soybean (G. max Merr.) from different latitudes in China. Pak. J. Bot. 2009, 41, 2229–2242. [Google Scholar]
- Sun, B.; Fu, C.; Yang, C.; Ma, Q.; Pan, D.; Nian, H. Genetic diversity of wild soybeans from some regions of Southern China based on SSR and SRAP markers. Am. J. Plant Sci. 2013, 4, 257–268. [Google Scholar] [CrossRef]
- Wang, L.; Lin, F.; Li, L.; Li, W.; Yan, Z.; Luan, W.; Piao, R.; Guan, Y.; Ning, X.; Zhu, L.; et al. Genetic diversity center of cultivated soybean (Glycine max) in China—New insight and evidence for the diversity center of Chinese cultivated soybean. J. Integr. Agric. 2016, 15, 2481–2487. [Google Scholar] [CrossRef]
- Dong, D.; Fu, X.; Yuan, F.; Chen, P.; Zhu, S.; Li, B.; Yang, Q.; Yu, X.; Zhu, D. Genetic diversity and population structure of vegetable soybean (Glycine max (L.) Merr.) in China as revealed by SSR markers. Genet. Resour. Crop Evol. 2014, 61, 173–183. [Google Scholar] [CrossRef]
- Yu, X.; Fu, X.; Yang, Q.; Jin, H.; Zhu, L.; Yuan, F. Genome-wide variation analysis of four vegetable soybean cultivars based on re-sequencing. Plants 2022, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Jia, G.; Zhu, Y.; Grant, D.; Nelson, R.T.; Hwang, E.Y.; Hyten, D.L.; Cregan, P.B. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop. Sci. 2010, 50, 1950–1960. [Google Scholar] [CrossRef]
- Kachare, S.; Tiwari, S.; Tripathi, N.; Thakur, V.V. Assessment of genetic diversity of soybean (Glycine max) genotypes using qualitative traits and microsatellite markers. Agric. Res. 2020, 9, 23–34. [Google Scholar] [CrossRef]
- Dong, L.; Li, S.; Wang, L.; Su, T.; Zhang, C.; Bi, Y.; Lai, Y.; Kong, L.; Wang, F.; Pei, X.; et al. The genetic basis of high-latitude adaptation in wild soybean. Curr. Biol. 2023, 33, 252–262. [Google Scholar] [CrossRef]
- Xie, M.; Chung, C.; Li, M.; Wong, F.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.; Wong, T.; Tong, S.; et al. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Wang, X.; Li, X.; Hu, J.; Fan, L.; Landis, J.B.; Cannon, S.B.; Grimwood, J.; Schmutz, J.; Jackson, S.A.; et al. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life strategy transition. Nat. Plants 2022, 8, 233–244. [Google Scholar] [CrossRef]
- Mihelich, T.N.; Mulkey, E.S.; Stec, O.A.; Stupar, R.M. Characterization of genetic heterogeneity within accessions in the USDA soybean germplasm collection. Plant Genome 2020, 13, e20000. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Li, Y.; Zhang, S.; Ge, T.; Wang, C.; Zhang, F.; Faruquee, M.; Zhang, L.; Wu, X.; et al. A general model for “germplasm-omics” data sharing and mining: A case study of SoyFGB v2.0. Sci. Bull. 2022, 67, 1716–1719. [Google Scholar] [CrossRef]
- Tiwari, S.; Tripathi, N.; Tsuji, K.; Tantwai, K. Genetic diversity and population structure of Indian soybean (Glycine max (L.) Merr.) as revealed by microsatellite markers. Physiol. Mol. Biol. Plant. 2019, 25, 953–964. [Google Scholar] [CrossRef]
- Gupta, S.K.; Manjaya, J.G. Genetic diversity and population structure of Indian soybean [Glycine max (L.) Merr.] revealed by simple sequence repeat markers. J. Crop Sci. Biotech. 2017, 20, 221–231. [Google Scholar] [CrossRef]
- Xie, H.; Chang, R.; Cao, Y.; Zhang, M.; Feng, Z.; Qiu, L. Selection of core SSR loci by using Chinese autumn soybean. Sci. Agric. Sin. 2003, 36, 360–366. [Google Scholar]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Lam, H.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.; Li, M.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Do, T.D.; Chen, H.T.; Hien, V.T.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.L.; Cong, H.; Shono, M.; Suenaga, K.; et al. Ncl synchronously regulates Na+, K+, and Cl− in soybean and greatly increases the grain yield in saline field conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef]
- Ning, W.; Zhai, H.; Yu, J.; Liang, S.; Yang, X.; Xing, X.; Hou, J.; Tian, P.; Yang, Y.; Bai, X. Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 2017, 37, 19. [Google Scholar] [CrossRef]
- Zhang, H.; Goettel, W.; Song, Q.; Jiang, H.; Hu, Z.; Wang, M.; An, Y.C. Selection of GmSWEET39 for oil and protein improvement in soybean. PLoS Genet. 2020, 16, 1009114. [Google Scholar] [CrossRef]
- Xun, H.; Qian, X.; Wang, M.; Yu, J.; Zhang, X.; Pang, J.; Wang, S.; Jiang, L.; Dong, Y.; Liu, B. Overexpression of a cinnamyl alcohol dehydrogenase-coding gene, GsCAD1, from wild soybean enhances resistance to soybean mosaic virus. Int. J. Mol. Sci. 2022, 23, 15206. [Google Scholar] [CrossRef]
- Usovsky, M.; Gamage, V.A.; Meinhardt, C.G.; Dietz, N.; Triller, M.; Basnet, P.; Gillman, J.D.; Bilyeu, K.D.; Song, Q.; Dhital, B.; et al. Loss-of-function of an α-SNAP gene confers resistance to soybean cyst nematode. Nat. Commun. 2023, 14, 7629. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef]


| Primer Name | Linkage Group | Position (cM) | Chromosome Number | No. of Alleles | Gene Diversity | Heterozygosity | PIC |
|---|---|---|---|---|---|---|---|
| satt300 | A1 | 30.93 | 5 | 5 | 0.255 | 0.790 | 0.947 |
| satt236 | A1 | 80.81 | 5 | 3 | 0.538 | 0.600 | 0.272 |
| satt429 | A2 | 162.02 | 8 | 4 | 0.375 | 0.715 | 0.238 |
| satt197 | B1 | 49.07 | 11 | 4 | 0.691 | 0.489 | 0.245 |
| satt453 | B1 | 123.95 | 11 | 4 | 0.447 | 0.690 | 0.947 |
| satt577 | B2 | 6.05 | 14 | 5 | 0.269 | 0.783 | 0.650 |
| satt168 | B2 | 46.87 | 14 | 5 | 0.353 | 0.731 | 0.147 |
| satt556 | B2 | 63.25 | 14 | 6 | 0.326 | 0.789 | 0.926 |
| satt565 | C1 | 5.74 | 4 | 3 | 0.423 | 0.644 | 0.321 |
| satt194 | C1 | 13.37 | 4 | 2 | 0.663 | 0.447 | 0.410 |
| satt281 | C2 | 38.90 | 6 | 4 | 0.494 | 0.651 | 0.517 |
| satt371 | C2 | 145.47 | 6 | 4 | 0.432 | 0.689 | 0.874 |
| satt184 | D1a | 16.13 | 1 | 5 | 0.408 | 0.743 | 0.293 |
| satt267 | D1a | 57.34 | 1 | 3 | 0.467 | 0.639 | 0.189 |
| satt005 | D1b | 83.41 | 2 | 5 | 0.296 | 0.768 | 0.806 |
| satt216 | D1b | n/a | 2 | 3 | 0.413 | 0.647 | 0.500 |
| satt002 | D2 | 42.69 | 17 | 5 | 0.429 | 0.722 | 0.152 |
| satt226 | D2 | 85.15 | 17 | 5 | 0.342 | 0.759 | 0.400 |
| satt386 | D2 | 125.00 | 17 | 3 | 0.483 | 0.607 | 0.144 |
| sat-112 | E | 8.67 | 15 | 4 | 0.382 | 0.716 | 0.892 |
| satt268 | E | 44.27 | 15 | 4 | 0.407 | 0.713 | 0.231 |
| satt230 | E | 71.30 | 15 | 3 | 0.634 | 0.529 | 0.268 |
| satt586 | F | 33.70 | 13 | 5 | 0.324 | 0.732 | 0.920 |
| satt146 | F | 37.14 | 13 | 4 | 0.330 | 0.736 | 0.585 |
| satt334 | F | 51.20 | 13 | 4 | 0.390 | 0.723 | 0.588 |
| sct-188 | F | 85.33 | 13 | 4 | 0.452 | 0.589 | 0.938 |
| satt130 | G | 23.10 | 18 | 2 | 0.726 | 0.398 | 0.159 |
| satt352 | G | 50.52 | 18 | 3 | 0.482 | 0.616 | 0.500 |
| satt442 | H | 43.39 | 12 | 4 | 0.326 | 0.736 | 0.809 |
| satt434 | H | 105.73 | 12 | 3 | 0.542 | 0.589 | 0.281 |
| satt571 | I | 14.97 | 20 | 3 | 0.489 | 0.603 | 0.301 |
| satt239 | I | 29.61 | 20 | 5 | 0.389 | 0.750 | 0.522 |
| sct_189 | I | 109.27 | 20 | 5 | 0.484 | 0.666 | 1.000 |
| satt414 | J | 37.04 | 16 | 3 | 0.410 | 0.658 | 0.506 |
| satt596 | J | 39.63 | 16 | 3 | 0.474 | 0.628 | 0.295 |
| satt431 | J | 82.03 | 16 | 4 | 0.375 | 0.708 | 0.938 |
| satt242 | K | 14.73 | 9 | 3 | 0.440 | 0.647 | 0.560 |
| satt588 | K | 93.44 | 9 | 4 | 0.399 | 0.720 | 0.819 |
| sat-099 | L | 69.12 | 19 | 4 | 0.339 | 0.711 | 0.736 |
| satt373 | L | 93.95 | 19 | 4 | 0.397 | 0.695 | 0.598 |
| satt590 | M | 7.75 | 7 | 3 | 0.356 | 0.666 | 0.200 |
| satt346 | M | 106.12 | 7 | 4 | 0.393 | 0.714 | 0.663 |
| satt530 | N | 32.84 | 3 | 4 | 0.300 | 0.746 | 0.756 |
| satt387 | N | 53.25 | 3 | 3 | 0.581 | 0.571 | 0.353 |
| satt339 | N | 60.17 | 3 | 6 | 0.323 | 0.765 | 0.948 |
| satt022 | N | 84.45 | 3 | 4 | 0.402 | 0.713 | 0.141 |
| satt487 | O | 9.53 | 10 | 4 | 0.349 | 0.731 | 0.914 |
| satt173 | O | 58.40 | 10 | 5 | 0.259 | 0.792 | 0.805 |
| satt345 | O | 59.43 | 10 | 3 | 0.617 | 0.533 | 0.255 |
| satt243 | O | 107.31 | 10 | 6 | 0.354 | 0.763 | 0.831 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Fu, X.; Yang, Q.; Jin, H.; Zhu, L. Genetic Characterization of Wild Soybean Collected from Zhejiang Province in China. Genes 2025, 16, 776. https://doi.org/10.3390/genes16070776
Yu X, Fu X, Yang Q, Jin H, Zhu L. Genetic Characterization of Wild Soybean Collected from Zhejiang Province in China. Genes. 2025; 16(7):776. https://doi.org/10.3390/genes16070776
Chicago/Turabian StyleYu, Xiaomin, Xujun Fu, Qinghua Yang, Hangxia Jin, and Longming Zhu. 2025. "Genetic Characterization of Wild Soybean Collected from Zhejiang Province in China" Genes 16, no. 7: 776. https://doi.org/10.3390/genes16070776
APA StyleYu, X., Fu, X., Yang, Q., Jin, H., & Zhu, L. (2025). Genetic Characterization of Wild Soybean Collected from Zhejiang Province in China. Genes, 16(7), 776. https://doi.org/10.3390/genes16070776







